G-Quadruplexes and Their Ligands: Biophysical Methods to Unravel G-Quadruplex/Ligand Interactions
Abstract
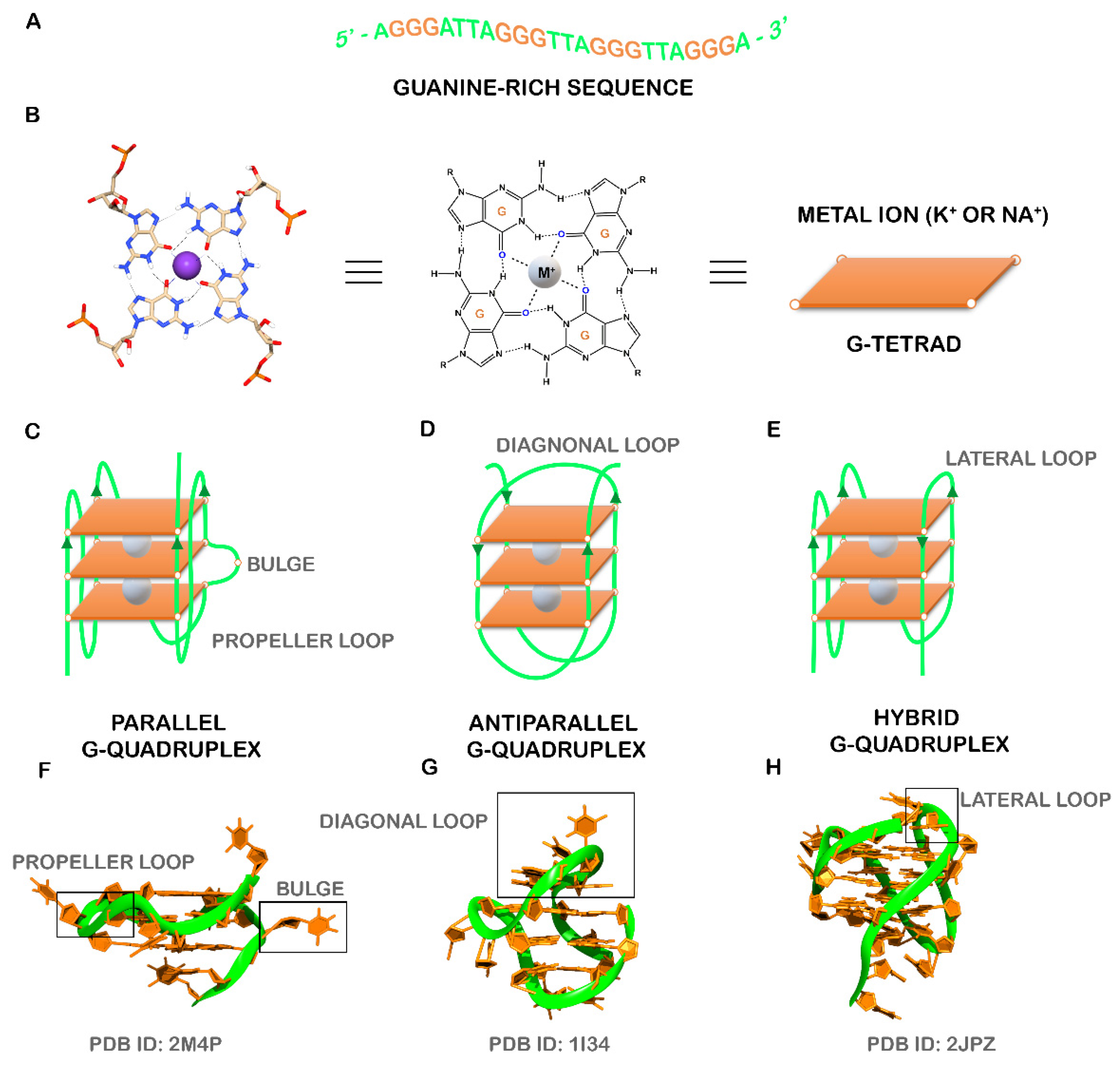
:1. Introduction
2. Overview of G4-Interacting Ligands
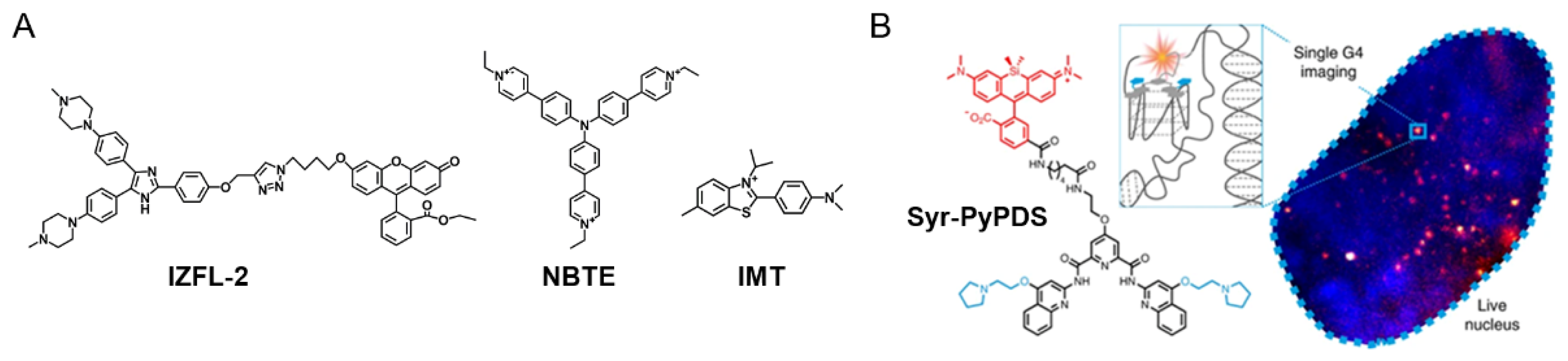
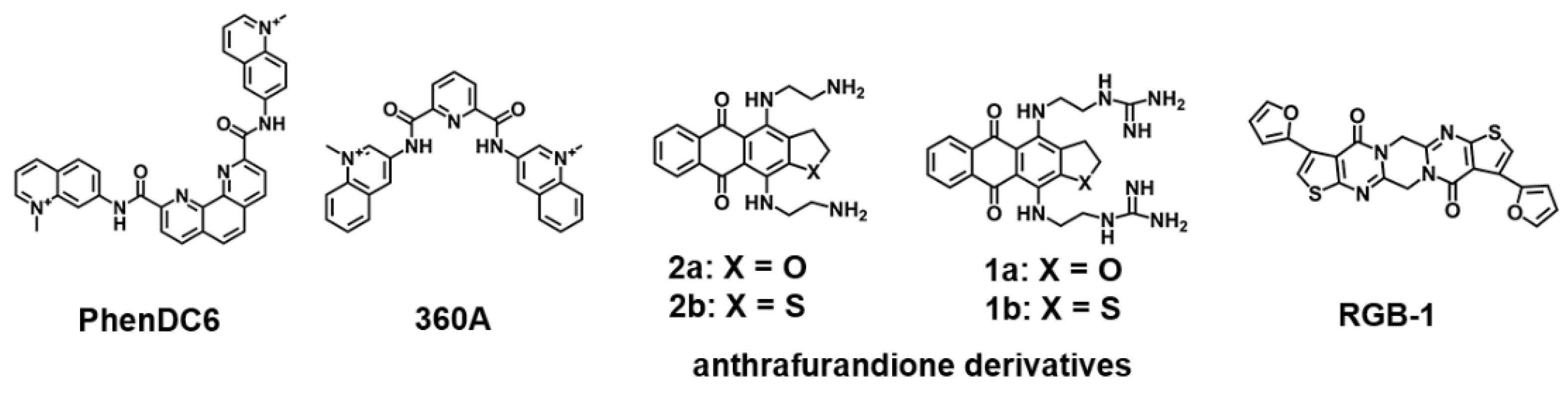
2.1. DNA G4-Interacting Ligands
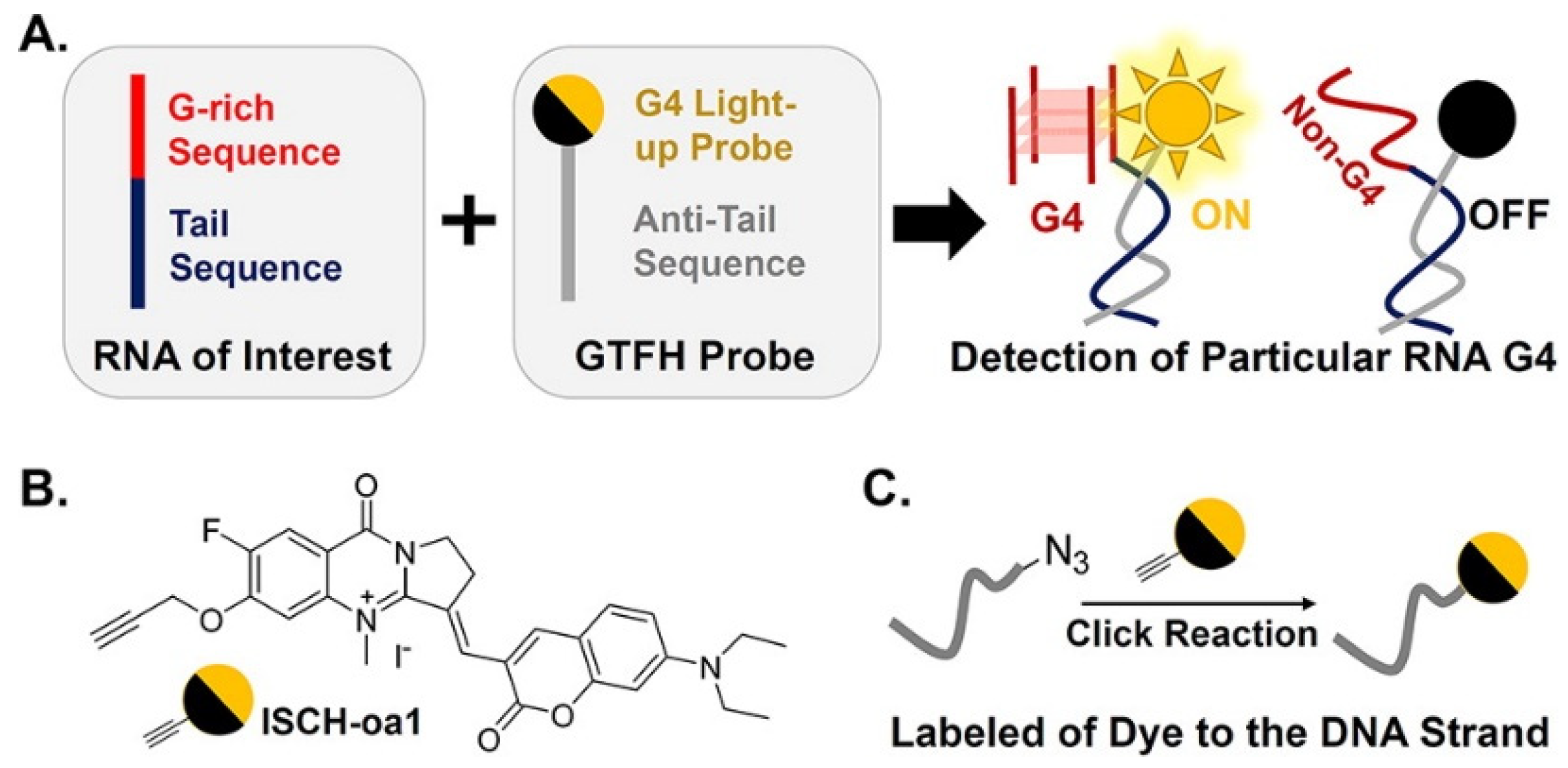
2.2. RNA G4-Interacting Ligands
3. Methods to Characterize G4/Ligand Interactions
3.1. Structure-Based Methods to Investigate G4/Ligand Interactions
3.1.1. Circular Dichroism (CD)
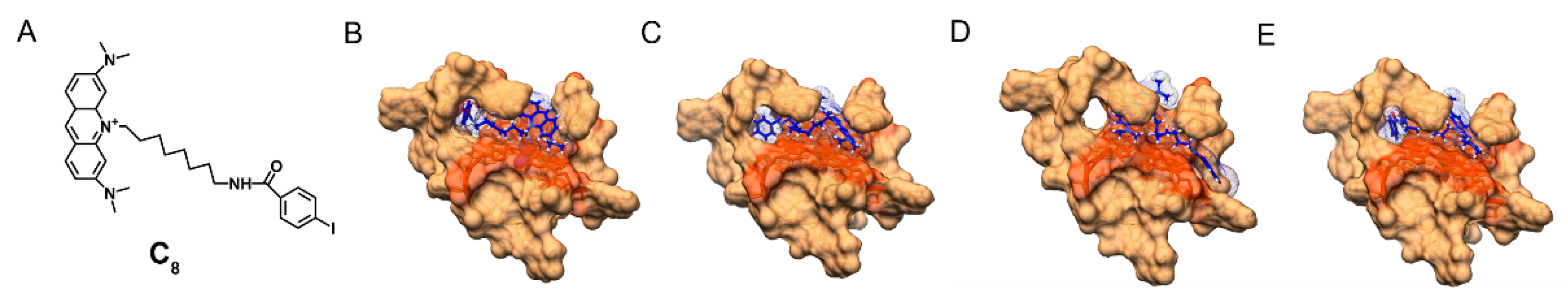
3.1.2. Nuclear Magnetic Resonance (NMR)
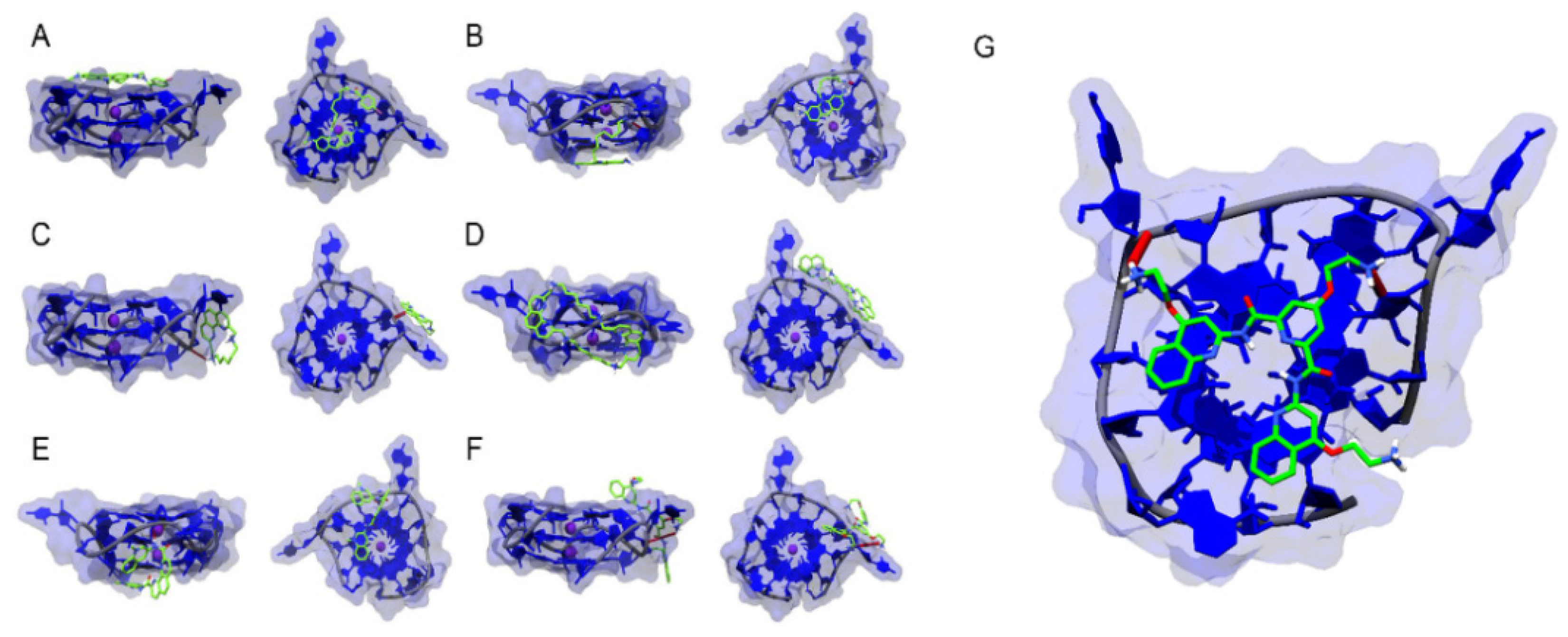
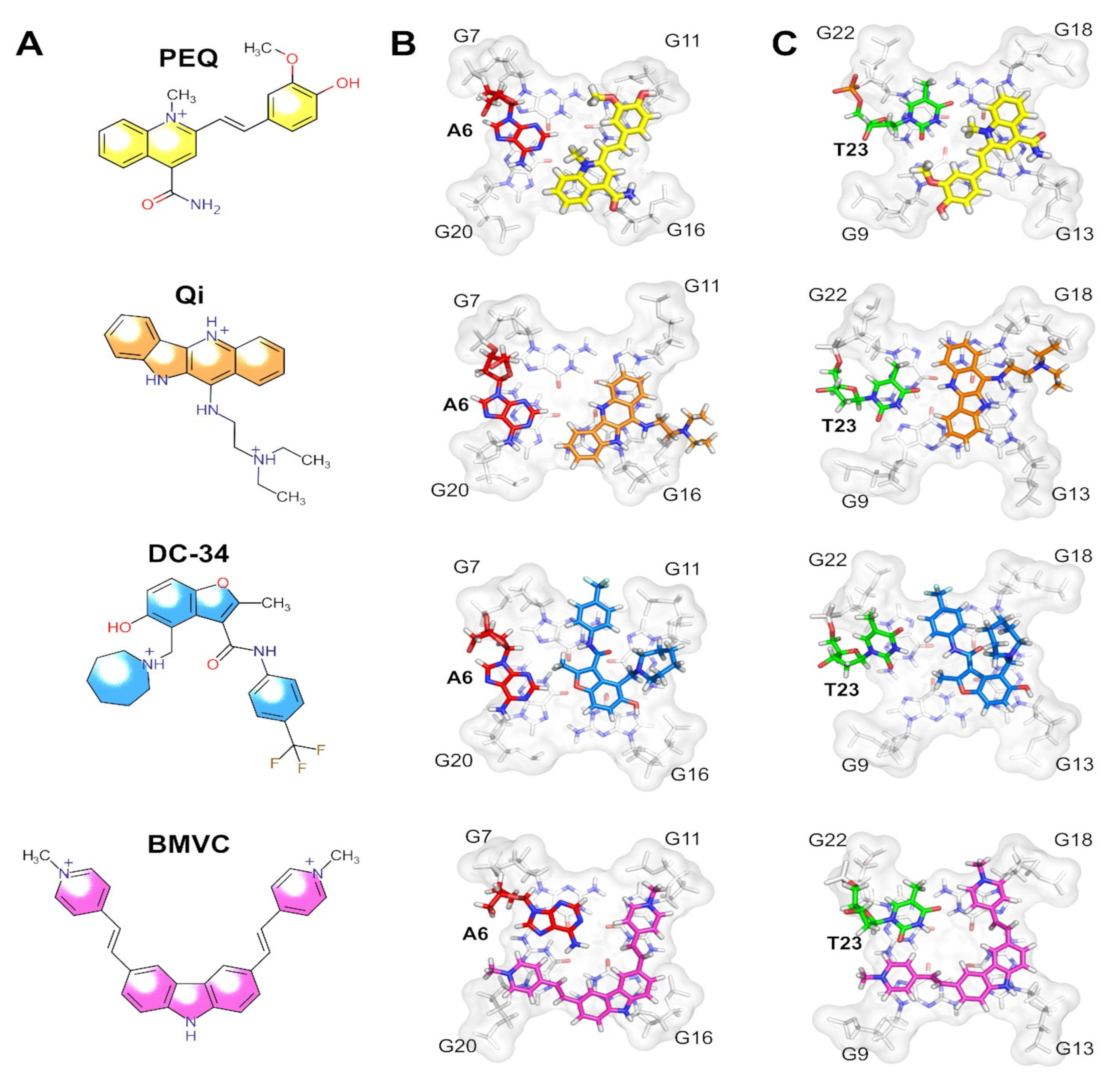
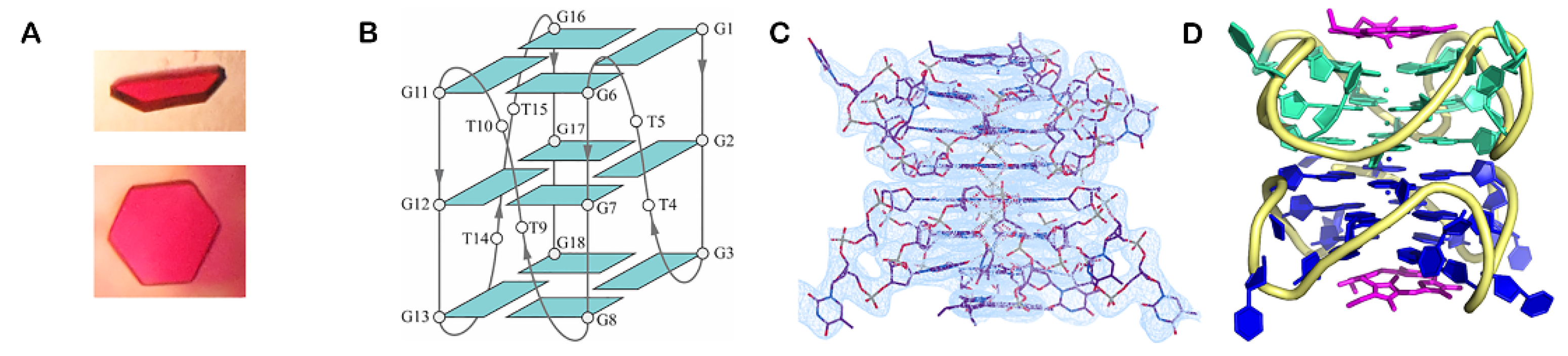
3.1.3. X-ray Crystallography
3.2. Affinity- and Apparent Affinity-Based Methods to Investigate G4/Ligand Interactions
3.2.1. Surface Plasmon Resonance (SPR)
3.2.2. Isothermal Titration Calorimetry (ITC)
3.2.3. Mass Spectrometry (MS)
3.3. High-Throughput Methods to Investigate G4/Ligand Interactions
3.3.1. FRET-Melting
3.3.2. G4-FID Screening
3.3.3. Affinity Chromatography Screening
3.3.4. Microarrays-Based Screening
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [Green Version]
- Arnott, S.; Chandrasekaran, K.; Marttila, C.M. Structures for polyinosinic acid and polyguanylic acid. Biochem. J. 1974, 141, 537–543. [Google Scholar] [CrossRef] [Green Version]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [Green Version]
- Largy, E.; Mergny, J.L.; Gabelica, V. Role of Alkali Metal Ions in G-Quadruplex Nucleic Acid Structure and Stability. In Metal Ions in Life Sciences; Springer: Berlin, Germany, 2016; Volume 16, pp. 203–258. [Google Scholar]
- Ma, Y.; Iida, K.; Nagasawa, K. Topologies of G-quadruplex: Biological functions and regulation by ligands. Biochem. Biophys. Res. Commun. 2020, 531, 3–17. [Google Scholar] [CrossRef]
- Ngoc Nguyen, T.Q.; Lim, K.W.; Phan, A.T. Duplex formation in a G-quadruplex bulge. Nucleic Acids Res. 2020, 48, 10567–10575. [Google Scholar] [CrossRef]
- Meier, M.; Moya-Torres, A.; Krahn, N.J.; McDougall, M.D.; Orriss, G.L.; McRae, E.K.S.; Booy, E.P.; McEleney, K.; Patel, T.R.; McKenna, S.A.; et al. Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge. Nucleic Acids Res. 2018, 46, 5319–5331. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Maizels, N.; Gray, L.T. The G4 Genome. PLoS Genet. 2013, 9, e1003468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedrat, A.; Lacroix, L.; Mergny, J.-L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef]
- Brázda, V.; Kolomazník, J.; Lýsek, J.; Bartas, M.; Fojta, M.; Šťastný, J.; Mergny, J.L. G4Hunter web application: A web server for G-quadruplex prediction. Bioinformatics 2019, 35, 3493–3495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hon, J.; Martínek, T.; Zendulka, J.; Lexa, M. Pqsfinder: An exhaustive and imperfection-tolerant search tool for potential quadruplex-forming sequences in R. Bioinformatics 2017, 33, 3373–3379. [Google Scholar] [CrossRef]
- Klimentova, E.; Polacek, J.; Simecek, P.; Alexiou, P. Penguinn: Precise Exploration of Nuclear G-Quadruplexes Using Interpretable Neural Networks. Front. Genet. 2020, 11, 1287. [Google Scholar] [CrossRef]
- Garant, J.M.; Perreault, J.P.; Scott, M.S. Motif independent identification of potential RNA G-quadruplexes by G4RNA screener. Bioinformatics 2017, 33, 3532–3537. [Google Scholar] [CrossRef] [Green Version]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef] [Green Version]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M. G-quadruplexes at telomeres: Friend or foe? Molecules 2020, 25, 3686. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hänsel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiegel, J.; Cuesta, S.M.; Adhikari, S.; Hänsel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol. 2021, 22, 117. [Google Scholar] [CrossRef] [PubMed]
- Lago, S.; Nadai, M.; Cernilogar, F.M.; Kazerani, M.; Domíniguez Moreno, H.; Schotta, G.; Richter, S.N. Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat. Commun. 2021, 12, 3885. [Google Scholar] [CrossRef] [PubMed]
- Jara-Espejo, M.; Peres Line, S.R. DNA G-quadruplex stability, position and chromatin accessibility are associated with CpG island methylation. FEBS J. 2020, 287, 483–495. [Google Scholar] [CrossRef]
- Shen, J.; Varshney, D.; Simeone, A.; Zhang, X.; Adhikari, S.; Tannahill, D.; Balasubramanian, S. Promoter G-quadruplex folding precedes transcription and is controlled by chromatin. Genome Biol. 2021, 22, 143. [Google Scholar] [CrossRef]
- Komůrková, D.; Kovaříková, A.S.; Bártová, E. G-quadruplex structures colocalize with transcription factories and nuclear speckles surrounded by acetylated and dimethylated histones H3. Int. J. Mol. Sci. 2021, 22, 1995. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, R.F.; Moshkin, Y.M.; Mouton, S.; Grzeschik, N.A.; Kalicharan, R.D.; Kuipers, J.; Wolters, A.H.G.; Nishida, K.; Romashchenko, A.V.; Postberg, J.; et al. Guanine quadruplex structures localize to heterochromatin. Nucleic Acids Res. 2016, 44, 152–163. [Google Scholar] [CrossRef] [Green Version]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Lee, W.T.C.; Yin, Y.; Morten, M.J.; Tonzi, P.; Gwo, P.P.; Odermatt, D.C.; Modesti, M.; Cantor, S.B.; Gari, K.; Huang, T.T.; et al. Single-molecule imaging reveals replication fork coupled formation of G-quadruplex structures hinders local replication stress signaling. Nat. Commun. 2021, 12, 2525. [Google Scholar] [CrossRef]
- Tran, P.L.T.; Rieu, M.; Hodeib, S.; Joubert, A.; Ouellet, J.; Alberti, P.; Bugaut, A.; Allemand, J.F.; Boulé, J.B.; Croquette, V. Folding and persistence times of intramolecular G-quadruplexes transiently embedded in a DNA duplex. Nucleic Acids Res. 2021, 49, 5189–5201. [Google Scholar] [CrossRef]
- Murat, P.; Marsico, G.; Herdy, B.; Ghanbarian, A.; Portella, G.; Balasubramanian, S. RNA G-quadruplexes at upstream open reading frames cause DHX36- and DHX9-dependent translation of human mRNAs. Genome Biol. 2018, 19, 229. [Google Scholar] [CrossRef]
- Dutta, A.; Maji, N.; Sengupta, P.; Banerjee, N.; Kar, S.; Mukherjee, G.; Chatterjee, S.; Basu, M. Promoter G-quadruplex favours epigenetic reprogramming-induced atypical expression of ZEB1 in cancer cells. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129899. [Google Scholar] [CrossRef] [PubMed]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; McCauley, P.; Boutell, J.M.; Antonio, M.D.; Balasubramanian, S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef] [Green Version]
- Zaccaria, F.; Fonseca Guerra, C. RNA versus DNA G-Quadruplex: The Origin of Increased Stability. Chem. Eur. J. 2018, 24, 16315–16322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vannutelli, A.; Belhamiti, S.; Garant, J.-M.; Ouangraoua, A.; Perreault, J.-P. Where are G-quadruplexes located in the human transcriptome? NAR Genomics Bioinform. 2020, 2, lqaa035. [Google Scholar] [CrossRef]
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014, 6, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Tassinari, M.; Richter, S.N.; Gandellini, P. Biological relevance and therapeutic potential of G-quadruplex structures in the human noncoding transcriptome. Nucleic Acids Res. 2021, 49, 3617–3633. [Google Scholar] [CrossRef]
- Kwok, C.K.; Marsico, G.; Sahakyan, A.B.; Chambers, V.S.; Balasubramanian, S. RG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods 2016, 13, 841–844. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Lejault, P.; Chevrier, S.; Boidot, R.; Robertson, A.G.; Wong, J.M.Y.; Monchaud, D. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat. Commun. 2018, 9, 4730. [Google Scholar] [CrossRef]
- Yang, X.; Cheema, J.; Zhang, Y.; Deng, H.; Duncan, S.; Umar, M.I.; Zhao, J.; Liu, Q.; Cao, X.; Kwok, C.K.; et al. RNA G-quadruplex structures exist and function in vivo in plants. Genome Biol. 2020, 21, 226. [Google Scholar] [CrossRef]
- Shao, X.; Zhang, W.; Umar, M.I.; Wong, H.Y.; Seng, Z.; Xie, Y.; Zhang, Y.; Yang, L.; Kwok, C.K.; Deng, X. RNA G-quadruplex structures mediate gene regulation in bacteria. MBio 2020, 11, e02926-19. [Google Scholar] [CrossRef] [Green Version]
- Renard, I.; Grandmougin, M.; Roux, A.; Yang, S.Y.; Lejault, P.; Pirrotta, M.; Wong, J.M.Y.; Monchaud, D. Small-molecule affinity capture of DNA/RNA quadruplexes and their identification in vitro and in vivo through the G4RP protocol. Nucleic Acids Res. 2019, 47, 502–510. [Google Scholar] [CrossRef]
- Maizels, N. G4-associated human diseases. EMBO Rep. 2015, 16, 910–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Magis, A.; Manzo, S.G.; Russo, M.; Marinello, J.; Morigi, R.; Sordet, O.; Capranico, G. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 816–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zell, J.; Sperti, F.R.; Britton, S.; Monchaud, D. DNA folds threaten genetic stability and can be leveraged for chemotherapy. RSC Chem. Biol. 2021, 2, 47–76. [Google Scholar] [CrossRef]
- Maffia, A.; Ranise, C.; Sabbioneda, S. From R-loops to G-quadruplexes: Emerging new threats for the replication fork. Int. J. Mol. Sci. 2020, 21, 1506. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, R.; Miller, K.M.; Forment, J.V.; Bradshaw, C.R.; Nikan, M.; Britton, S.; Oelschlaegel, T.; Xhemalce, B.; Balasubramanian, S.; Jackson, S.P. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012, 8, 301–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, P.; Kim, N.; Kumari, M.; Verma, S.; Sharma, T.K.; Yadav, V.; Kumar, A. G-quadruplex structures in bacteria: Biological relevance and potential as an antimicrobial target. J. Bacteriol. 2021, 203, e0057720. [Google Scholar] [CrossRef]
- Ruggiero, E.; Richter, S.N. Viral G-quadruplexes: New frontiers in virus pathogenesis and antiviral therapy. In Annual Reports in Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 2020; Volume 54, pp. 101–131. [Google Scholar]
- Abiri, A.; Lavigne, M.; Rezaei, M.; Nikzad, S.; Zare, P.; Mergny, J.L.; Rahimi, H.R. Unlocking G-quadruplexes as antiviral targets. Pharmacol. Rev. 2021, 73, 897–923. [Google Scholar] [CrossRef]
- Saranathan, N.; Vivekanandan, P. G-Quadruplexes: More Than Just a Kink in Microbial Genomes. Trends Microbiol. 2019, 27, 148–163. [Google Scholar] [CrossRef] [Green Version]
- Seifert, H.S. Above and beyond Watson and Crick: Guanine Quadruplex Structures and Microbes. Annu. Rev. Microbiol. 2018, 72, 49–69. [Google Scholar] [CrossRef]
- Metifiot, M.; Amrane, S.; Litvak, S.; Andreola, M.L. G-quadruplexes in viruses: Function and potential therapeutic applications. Nucleic Acids Res. 2014, 42, 12352–12366. [Google Scholar] [CrossRef] [Green Version]
- Perrone, R.; Lavezzo, E.; Riello, E.; Manganelli, R.; Palù, G.; Toppo, S.; Provvedi, R.; Richter, S.N. Mapping and characterization of G-quadruplexes in Mycobacterium tuberculosis gene promoter regions. Sci. Rep. 2017, 7, 5743. [Google Scholar] [CrossRef] [Green Version]
- Tlučková, K.; Marušič, M.; Tóthová, P.; Bauer, L.; Šket, P.; Plavec, J.; Viglasky, V. Human papillomavirus G-quadruplexes. Biochemistry 2013, 52, 7207–7216. [Google Scholar] [CrossRef]
- Zhao, C.; Qin, G.; Niu, J.; Wang, Z.; Wang, C.; Ren, J.; Qu, X. Targeting RNA G-Quadruplex in SARS-CoV-2: A Promising Therapeutic Target for COVID-19? Angew. Chem. Int. Ed. 2021, 60, 432–438. [Google Scholar] [CrossRef]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent progress of targeted G-quadruplex-preferred ligands toward cancer therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef] [Green Version]
- Sun, D.; Thompson, B.; Cathers, B.E.; Salazar, M.; Kerwin, S.M.; Trent, J.O.; Jenkins, T.C.; Neidle, S.; Hurley, L.H. Inhibition of human telomerase by a G-Quadruplex-Interactive compound. J. Med. Chem. 1997, 40, 2113–2116. [Google Scholar] [CrossRef]
- Duarte, A.R.; Cadoni, E.; Ressurreição, A.S.; Moreira, R.; Paulo, A. Design of Modular G-quadruplex Ligands. Chem. Med. Chem. 2018, 13, 869–893. [Google Scholar] [CrossRef] [PubMed]
- Umar, M.I.; Ji, D.; Chan, C.Y.; Kwok, C.K. G-quadruplex-based fluorescent turn-on ligands and aptamers: From development to applications. Molecules 2019, 24, 2416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Xiang, J.-F.; Yang, Q.-F.; Sun, H.-X.; Guan, A.-J.; Tang, Y.-L. G4LDB: A database for discovering and studying G-quadruplex ligands. Nucleic Acids Res. 2013, 41, D1115–D1123. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.Y.; Wang, X.N.; Cheng, S.Q.; Su, X.X.; Ou, T.M. Developing novel G-quadruplex ligands: From interaction with nucleic acids to interfering with nucleic acid–protein interaction. Molecules 2019, 24, 396. [Google Scholar] [CrossRef] [Green Version]
- Martino, L.; Pagano, B.; Fotticchia, I.; Neidle, S.; Giancola, C. Shedding light on the interaction between TMPyP4 and human telomeric quadruplexes. J. Phys. Chem. B 2009, 113, 14779–14786. [Google Scholar] [CrossRef]
- Read, M.; Harrison, R.J.; Romagnoli, B.; Tanious, F.A.; Gowan, S.H.; Reszka, A.P.; Wilson, W.D.; Kelland, L.R.; Neidle, S. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl. Acad. Sci. USA 2001, 98, 4844–4849. [Google Scholar] [CrossRef] [Green Version]
- Zuffo, M.; Guédin, A.; Leriche, E.D.; Doria, F.; Pirota, V.; Gabelica, V.; Mergny, J.L.; Freccero, M. More is not always better: Finding the right trade-off between affinity and selectivity of a G-quadruplex ligand. Nucleic Acids Res. 2018, 46, e115. [Google Scholar] [CrossRef] [PubMed]
- Asamitsu, S.; Bando, T.; Sugiyama, H. Ligand Design to Acquire Specificity to Intended G-Quadruplex Structures. Chem. Eur. J. 2019, 25, 417–430. [Google Scholar] [CrossRef]
- Neidle, S. Challenges in developing small-molecule quadruplex therapeutics. In Annual Reports in Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 2020; Volume 54, pp. 517–546. [Google Scholar]
- Drygin, D.; Siddiqui-Jain, A.; O’Brien, S.; Schwaebe, M.; Lin, A.; Bliesath, J.; Ho, C.B.; Proffitt, C.; Trent, K.; Whitten, J.P.; et al. Anticancer activity of CX-3543: A direct inhibitor of rRNA biogenesis. Cancer Res. 2009, 69, 7653–7661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Antonio, M.D.; McKinney, S.; Mathew, V.; Ho, B.; O’Neil, N.J.; Santos, N.D.; Silvester, J.; Wei, V.; Garcia, J.; et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017, 8, 14432. [Google Scholar] [CrossRef]
- Masud, T.; Soong, C.; Xu, H.; Biele, J.; Bjornson, S.; McKinney, S.; Aparicio, S. Ubiquitin-mediated DNA damage response is synthetic lethal with G-quadruplex stabilizer CX-5461. Sci. Rep. 2021, 11, 9812. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Feng, H.; Dai, C.; Lu, W.; Zhang, J.; Guo, X.; Yin, Q.; Wang, J.; Cui, X.; Jiang, F. Therapeutic efficacy of the novel selective RNA polymerase I inhibitor CX-5461 on pulmonary arterial hypertension and associated vascular remodelling. Br. J. Pharmacol. 2021, 178, 1605–1619. [Google Scholar] [CrossRef]
- Neidle, S. Human telomeric G-quadruplex: The current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010, 277, 1118–1125. [Google Scholar] [CrossRef]
- Burger, A.M.; Dai, F.; Schultes, C.M.; Reszka, A.P.; Moore, M.J.; Double, J.A.; Neidle, S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005, 65, 1489–1496. [Google Scholar] [CrossRef] [Green Version]
- Machireddy, B.; Sullivan, H.J.; Wu, C. Binding of BRACO19 to a telomeric G-quadruplex DNA probed by all-atom molecular dynamics simulations with explicit solvent. Molecules 2019, 24, 1010. [Google Scholar] [CrossRef] [Green Version]
- Hamon, F.; Largy, E.; Guédin-Beaurepaire, A.; Rouchon-Dagois, M.; Sidibe, A.; Monchaud, D.; Mergny, J.L.; Riou, J.F.; Nguyen, C.H.; Teulade-Fichou, M.P. An acyclic oligoheteroaryle that discriminates strongly between diverse G-Quadruplex topologies. Angew. Chem. Int. Ed. 2011, 50, 8745–8749. [Google Scholar] [CrossRef] [PubMed]
- Răsădean, D.M.; Sheng, B.; Dash, J.; Pantoş, G.D. Amino-Acid-Derived Naphthalenediimides as Versatile G-Quadruplex Binders. Chem. Eur. J. 2017, 23, 8491–8499. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.H.; Chen, S.B.; Wang, B.; Ou, T.M.; Gu, L.Q.; Tan, J.H.; Huang, Z.S. Specific targeting of telomeric multimeric G-quadruplexes by a new triaryl-substituted imidazole. Nucleic Acids Res. 2017, 45, 1606–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigo, R.; Palumbo, M.; Sissi, C. G-quadruplexes in human promoters: A challenge for therapeutic applications. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1399–1413. [Google Scholar] [CrossRef]
- Micco, M.; Collie, G.W.; Dale, A.G.; Ohnmacht, S.A.; Pazitna, I.; Gunaratnam, M.; Reszka, A.P.; Neidle, S. Structure-based design and evaluation of naphthalene diimide G-quadruplex ligands as telomere targeting agents in pancreatic cancer cells. J. Med. Chem. 2013, 56, 2959–2974. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, C.; Zyner, K.G.; Ohnmacht, S.A.; Robson, M.; Haider, S.M.; Morton, J.P.; Marsico, G.; Vo, T.; Laughlin-Toth, S.; Ahmed, A.A.; et al. Targeting Multiple Effector Pathways in Pancreatic Ductal Adenocarcinoma with a G-Quadruplex-Binding Small Molecule. J. Med. Chem. 2018, 61, 2500–2517. [Google Scholar] [CrossRef]
- Carvalho, J.; Pereira, E.; Marquevielle, J.; Campello, M.P.C.; Mergny, J.L.; Paulo, A.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. Fluorescent light-up acridine orange derivatives bind and stabilize KRAS-22RT G-quadruplex. Biochimie 2018, 144, 144–152. [Google Scholar] [CrossRef]
- Shin-ya, K.; Wierzba, K.; Matsuo, K.; Ohtani, T.; Yamada, Y.; Furihata, K.; Hayakawa, Y.; Seto, H. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J. Am. Chem. Soc. 2001, 123, 1262–1263. [Google Scholar] [CrossRef]
- Sullivan, H.J.; Readmond, C.; Radicella, C.; Persad, V.; Fasano, T.J.; Wu, C. Binding of Telomestatin, TMPyP4, BSU6037, and BRACO19 to a Telomeric G-Quadruplex-Duplex Hybrid Probed by All-Atom Molecular Dynamics Simulations with Explicit Solvent. ACS Omega 2018, 3, 14788–14806. [Google Scholar] [CrossRef]
- Gavathiotis, E.; Heald, R.A.; Stevens, M.F.G.; Searle, M.S. Drug recognition and stabilisation of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat. J. Mol. Biol. 2003, 334, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Mulholland, K.; Siddiquei, F.; Wu, C. Binding modes and pathway of RHPS4 to human telomeric G-quadruplex and duplex DNA probed by all-atom molecular dynamics simulations with explicit solvent. Phys. Chem. Chem. Phys. 2017, 19, 18685–18694. [Google Scholar] [CrossRef] [PubMed]
- Muoio, D.; Berardinelli, F.; Leone, S.; Coluzzi, E.; di Masi, A.; Doria, F.; Freccero, M.; Sgura, A.; Folini, M.; Antoccia, A. Naphthalene diimide-derivatives G-quadruplex ligands induce cell proliferation inhibition, mild telomeric dysfunction and cell cycle perturbation in U251MG glioma cells. FEBS J. 2018, 285, 3769–3785. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, C.; Minarini, A.; Tumiatti, V.; Moraca, F.; Parrotta, L.; Alcaro, S.; Rigo, R.; Sissi, C.; Gunaratnam, M.; Ohnmacht, S.A.; et al. Macrocyclic naphthalene diimides as G-quadruplex binders. Bioorg. Med. Chem. 2015, 23, 3819–3830. [Google Scholar] [CrossRef]
- Hu, M.H.; Zhou, J.; Luo, W.H.; Chen, S.B.; Huang, Z.S.; Wu, R.; Tan, J.H. Development of a Smart Fluorescent Sensor That Specifically Recognizes the c-MYC G-Quadruplex. Anal. Chem. 2019, 91, 2480–2487. [Google Scholar] [CrossRef]
- Vummidi, B.R.; Alzeer, J.; Luedtke, N.W. Fluorescent Probes for G-Quadruplex Structures. ChemBioChem 2013, 14, 540–558. [Google Scholar] [CrossRef]
- Largy, E.; Granzhan, A.; Hamon, F.; Verga, D.; Teulade-Fichou, M.-P. Visualizing the Quadruplex: From Fluorescent Ligands to Light-Up Probes. In Quadruplex Nucleic Acids; Royal Society of Chemistry: London, UK, 2012; pp. 111–177. [Google Scholar]
- Kwok, C.K.; Merrick, C.J. G-Quadruplexes: Prediction, Characterization, and Biological Application. Trends Biotechnol. 2017, 35, 997–1013. [Google Scholar] [CrossRef]
- Chilka, P.; Desai, N.; Datta, B. Small molecule fluorescent probes for G-quadruplex visualization as potential cancer theranostic agents. Molecules 2019, 24, 752. [Google Scholar] [CrossRef] [Green Version]
- Shivalingam, A.; Izquierdo, M.A.; Marois, A.L.; Vyšniauskas, A.; Suhling, K.; Kuimova, M.K.; Vilar, R. The interactions between a small molecule and G-quadruplexes are visualized by fluorescence lifetime imaging microscopy. Nat. Commun. 2015, 6, 8178. [Google Scholar] [CrossRef] [Green Version]
- Kotar, A.; Wang, B.; Shivalingam, A.; Gonzalez-Garcia, J.; Vilar, R.; Plavec, J. NMR Structure of a Triangulenium-Based Long-Lived Fluorescence Probe Bound to a G-Quadruplex. Angew. Chem. Int. Ed. 2016, 55, 12508–12511. [Google Scholar] [CrossRef]
- Liu, L.Y.; Liu, W.; Wang, K.N.; Zhu, B.C.; Xia, X.Y.; Ji, L.N.; Mao, Z.W. Quantitative Detection of G-Quadruplex DNA in Live Cells Based on Photon Counts and Complex Structure Discrimination. Angew. Chem. Int. Ed. 2020, 59, 9719–9726. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, H.; Wang, L.; Liu, Y.; Chen, H.; Li, Q.; Guan, A.; Liu, M.; Tang, Y. Real-time monitoring of DNA G-quadruplexes in living cells with a small-molecule fluorescent probe. Nucleic Acids Res. 2018, 46, 7522–7532. [Google Scholar] [CrossRef] [Green Version]
- Di Antonio, M.; Ponjavic, A.; Radzevičius, A.; Ranasinghe, R.T.; Catalano, M.; Zhang, X.; Shen, J.; Needham, L.M.; Lee, S.F.; Klenerman, D.; et al. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat. Chem. 2020, 12, 832–837. [Google Scholar] [CrossRef]
- Ofer, N.; Weisman-Shomer, P.; Shklover, J.; Fry, M. The quadruplex r(CGG)n destabilizing cationic porphyrin TMPyP4 cooperates with hnRNPs to increase the translation efficiency of fragile X premutation mRNA. Nucleic Acids Res. 2009, 37, 2712–2722. [Google Scholar] [CrossRef] [Green Version]
- Morris, M.J.; Wingate, K.L.; Silwal, J.; Leeper, T.C.; Basu, S. The porphyrin TmPyP4 unfolds the extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its repressive effect to enhance translation in eukaryotic cells. Nucleic Acids Res. 2012, 40, 4137–4145. [Google Scholar] [CrossRef]
- Zamiri, B.; Reddy, K.; Macgregor, R.B.; Pearson, C.E. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNAbinding proteins. J. Biol. Chem. 2014, 289, 4653–4659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Zhang, J.; Harvey, S.E.; Hu, X.; Cheng, C. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017, 31, 2296–2309. [Google Scholar] [CrossRef]
- Ghosh, A.; Ekka, M.K.; Tawani, A.; Kumar, A.; Chakraborty, D.; Maiti, S. Restoration of miRNA-149 Expression by TmPyP4 Induced Unfolding of Quadruplex within Its Precursor. Biochemistry 2019, 58, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Banco, M.T.; Ferré-D’Amaré, A.R. The emerging structural complexity of G-quadruplex RNAs. RNA 2021, 27, 390–402. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zheng, Y.; Zhai, Q.; Wei, D. Recent advances in the development of small molecules targeting RNA G-quadruplexes for drug discovery. Bioorg. Chem. 2021, 110, 104804. [Google Scholar] [CrossRef]
- Song, J.; Perreault, J.-P.; Topisirovic, I.; Richard, S. RNA G-quadruplexes and their potential regulatory roles in translation. Translation 2016, 4, e1244031. [Google Scholar] [CrossRef] [Green Version]
- Fay, M.M.; Lyons, S.M.; Ivanov, P. RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J. Mol. Biol. 2017, 429, 2127–2147. [Google Scholar] [CrossRef] [PubMed]
- Halder, K.; Largy, E.; Benzler, M.; Teulade-Fichou, M.P.; Hartig, J.S. Efficient Suppression of Gene Expression by Targeting 5′-UTR-Based RNA Quadruplexes with Bisquinolinium Compounds. ChemBioChem 2011, 12, 1663–1668. [Google Scholar] [CrossRef]
- Miglietta, G.; Cogoi, S.; Marinello, J.; Capranico, G.; Tikhomirov, A.S.; Shchekotikhin, A.; Xodo, L.E. RNA G-Quadruplexes in Kirsten Ras (KRAS) Oncogene as Targets for Small Molecules Inhibiting Translation. J. Med. Chem. 2017, 60, 9448–9461. [Google Scholar] [CrossRef] [PubMed]
- Katsuda, Y.; Sato, S.I.; Asano, L.; Morimura, Y.; Furuta, T.; Sugiyama, H.; Hagihara, M.; Uesugi, M. A Small Molecule That Represses Translation of G-Quadruplex-Containing mRNA. J. Am. Chem. Soc. 2016, 138, 9037–9040. [Google Scholar] [CrossRef] [PubMed]
- Di Antonio, M.; Biffi, G.; Mariani, A.; Raiber, E.A.; Rodriguez, R.; Balasubramanian, S. Selective RNA versus DNA G-quadruplex targeting by situ click chemistry. Angew. Chem. Int. Ed. 2012, 51, 11073–11078. [Google Scholar] [CrossRef] [Green Version]
- Rocca, R.; Talarico, C.; Moraca, F.; Costa, G.; Romeo, I.; Ortuso, F.; Alcaro, S.; Artese, A. Molecular recognition of a carboxy pyridostatin toward G-quadruplex structures: Why does it prefer RNA? Chem. Biol. Drug Des. 2017, 90, 919–925. [Google Scholar] [CrossRef]
- Kwok, C.K.; Sahakyan, A.B.; Balasubramanian, S. Structural Analysis using SHALiPE to Reveal RNA G-Quadruplex Formation in Human Precursor MicroRNA. Angew. Chem. Int. Ed. 2016, 55, 8958–8961. [Google Scholar] [CrossRef]
- Santos, T.; Pereira, P.; Campello, M.P.C.; Paulo, A.; Queiroz, J.A.; Cabrita, E.; Cruz, C. RNA G-quadruplex as supramolecular carrier for cancer-selective delivery. Eur. J. Pharm. Biopharm. 2019, 142, 473–479. [Google Scholar] [CrossRef]
- Santos, T.; Miranda, A.; Campello, M.P.C.; Paulo, A.; Salgado, G.; Cabrita, E.J.; Cruz, C. Recognition of nucleolin through interaction with RNA G-quadruplex. Biochem. Pharmacol. 2020, 189, 114208. [Google Scholar] [CrossRef]
- Carvalho, J.; Santos, T.; Carrilho, R.; Sousa, F.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. Ligand screening to pre-miRNA 149 G-quadruplex investigated by molecular dynamics. J. Biomol. Struct. Dyn. 2020, 38, 2276–2286. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.C.; Chen, S.B.; Dai, J.; Yuan, J.H.; Ou, T.M.; Huang, Z.S.; Tan, J.H. Tracking the Dynamic Folding and Unfolding of RNA G-Quadruplexes in Live Cells. Angew. Chem. Int. Ed. 2018, 57, 4702–4706. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.B.; Hu, M.H.; Liu, G.C.; Wang, J.; Ou, T.M.; Gu, L.Q.; Huang, Z.S.; Tan, J.H. Visualization of NRAS RNA G-Quadruplex Structures in Cells with an Engineered Fluorogenic Hybridization Probe. J. Am. Chem. Soc. 2016, 138, 10382–10385. [Google Scholar] [CrossRef]
- Laguerre, A.; Stefan, L.; Larrouy, M.; Genest, D.; Novotna, J.; Pirrotta, M.; Monchaud, D. A Twice-As-smart synthetic G-quartet: PyroTASQ is both a smart quadruplex ligand and a smart fluorescent probe. J. Am. Chem. Soc. 2014, 136, 12406–12414. [Google Scholar] [CrossRef] [PubMed]
- Laguerre, A.; Hukezalie, K.; Winckler, P.; Katranji, F.; Chanteloup, G.; Pirrotta, M.; Perrier-Cornet, J.M.; Wong, J.M.Y.; Monchaud, D. Visualization of RNA-Quadruplexes in Live Cells. J. Am. Chem. Soc. 2015, 137, 8521–8525. [Google Scholar] [CrossRef]
- Laguerre, A.; Wong, J.M.Y.; Monchaud, D. Direct visualization of both DNA and RNA quadruplexes in human cells via an uncommon spectroscopic method. Sci. Rep. 2016, 6, 32141. [Google Scholar] [CrossRef] [Green Version]
- Murat, P.; Singh, Y.; Defrancq, E. Methods for investigating G-quadruplex DNA/ligand interactions. Chem. Soc. Rev. 2011, 40, 5293–5307. [Google Scholar] [CrossRef] [PubMed]
- Jaumot, J.; Gargallo, R. Experimental Methods for Studying the Interactions between G-Quadruplex Structures and Ligands. Curr. Pharm. Des. 2012, 18, 1900–1916. [Google Scholar] [CrossRef] [Green Version]
- Vorlíčková, M.; Kejnovská, I.; Bednářová, K.; Renčiuk, D.; Kypr, J. Circular dichroism spectroscopy of DNA: From duplexes to quadruplexes. Chirality 2012, 24, 691–698. [Google Scholar] [CrossRef]
- Carvalho, J.; Queiroz, J.A.; Cruz, C. Circular dichroism of G-Quadruplex: A laboratory experiment for the study of topology and ligand binding. J. Chem. Educ. 2017, 94, 1547–1551. [Google Scholar] [CrossRef]
- Villar-Guerra, R.D.; Trent, J.O.; Chaires, J.B. G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy. Angew. Chem. Int. Ed. 2018, 57, 7171–7175. [Google Scholar] [CrossRef]
- Eriksson, M.; Nordén, B. Linear and circular dichroism of drug-nucleic acid complexes. Methods Enzymol. 2001, 340, 68–98. [Google Scholar] [CrossRef]
- Garbett, N.C.; Ragazzon, P.A.; Chaires, J.O.B. Circular dichroism to determine binding mode and affinity of ligand-dna interactions. Nat. Protoc. 2007, 2, 3166–3172. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, M.P.; Morales, J.C.; Galan, M.C. Binding and Beyond: What Else Can G-Quadruplex Ligands Do? Eur. J. Org. Chem. 2019, 2019, 4995–5017. [Google Scholar] [CrossRef]
- Rodriguez, R.; Pantoş, G.D.; Gonçalves, D.P.N.; Sanders, J.K.M.; Balasubramanian, S. Ligand-driven G-quadruplex conformational switching by using an unusual mode of interaction. Angew. Chem. Int. Ed. 2007, 46, 5405–5407. [Google Scholar] [CrossRef] [PubMed]
- He, J.H.; Liu, H.Y.; Li, Z.; Tan, J.H.; Ou, T.M.; Huang, S.L.; An, L.K.; Li, D.; Gu, L.Q.; Huang, Z.S. New quinazoline derivatives for telomeric G-quadruplex DNA: Effects of an added phenyl group on quadruplex binding ability. Eur. J. Med. Chem. 2013, 63, 1–13. [Google Scholar] [CrossRef]
- Cousins, A.R.O.; Ritson, D.; Sharma, P.; Stevens, M.F.G.; Moses, J.E.; Searle, M.S. Ligand selectivity in stabilising tandem parallel folded G-quadruplex motifs in human telomeric DNA sequences. Chem. Commun. 2014, 50, 15202–15205. [Google Scholar] [CrossRef] [Green Version]
- Xing, X.; Wang, X.; Xu, L.; Tai, Y.; Dai, L.; Zheng, X.; Mao, W.; Xu, X.; Zhou, X. Light-driven conformational regulation of human telomeric G-quadruplex DNA in physiological conditions. Org. Biomol. Chem. 2011, 9, 6639–6645. [Google Scholar] [CrossRef]
- Wang, Z.F.; Li, M.H.; Chen, W.W.; Hsu, S.T.D.; Chang, T.C. A novel transition pathway of ligand-induced topological conversion from hybrid forms to parallel forms of human telomeric G-quadruplexes. Nucleic Acids Res. 2016, 44, 3958–3968. [Google Scholar] [CrossRef] [Green Version]
- Marchand, A.; Granzhan, A.; Iida, K.; Tsushima, Y.; Ma, Y.; Nagasawa, K.; Teulade-Fichou, M.-P.; Gabelica, V. Ligand-Induced Conformational Changes with Cation Ejection upon Binding to Human Telomeric DNA G-Quadruplexes. J. Am. Chem. Soc. 2015, 137, 750–756. [Google Scholar] [CrossRef] [Green Version]
- Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Polarization spectroscopy methods in the determination of interactions of small molecules with nucleic acids-Tutorial. Beilstein J. Org. Chem. 2017, 14, 84–105. [Google Scholar] [CrossRef]
- Nanjunda, R.; Musetti, C.; Kumar, A.; Ismail, M.A.; Farahat, A.A.; Wang, S.; Sissi, C.; Palumbo, M.; Boykin, D.W.; Wilson, W.D. Heterocyclic Dications as a New Class of Telomeric G-Quadruplex Targeting Agents. Curr. Pharm. Des. 2012, 18, 1934–1947. [Google Scholar] [CrossRef] [Green Version]
- Becher, J.; Berdnikova, D.V.; Ihmels, H.; Stremmel, C. Synthesis and investigation of quadruplex-DNA-binding, 9-O-substituted berberine derivatives. Beilstein J. Org. Chem. 2020, 16, 2795–2806. [Google Scholar] [CrossRef]
- Wickhorst, P.J.; Ihmels, H. Berberrubine phosphate: A selective fluorescent probe for quadruplex dna. Molecules 2021, 26, 2566. [Google Scholar] [CrossRef]
- Zuffo, M.; Doria, F.; Botti, S.; Bergamaschi, G.; Freccero, M. G-quadruplex fluorescence sensing by core-extended naphthalene diimides. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1303–1311. [Google Scholar] [CrossRef]
- Głuszyńska, A.; Juskowiak, B.; Rubiś, B. Binding study of the fluorescent carbazole derivative with human telomeric G-quadruplexes. Molecules 2018, 23, 3154. [Google Scholar] [CrossRef] [Green Version]
- Yaku, H.; Murashima, T.; Tateishi-Karimata, H.; Nakano, S.i.; Miyoshi, D.; Sugimoto, N. Study on effects of molecular crowding on G-quadruplex-ligand binding and ligand-mediated telomerase inhibition. Methods 2013, 64, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Krafcikova, M.; Dzatko, S.; Caron, C.; Granzhan, A.; Fiala, R.; Loja, T.; Teulade-Fichou, M.P.; Fessl, T.; Hänsel-Hertsch, R.; Mergny, J.L.; et al. Monitoring DNA-Ligand Interactions in Living Human Cells Using NMR Spectroscopy. J. Am. Chem. Soc. 2019, 141, 13281–13285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salgado, G.F.; Cazenave, C.; Kerkour, A.; Mergny, J.L. G-quadruplex DNA and ligand interaction in living cells using NMR spectroscopy. Chem. Sci. 2015, 6, 3314–3320. [Google Scholar] [CrossRef] [PubMed]
- Krafcikova, M.; Hänsel-Hertsch, R.; Trantirek, L.; Foldynova-Trantirkova, S. In Cell NMR Spectroscopy: Investigation of G-Quadruplex Structures Inside Living Xenopus laevis Oocytes. In Methods in Molecular Biology; Springer: Berlin, Germany, 2019; Volume 2035, pp. 397–405. [Google Scholar]
- Carver, T.R.; Slichter, C.P. Polarization of nuclear spins in metals. Phys. Rev. 1953, 92, 212–213. [Google Scholar] [CrossRef]
- Ni, F. Recent developments in transferred NOE methods. Prog. Nucl. Magn. Reson. Spectrosc. 1994, 26, 517–606. [Google Scholar] [CrossRef]
- Mayer, M.; Meyer, B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew. Chem. Int. Ed. 1999, 38, 1784–1788. [Google Scholar] [CrossRef]
- Dalvit, C.; Fogliatto, G.P.; Stewart, A.; Veronesi, M.; Stockman, B. WaterLOGSY as a method for primary NMR screening: Practical aspects and range of applicability. J. Biomol. NMR 2001, 21, 349–359. [Google Scholar] [CrossRef]
- Liu, W.; Lin, C.; Wu, G.; Dai, J.; Chang, T.C.; Yang, D. Structures of 1:1 and 2:1 complexes of BMVC and MYC promoter G-quadruplex reveal a mechanism of ligand conformation adjustment for G4-recognition. Nucleic Acids Res. 2019, 47, 11931–11942. [Google Scholar] [CrossRef]
- Kerkour, A.; Mergny, J.L.; Salgado, G.F. NMR based model of human telomeric repeat G-quadruplex in complex with 2,4,6-triarylpyridine family ligand. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1293–1302. [Google Scholar] [CrossRef]
- Kerkour, A.; Marquevielle, J.; Ivashchenko, S.; Yatsunyk, L.A.; Mergny, J.L.; Salgado, G.F. High-resolution three-dimensional NMR structure of the KRAS proto-oncogene promoter reveals key features of a G-quadruplex involved in transcriptional regulation. J. Biol. Chem. 2017, 292, 8082–8091. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, R.; Artali, R.; Benoit, A.; Gargallo, R.; Eritja, R.; Ferguson, D.M.; Sham, Y.Y.; Mazzini, S. Structure and Stability of Human Telomeric G-Quadruplex with Preclinical 9-Amino Acridines. PLoS ONE 2013, 8, e57701. [Google Scholar] [CrossRef]
- Dickerhoff, J.; Dai, J.; Yang, D. Structural recognition of the MYC promoter G-quadruplex by a quinoline derivative: Insights into molecular targeting of parallel G-quadruplexes. Nucleic Acids Res. 2021, 49, 5905–5915. [Google Scholar] [CrossRef] [PubMed]
- Tawani, A.; Mishra, S.K.; Kumar, A. Structural insight for the recognition of G-quadruplex structure at human c-myc promoter sequence by flavonoid Quercetin. Sci. Rep. 2017, 7, 3600. [Google Scholar] [CrossRef] [Green Version]
- Chung, W.J.; Heddi, B.; Hamon, F.; Teulade-Fichou, M.P.; Phan, A.T. Solution structure of a G-quadruplex bound to the bisquinolinium compound phen-DC3. Angew. Chem. Int. Ed. 2014, 53, 999–1002. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhong, Y.-F.; Liu, L.-Y.; Shen, C.-T.; Zeng, W.; Wang, F.; Yang, D.; Mao, Z.-W. Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding. Nat. Commun. 2018, 9, 3496. [Google Scholar] [CrossRef] [PubMed]
- Gimenez, D.; Phelan, A.; Murphy, C.D.; Cobb, S.L. 19F NMR as a tool in chemical biology. Beilstein J. Org. Chem. 2021, 17, 293–318. [Google Scholar] [CrossRef]
- Ishizuka, T.; Bao, H.L.; Xu, Y. 19F NMR Spectroscopy for the Analysis of DNA G-Quadruplex Structures Using 19F-Labeled Nucleobase. In Methods in Molecular Biology; Springer: Berlin, Germany, 2019; Volume 2035, pp. 407–433. [Google Scholar]
- Bao, H.L.; Ishizuka, T.; Iwanami, A.; Oyoshi, T.; Xu, Y. A Simple and Sensitive 19F NMR Approach for Studying the Interaction of RNA G-Quadruplex with Ligand Molecule and Protein. Chem. Sel. 2017, 2, 4170–4175. [Google Scholar] [CrossRef]
- Bao, H.L.; Ishizuka, T.; Sakamoto, T.; Fujimoto, K.; Uechi, T.; Kenmochi, N.; Xu, Y. Characterization of human telomere RNA G-quadruplex structures in vitro and in living cells using 19F NMR spectroscopy. Nucleic Acids Res. 2017, 45, 5501–5511. [Google Scholar] [CrossRef]
- Bao, H.L.; Liu, H.S.; Xu, Y. Hybrid-type and two-tetrad antiparallel telomere DNA G-quadruplex structures in living human cells. Nucleic Acids Res. 2019, 47, 4940–4947. [Google Scholar] [CrossRef]
- Bao, H.L.; Xu, Y. Telomeric DNA-RNA-hybrid G-quadruplex exists in environmental conditions of HeLa cells. Chem. Commun. 2020, 56, 6547–6550. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Collie, G.W. X-ray Crystallographic Studies of G-Quadruplex Structures. In Methods in Molecular Biology; Springer: Berlin, Germany, 2019; Volume 2035, pp. 131–155. [Google Scholar]
- Clark, G.R.; Pytel, P.D.; Squire, C.J.; Neidle, S. Structure of the first parallel DNA quadruplex-drug complex. J. Am. Chem. Soc. 2003, 125, 4066–4067. [Google Scholar] [CrossRef]
- Haider, S.M.; Parkinson, G.N.; Neidle, S. Structure of a G-quadruplex-ligand complex. J. Mol. Biol. 2003, 326, 117–125. [Google Scholar] [CrossRef]
- Lin, L.Y.; McCarthy, S.; Powell, B.M.; Manurung, Y.; Xiang, I.M.; Dean, W.L.; Chaires, B.; Yatsunyk, L.A. Biophysical and X-ray structural studies of the (GGGTT)3GGG G-quadruplex in complex with N-methyl mesoporphyrin IX. PLoS ONE 2020, 15, e0241513. [Google Scholar] [CrossRef] [PubMed]
- McQuaid, K.; Abell, H.; Gurung, S.P.; Allan, D.R.; Winter, G.; Sorensen, T.; Cardin, D.J.; Brazier, J.A.; Cardin, C.J.; Hall, J.P. Structural Studies Reveal Enantiospecific Recognition of a DNA G-Quadruplex by a Ruthenium Polypyridyl Complex. Angew. Chem. Int. Ed. 2019, 58, 9881–9885. [Google Scholar] [CrossRef] [PubMed]
- Guarra, F.; Marzo, T.; Ferraroni, M.; Papi, F.; Bazzicalupi, C.; Gratteri, P.; Pescitelli, G.; Messori, L.; Biver, T.; Gabbiani, C. Interaction of a gold(i) dicarbene anticancer drug with human telomeric DNA G-quadruplex: Solution and computationally aided X-ray diffraction analysis. Dalt. Trans. 2018, 47, 16132–16138. [Google Scholar] [CrossRef] [PubMed]
- Bazzicalupi, C.; Ferraroni, M.; Bilia, A.R.; Scheggi, F.; Gratteri, P. The crystal structure of human telomeric DNA complexed with berberine: An interesting case of stacked ligand to G-tetrad ratio higher than 1:1. Nucleic Acids Res. 2013, 41, 632–638. [Google Scholar] [CrossRef]
- Prado, E.; Bonnat, L.; Bonnet, H.; Lavergne, T.; Van Der Heyden, A.; Pratviel, G.; Dejeu, J.; Defrancq, E. Influence of the SPR Experimental Conditions on the G-Quadruplex DNA Recognition by Porphyrin Derivatives. Langmuir 2018, 34, 13057–13064. [Google Scholar] [CrossRef]
- Perenon, M.; Bonnet, H.; Lavergne, T.; Dejeu, J.; Defrancq, E. Surface plasmon resonance study of the interaction of: N -methyl mesoporphyrin IX with G-quadruplex DNA. Phys. Chem. Chem. Phys. 2020, 22, 4158–4164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda, A.; Santos, T.; Largy, E.; Cruz, C. Locking up the as1411 aptamer with a flanking duplex: Towards an improved nucleolin-targeting. Pharmaceuticals 2021, 14, 121. [Google Scholar] [CrossRef] [PubMed]
- Vo, T.; Paul, A.; Kumar, A.; Boykin, D.W.; Wilson, W.D. Biosensor-surface plasmon resonance: A strategy to help establish a new generation RNA-specific small molecules. Methods 2019, 167, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Zhou, J.; Gu, J.; Xu, M.; Xu, X.; Yuan, G. Probing the G-quadruplex from hsa-miR-3620-5p and inhibition of its interaction with the target sequence. Talanta 2016, 154, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Pagano, B.; Mattia, C.A.; Giancola, C. Applications of isothermal titration calorimetry in biophysical studies of G-quadruplexes. Int. J. Mol. Sci. 2009, 10, 2935–2957. [Google Scholar] [CrossRef] [Green Version]
- Giancola, C.; Pagano, B. Energetics of ligand binding to G-quadruplexes. Top. Curr. Chem. 2013, 330, 211–242. [Google Scholar] [CrossRef]
- Funke, A.; Weisz, K. Revealing the Energetics of Ligand-Quadruplex Interactions Using Isothermal Titration Calorimetry. In Methods in Molecular Biology; Springer: Berlin, Germany, 2019; Volume 2035, pp. 45–61. [Google Scholar]
- Funke, A.; Dickerhoff, J.; Weisz, K. Towards the Development of Structure-Selective G-Quadruplex-Binding Indolo[3,2-b]quinolines. Chem. Eur. J. 2016, 22, 3170–3181. [Google Scholar] [CrossRef]
- Funke, A.; Weisz, K. Comprehensive Thermodynamic Profiling for the Binding of a G-Quadruplex Selective Indoloquinoline. J. Phys. Chem. 2017, 121, 5735–5743. [Google Scholar] [CrossRef]
- Funke, A.; Karg, B.; Dickerhoff, J.; Balke, D.; Müller, S.; Weisz, K. Ligand-Induced Dimerization of a Truncated Parallel MYC G-Quadruplex. ChemBioChem 2018, 19, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Arnaiz, C.; Busto, N.; Santolaya, J.; Leal, J.M.; Barone, G.; García, B. Kinetic evidence for interaction of TMPyP4 with two different G-quadruplex conformations of human telomeric DNA. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 522–531. [Google Scholar] [CrossRef]
- Dupont, J.I.; Henderson, K.L.; Metz, A.; Le, V.H.; Emerson, J.P.; Lewis, E.A. Calorimetric and spectroscopic investigations of the binding of metallated porphyrins to G-quadruplex DNA. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Bončina, M.; Podlipnik, Č.; Piantanida, I.; Eilmes, J.; Teulade-Fichou, M.P.; Vesnaver, G.; Lah, J. Thermodynamic fingerprints of ligand binding to human telomeric G-quadruplexes. Nucleic Acids Res. 2015, 43, 10376–10386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bončina, M.; Hamon, F.; Islam, B.; Teulade-Fichou, M.P.; Vesnaver, G.; Haider, S.; Lah, J. Dominant Driving Forces in Human Telomere Quadruplex Binding-Induced Structural Alterations. Biophys. J. 2015, 108, 2903–2911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alniss, H.; Zamiri, B.; Khalaj, M.; Pearson, C.E.; Macgregor, R.B. Thermodynamic and spectroscopic investigations of TMPyP4 association with guanine- and cytosine-rich DNA and RNA repeats of C9orf72. Biochem. Biophys. Res. Commun. 2018, 495, 2410–2417. [Google Scholar] [CrossRef]
- Yuan, G.; Zhang, Q.; Zhou, J.; Li, H. Mass spectrometry of G-quadruplex DNA: Formation, recognition, property, conversion, and conformation. Mass Spectrom. Rev. 2011, 30, 1121–1142. [Google Scholar] [CrossRef]
- Li, H. Mass Spectroscopic Study of G-Quadruplex. In Methods in Molecular Biology; Springer: Berlin, Germany, 2019; Volume 2035, pp. 105–116. [Google Scholar]
- Lecours, M.J.; Marchand, A.; Anwar, A.; Guetta, C.; Hopkins, W.S.; Gabelica, V. What stoichiometries determined by mass spectrometry reveal about the ligand binding mode to G-quadruplex nucleic acids. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1353–1361. [Google Scholar] [CrossRef] [Green Version]
- Marchand, A.; Strzelecka, D.; Gabelica, V. Selective and Cooperative Ligand Binding to Antiparallel Human Telomeric DNA G-Quadruplexes. Chem. Eur. J. 2016, 22, 9551–9555. [Google Scholar] [CrossRef]
- Ceschi, S.; Largy, E.; Gabelica, V.; Sissi, C. A two-quartet G-quadruplex topology of human KIT2 is conformationally selected by a perylene derivative. Biochimie 2020, 179, 77–84. [Google Scholar] [CrossRef]
- Marchand, A.; Rosu, F.; Zenobi, R.; Gabelica, V. Thermal Denaturation of DNA G-Quadruplexes and Their Complexes with Ligands: Thermodynamic Analysis of the Multiple States Revealed by Mass Spectrometry. J. Am. Chem. Soc. 2018, 140, 12553–12565. [Google Scholar] [CrossRef]
- Paul, D.; Marchand, A.; Verga, D.; Teulade-Fichou, M.P.; Bombard, S.; Rosu, F.; Gabelica, V. Probing ligand and cation binding sites in G-quadruplex nucleic acids by mass spectrometry and electron photodetachment dissociation sequencing. Analyst 2019, 144, 3518–3524. [Google Scholar] [CrossRef] [PubMed]
- Scalabrin, M.; Palumbo, M.; Richter, S.N. Highly Improved Electrospray Ionization-Mass Spectrometry Detection of G-Quadruplex-Folded Oligonucleotides and Their Complexes with Small Molecules. Anal. Chem. 2017, 89, 8632–8637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, J.; Cruz, C. Forster resonance energy transfer for studying nucleic acids denaturation: A chemical and biological sciences laboratory experiment. Biochem. Mol. Biol. Educ. 2020, 48, 329–336. [Google Scholar] [CrossRef]
- Carvalho, J.; Quintela, T.; Gueddouda, N.M.; Bourdoncle, A.; Mergny, J.L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. Phenanthroline polyazamacrocycles as G-quadruplex DNA binders. Org. Biomol. Chem. 2018, 16, 2776–2786. [Google Scholar] [CrossRef]
- Lavrado, J.; Borralho, P.M.; Ohnmacht, S.A.; Castro, R.E.; Rodrigues, C.M.P.; Moreira, R.; Dos Santos, D.J.V.A.; Neidle, S.; Paulo, A. Synthesis, G-quadruplex stabilisation, docking studies, and effect on cancer cells of indolo[3,2-b]quinolines with one, two, or three basic side chains. Chem. Med. Chem. 2013, 8, 1648–1661. [Google Scholar] [CrossRef] [Green Version]
- Lavrado, J.; Brito, H.; Borralho, P.M.; Ohnmacht, S.A.; Kim, N.S.; Leitão, C.; Pisco, S.; Gunaratnam, M.; Rodrigues, C.M.P.; Moreira, R.; et al. KRAS oncogene repression in colon cancer cell lines by G-quadruplex binding indolo[3,2-c]quinolines. Sci. Rep. 2015, 5, 9696. [Google Scholar] [CrossRef] [Green Version]
- Cadoni, E.; Magalh, P.R.; Em, R.M.; Mendes, E.; Jorge, V.; Carvalho, J.; Cruz, C.; Victor, B.L.; Paulo, A. New (Iso) quinolinyl-pyridine-2,6-dicarboxamide G-Quadruplex Stabilizers. A Structure-Activity Relationship Study. Pharmaceuticals 2021, 14, 669. [Google Scholar] [CrossRef]
- Noureini, S.K.; Esmaeili, H.; Abachi, F.; Khiali, S.; Islam, B.; Kuta, M.; Saboury, A.A.; Hoffmann, M.; Sponer, J.; Parkinson, G.; et al. Selectivity of major isoquinoline alkaloids from Chelidonium majus towards telomeric G-quadruplex: A study using a transition-FRET (t-FRET) assay. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2020–2030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakers, V.; Cadinu, P.; Edel, J.B.; Vilar, R. Development of microfluidic platforms for the synthesis of metal complexes and evaluation of their DNA affinity using online FRET melting assays. Chem. Sci. 2018, 9, 3459–3469. [Google Scholar] [CrossRef] [Green Version]
- De Cian, A.; Guittat, L.; Shin-ya, K.; Riou, J.F.; Mergny, J.L. Affinity and selectivity of G4 ligands measured by FRET. Nucleic Acids Symp. Ser. 2005, 49, 235–236. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Granzhan, A.; Verga, D.; Mergny, J.L. FRET-MC: A fluorescence melting competition assay for studying G4 structures in vitro. Biopolymers 2021, 112, e23415. [Google Scholar] [CrossRef] [PubMed]
- Monchaud, D.; Allain, C.; Teulade-Fichou, M.P. Development of a fluorescent intercalator displacement assay (G4-FID) for establishing quadruplex-DNA affinity and selectivity of putative ligands. Bioorg. Med. Chem. Lett. 2006, 16, 4842–4845. [Google Scholar] [CrossRef]
- Monchaud, D.; Teulade-Fichou, M.P. G4-FID: A fluorescent DNA probe displacement assay for rapid evaluation of quadruplex ligands. Methods Mol. Biol. 2010, 608, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Monchaud, D.; Allain, C.; Bertrand, H.; Smargiasso, N.; Rosu, F.; Gabelica, V.; De Cian, A.; Mergny, J.L.; Teulade-Fichou, M.P. Ligands playing musical chairs with G-quadruplex DNA: A rapid and simple displacement assay for identifying selective G-quadruplex binders. Biochimie 2008, 90, 1207–1223. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; Hamon, F.; Teulade-Fichou, M.P. Development of a high-throughput G4-FID assay for screening and evaluation of small molecules binding quadruplex nucleic acid structures. Anal. Bioanal. Chem. 2011, 400, 3419–3427. [Google Scholar] [CrossRef]
- Tran, P.L.T.; Largy, E.; Hamon, F.; Teulade-Fichou, M.P.; Mergny, J.L. Fluorescence intercalator displacement assay for screening G4 ligands towards a variety of G-quadruplex structures. Biochimie 2011, 93, 1288–1296. [Google Scholar] [CrossRef]
- Beauvineau, C.; Guetta, C.; Teulade-Fichou, M.P.; Mahuteau-Betzer, F. PhenDV, a turn-off fluorescent quadruplex DNA probe for improving the sensitivity of drug screening assays. Org. Biomol. Chem. 2017, 15, 7117–7121. [Google Scholar] [CrossRef]
- Desai, N.; Shah, V.; Datta, B. Assessing G4-binding ligands in vitro and in cellulo using dimeric carbocyanine dye displacement assay. Molecules 2021, 26, 1400. [Google Scholar] [CrossRef]
- del Villar-Guerra, R.; Gray, R.D.; Trent, J.O.; Chaires, J.B. A rapid fluorescent indicator displacement assay and principal component/cluster data analysis for determination of ligand–nucleic acid structural selectivity. Nucleic Acids Res. 2018, 46, e41. [Google Scholar] [CrossRef] [Green Version]
- Santos, T.; Pereira, P.; Sousa, F.; Queiroz, J.A.; Cruz, C. Purification of supercoiled G-quadruplex pDNA for in vitro transcription. Sep. Purif. Technol. 2016, 163, 59–71. [Google Scholar] [CrossRef]
- Smith, J.S.; Johnson, F.B. Isolation of G-quadruplex DNA using NMM-sepharose affinity chromatography. Methods Mol. Biol. 2010, 608, 207–221. [Google Scholar] [CrossRef]
- Ferreira, J.; Santos, T.; Pereira, P.; Corvo, M.C.; Queiroz, J.A.; Sousa, F.; Cruz, C. Naphthalene amine support for G-quadruplex isolation. Analyst 2017, 142, 2982–2994. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.; Liu, X.; Cheng, X.; Qi, C.; Mei, H.; Shangguan, D. Selective isolation of G-quadruplexes by affinity chromatography. J. Chromatogr. 2012, 1246, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, D.; Amato, J.; Randazzo, A.; Novellino, E.; Giancola, C.; Montesarchio, D.; Pagano, B. G-quadruplex on oligo affinity support (G4-OAS): An easy affinity chromatography-based assay for the screening of G-quadruplex ligands. Anal. Chem. 2014, 86, 4126–4130. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, D.; Amato, J.; Zizza, P.; Platella, C.; Cosconati, S.; Cingolani, C.; Biroccio, A.; Novellino, E.; Randazzo, A.; Giancola, C.; et al. Tandem application of ligand-based virtual screening and G4-OAS assay to identify novel G-quadruplex-targeting chemotypes. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Musumeci, D.; Arciello, A.; Doria, F.; Freccero, M.; Randazzo, A.; Amato, J.; Pagano, B.; Montesarchio, D. Controlled Pore Glass-based oligonucleotide affinity support: Towards High Throughput Screening methods for the identification of conformation-selective G-quadruplex ligands. Anal. Chim. Acta 2018, 1030, 133–141. [Google Scholar] [CrossRef]
- Pirota, V.; Platella, C.; Musumeci, D.; Benassi, A.; Amato, J.; Pagano, B.; Colombo, G.; Freccero, M.; Doria, F.; Montesarchio, D. On the binding of naphthalene diimides to a human telomeric G-quadruplex multimer model. Int. J. Biol. Macromol. 2021, 166, 1320–1334. [Google Scholar] [CrossRef]
- Ray, S.; Tillo, D.; Boer, R.E.; Assad, N.; Barshai, M.; Wu, G.; Orenstein, Y.; Yang, D.; Schneekloth, J.S.; Vinson, C. Custom DNA Microarrays Reveal Diverse Binding Preferences of Proteins and Small Molecules to Thousands of G-Quadruplexes. ACS Chem. Biol. 2020, 15, 925–935. [Google Scholar] [CrossRef]
- Wu, G.; Tillo, D.; Ray, S.; Chang, T.C.; Schneekloth, J.S.; Vinson, C.; Yang, D. Custom G4 microarrays reveal selective G-quadruplex recognition of small molecule BMVC: A large-scale assessment of ligand binding selectivity. Molecules 2020, 25, 3465. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, K.M.; Saunders, L.B.; Simmons, J.K.; Leon, E.; Calabrese, D.R.; Zhang, S.; Michalowski, A.; Gareiss, P.; Mock, B.A.; Schneekloth, J.S. Small Molecule Microarrays Enable the Identification of a Selective, Quadruplex-Binding Inhibitor of MYC Expression. ACS Chem. Biol. 2016, 11, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, D.R.; Chen, X.; Leon, E.C.; Gaikwad, S.M.; Phyo, Z.; Hewitt, W.M.; Alden, S.; Hilimire, T.A.; He, F.; Michalowski, A.M.; et al. Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex. Nat. Commun. 2018, 9, 4229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gracie, K.; Dhamodharan, V.; Pradeepkumar, P.I.; Dhamodharan, V.; Graham, D. Qualitative SERS analysis of G-quadruplex DNAs using selective stabilising ligands. Analyst 2014, 139, 4458–4465. [Google Scholar] [CrossRef] [Green Version]
- Aznauryan, M.; Noer, S.L.; Pedersen, C.W.; Mergny, J.L.; Teulade-Fichou, M.P.; Birkedal, V. Ligand Binding to Dynamically Populated G-Quadruplex DNA. ChemBioChem 2021, 22, 1811–1817. [Google Scholar] [CrossRef]
- Rosu, F.; De Pauw, E.; Guittat, L.; Alberti, P.; Lacroix, L.; Mailliet, P.; Riou, J.F.; Mergny, J.L. Selective interaction of ethidium derivatives with quadruplexes: An equilibrium dialysis and electrospray ionization mass spectrometry analysis. Biochemistry 2003, 42, 10361–10371. [Google Scholar] [CrossRef]
- Saad, M.; Guédin, A.; Amor, S.; Bedrat, A.; Tourasse, N.J.; Fayyad-Kazan, H.; Pratviel, G.; Lacroix, L.; Mergny, J.L. Mapping and characterization of G-quadruplexes in the genome of the social amoeba Dictyostelium discoideum. Nucleic Acids Res. 2019, 47, 4363–4374. [Google Scholar] [CrossRef] [Green Version]
- Jamroskovic, J.; Obi, I.; Movahedi, A.; Chand, K.; Chorell, E.; Sabouri, N. Identification of putative G-quadruplex DNA structures in S. pombe genome by quantitative PCR stop assay. DNA Repair. 2019, 82, 102678. [Google Scholar] [CrossRef]
- Wu, G.; Han, H. A DNA Polymerase Stop Assay for Characterization of G-Quadruplex Formation and Identification of G-Quadruplex-Interactive Compounds. In Methods in Molecular Biology; Springer: Berlin, Germany, 2019; Volume 2035, pp. 223–231. [Google Scholar]
- Gomez, D.; Mergny, J.L.; Riou, J.F. Detection of telomerase inhibitors based on G-quadruplex ligands by a modified telomeric repeat amplification protocol assay. Cancer Res. 2002, 62, 3365–3368. [Google Scholar]
- Panda, D.; Saha, P.; Chaudhuri, R.; Prasanth, T.; Ravichandiran, V.; Dash, J. A Competitive Pull-Down Assay Using G-quadruplex DNA Linked Magnetic Nanoparticles to Determine Specificity of G-quadruplex Ligands. Anal. Chem. 2019, 91, 7705–7711. [Google Scholar] [CrossRef]
- Flusberg, D.A.; Rizvi, N.F.; Kutilek, V.; Andrews, C.; Saradjian, P.; Chamberlin, C.; Curran, P.; Swalm, B.; Kattar, S.; Smith, G.F.; et al. Identification of G-Quadruplex-Binding Inhibitors of Myc Expression through Affinity Selection–Mass Spectrometry. SLAS Discov. 2019, 24, 142–157. [Google Scholar] [CrossRef] [PubMed]












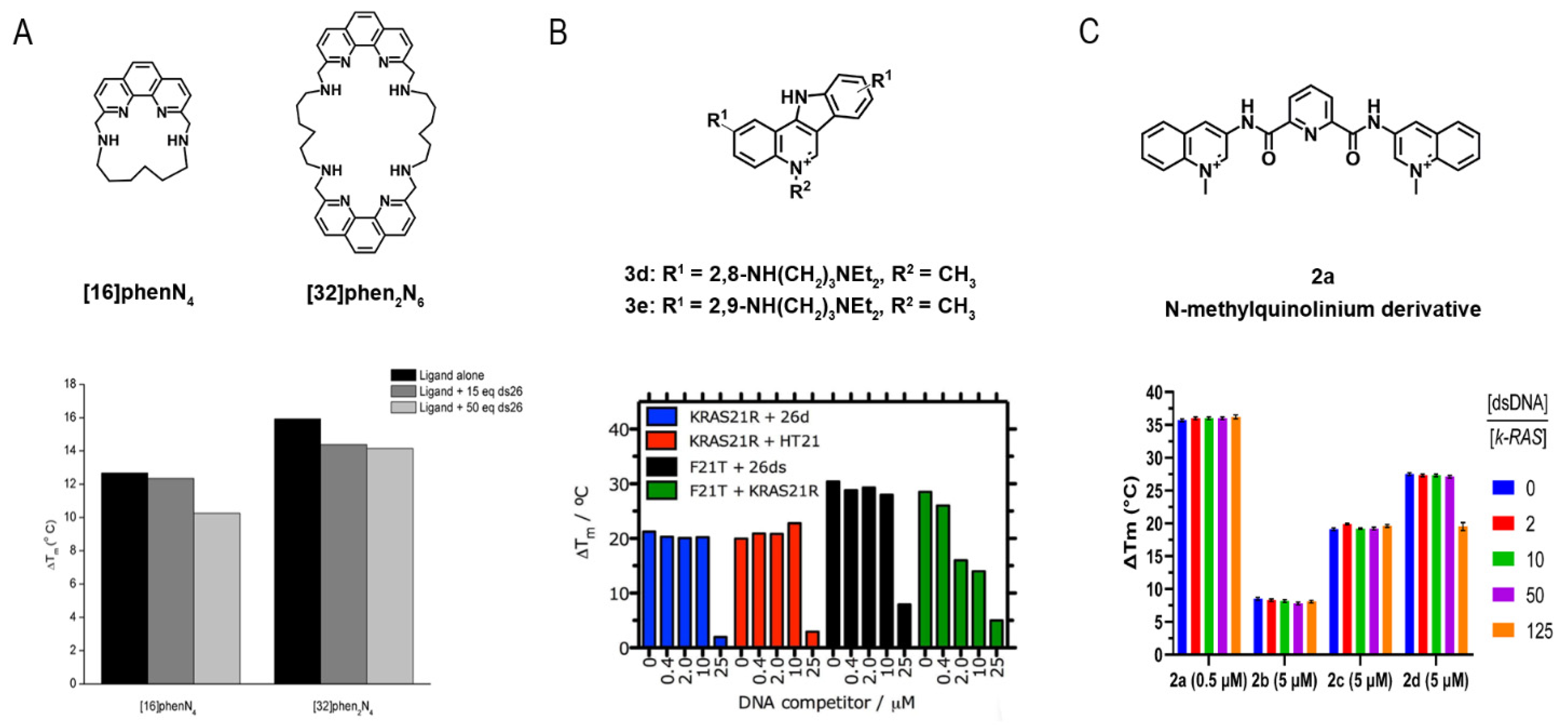
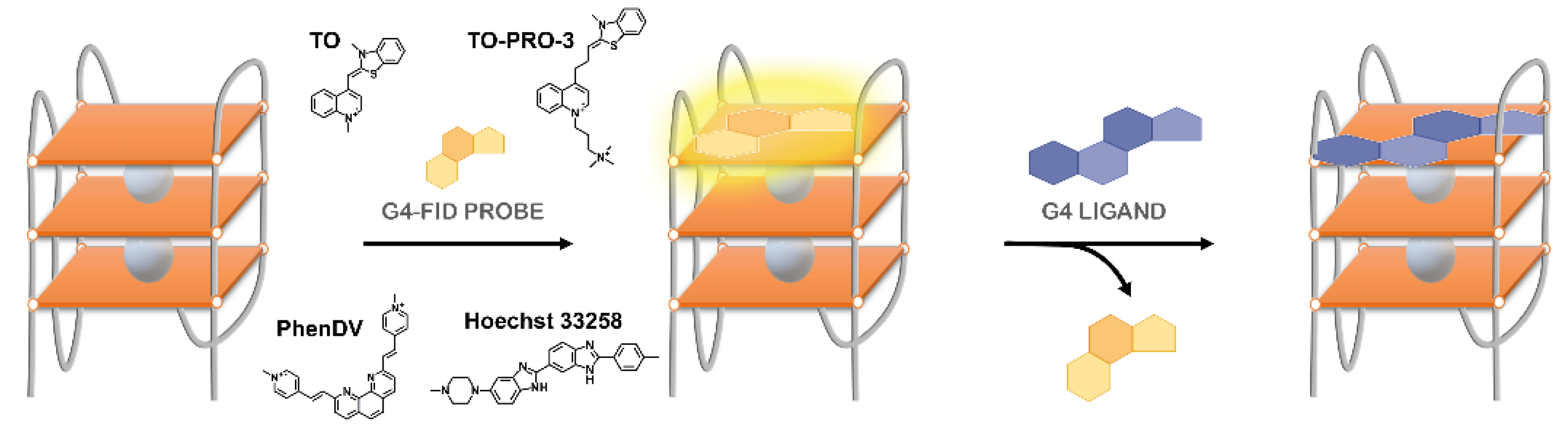

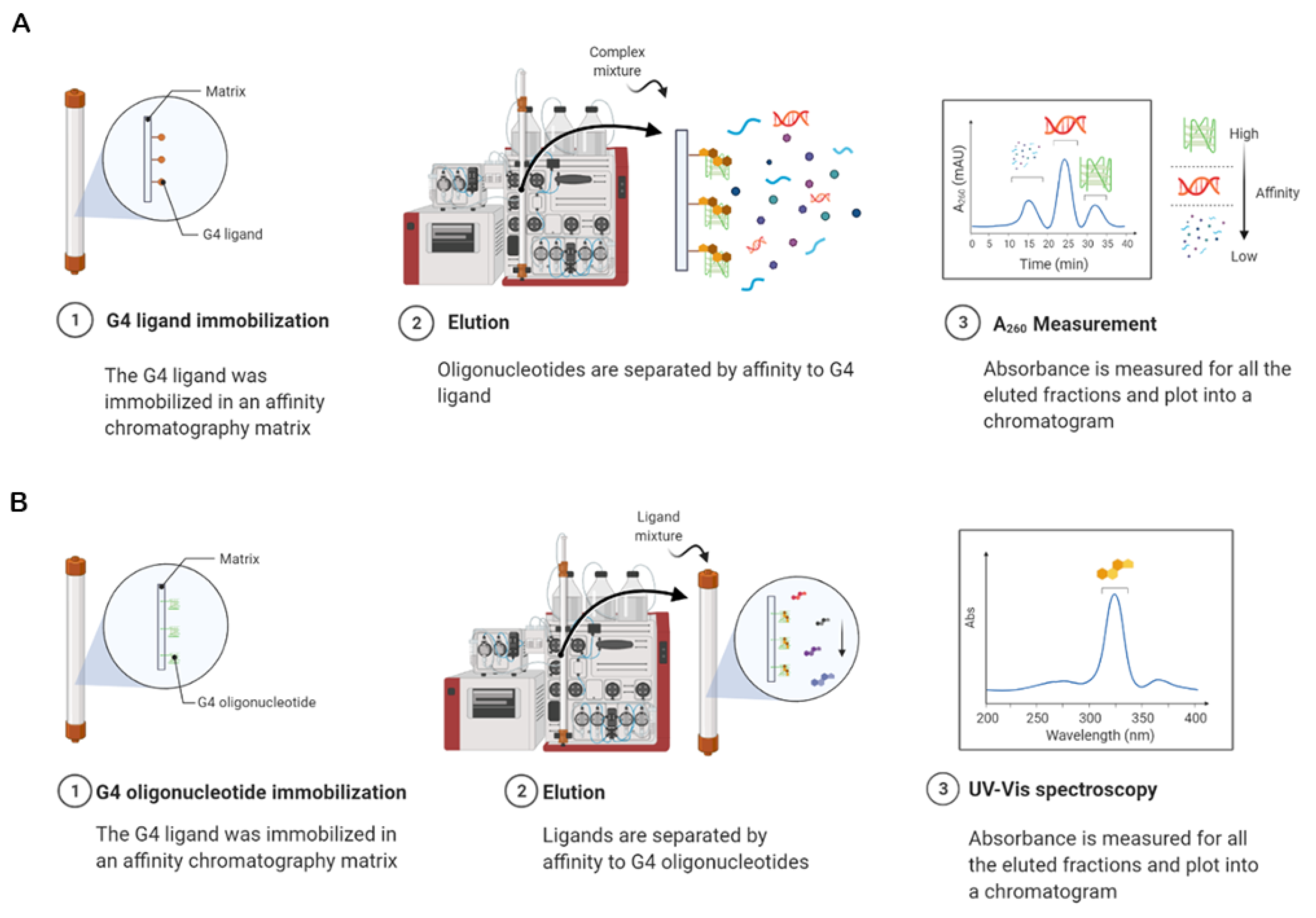
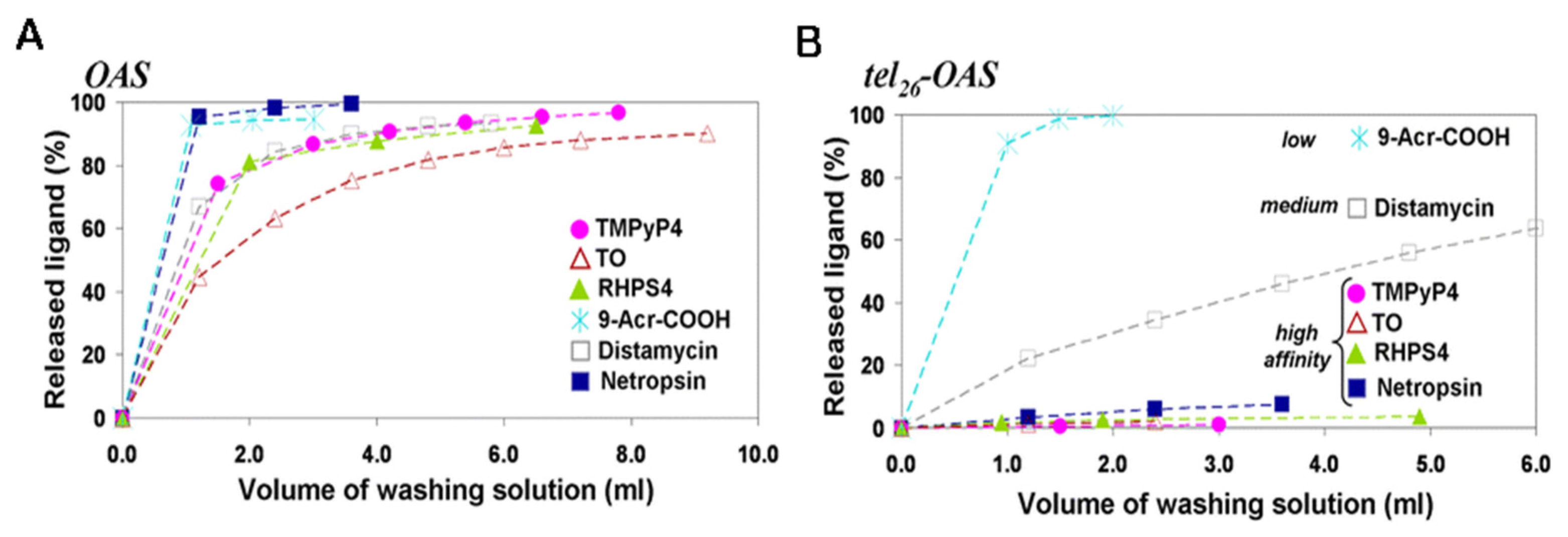
| Method | Advantages | Limitations |
|---|---|---|
| CD | Simplicity | Low-resolution |
| Small amount of sample | ||
| No need of sample labelling | ||
| Not limited by the molecular weight or size of the molecules | Most of the ligands are non-optically active | |
| Can easily provide melting temperature curves and global folding changes | ||
| Most suitable method for finding the polarity of chains | ||
| NMR | Provides atomic-resolution characterization of a G4/ligand complex | High amount of sample |
| Detailed pairs of atoms contacts between ligand and receptor | Time-consuming | |
| Three-dimensional structures in their natural state can be measured in solution | Limited by size or atomic weight | |
| Need of isotopic labelling | ||
| X-ray | Relatively cheap and simple | High amount of sample |
| Provides atomic-resolution characterization of a G4/ligand complex | Cryogenic temperature can induce altered contacts | |
| Provides void electronic areas in the receptor that can be used to improve ligands design | The sample must be crystallizable | |
| Not limited by size or atomic weight | Only provides static three-dimensional analysis |
| Method | Advantages | Limitations |
|---|---|---|
| SPR | Acquisition of data in real time | Requires sophisticated instrumentation and it is costly |
| Time efficiency | High dependence of experimental conditions | |
| High sensitivity | SPR often requires labeling with biotin | |
| Provides valuable kinetic and affinity information (association (Ka or Kon), dissociation (Kd or Koff) andequilibrium (KD) constants) | Requirement of maintaining the G4 structure intact after immobilization | |
| ITC | Provides insights of molecular forces that drive the interaction | High amount of sample |
| Provides kinetic and thermodynamically parameters | Ligands should be soluble in water | |
| MS | Provides information on formation, stoichiometry, and binding affinity of G4/ligand complex | Limitations regarding the media |
| Method | Advantages | Limitations |
|---|---|---|
| FRET-melting | Simplicity | Fluorescently labelled oligonucleotides |
| Small amount of sample | It only measures ligand-induced G4 stabilization, while other types of interactions are not detected | |
| Real-time monitoring | ||
| G4-FID | Simplicity | Ambiguous binding mode of used probes |
| Small amount of sample | Compatibility of the oligonucleotides with the fluorescent probe | |
| Affinity Chromatography | Selective and reversible interactions that undergo with the ligand and G4 | Unspecific binding of the ligand to the resin |
| Column chromatography allow real-time monitoring | The absence of structural information about G4 conformation | |
| Microarrays | Small amount of sample | Expensive |
| Massive parallel screening | Specialized equipment | |
| Fluorescently labeled molecules |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, T.; Salgado, G.F.; Cabrita, E.J.; Cruz, C. G-Quadruplexes and Their Ligands: Biophysical Methods to Unravel G-Quadruplex/Ligand Interactions. Pharmaceuticals 2021, 14, 769. https://doi.org/10.3390/ph14080769
Santos T, Salgado GF, Cabrita EJ, Cruz C. G-Quadruplexes and Their Ligands: Biophysical Methods to Unravel G-Quadruplex/Ligand Interactions. Pharmaceuticals. 2021; 14(8):769. https://doi.org/10.3390/ph14080769
Chicago/Turabian StyleSantos, Tiago, Gilmar F. Salgado, Eurico J. Cabrita, and Carla Cruz. 2021. "G-Quadruplexes and Their Ligands: Biophysical Methods to Unravel G-Quadruplex/Ligand Interactions" Pharmaceuticals 14, no. 8: 769. https://doi.org/10.3390/ph14080769
APA StyleSantos, T., Salgado, G. F., Cabrita, E. J., & Cruz, C. (2021). G-Quadruplexes and Their Ligands: Biophysical Methods to Unravel G-Quadruplex/Ligand Interactions. Pharmaceuticals, 14(8), 769. https://doi.org/10.3390/ph14080769








