Antimicrobial Activity of a Library of Thioxanthones and Their Potential as Efflux Pump Inhibitors
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Structure Elucidation
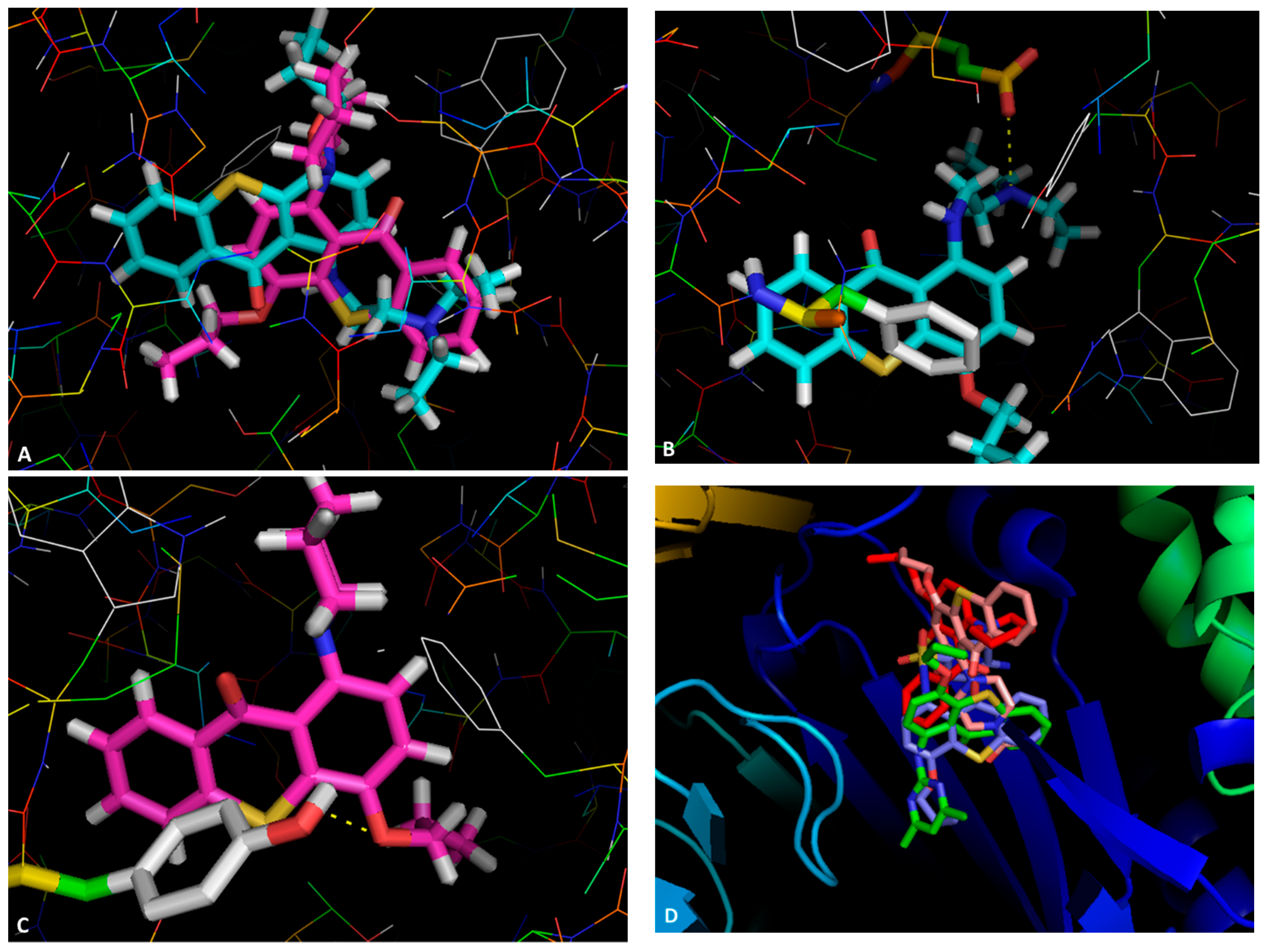
2.3. Docking Studies
2.4. Antibacterial Activity and Synergy with Antimicrobials
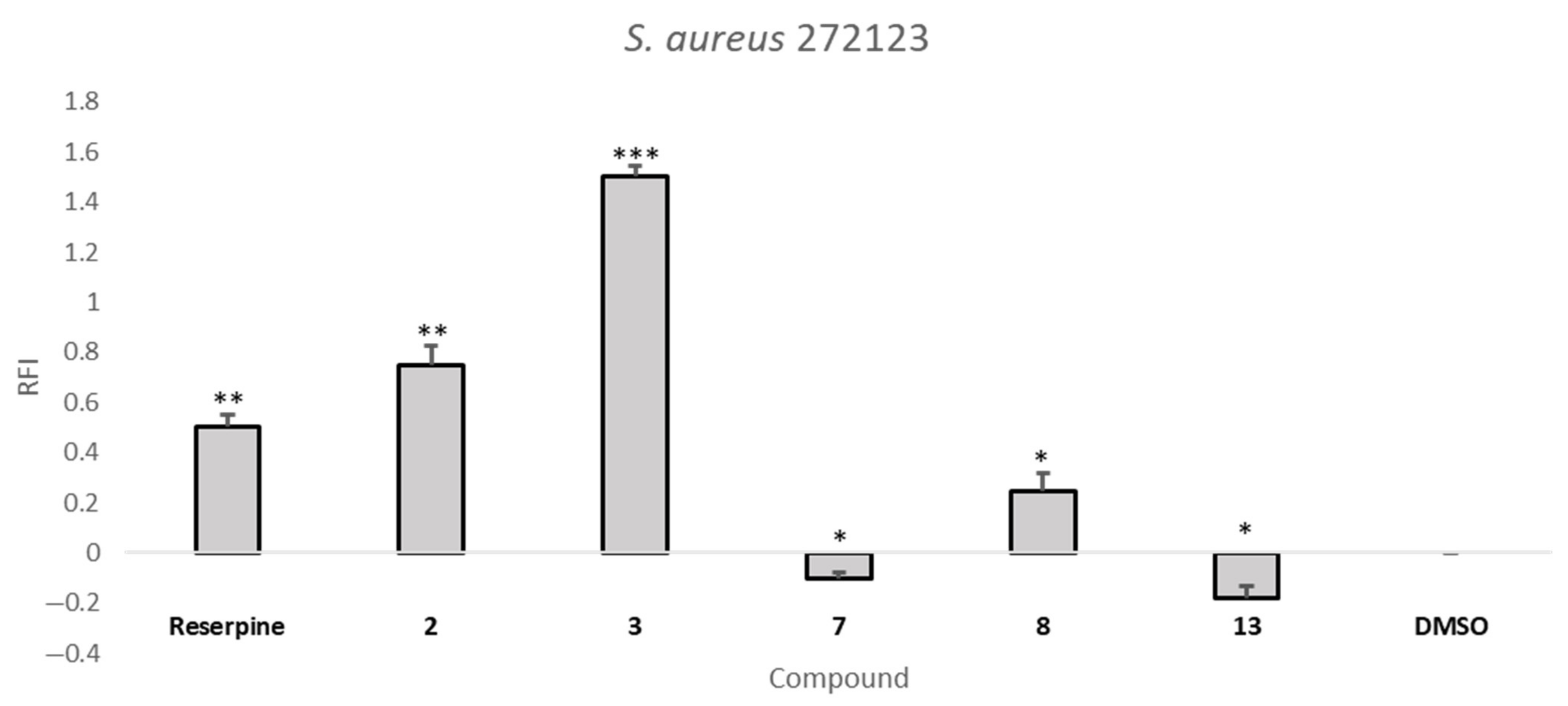
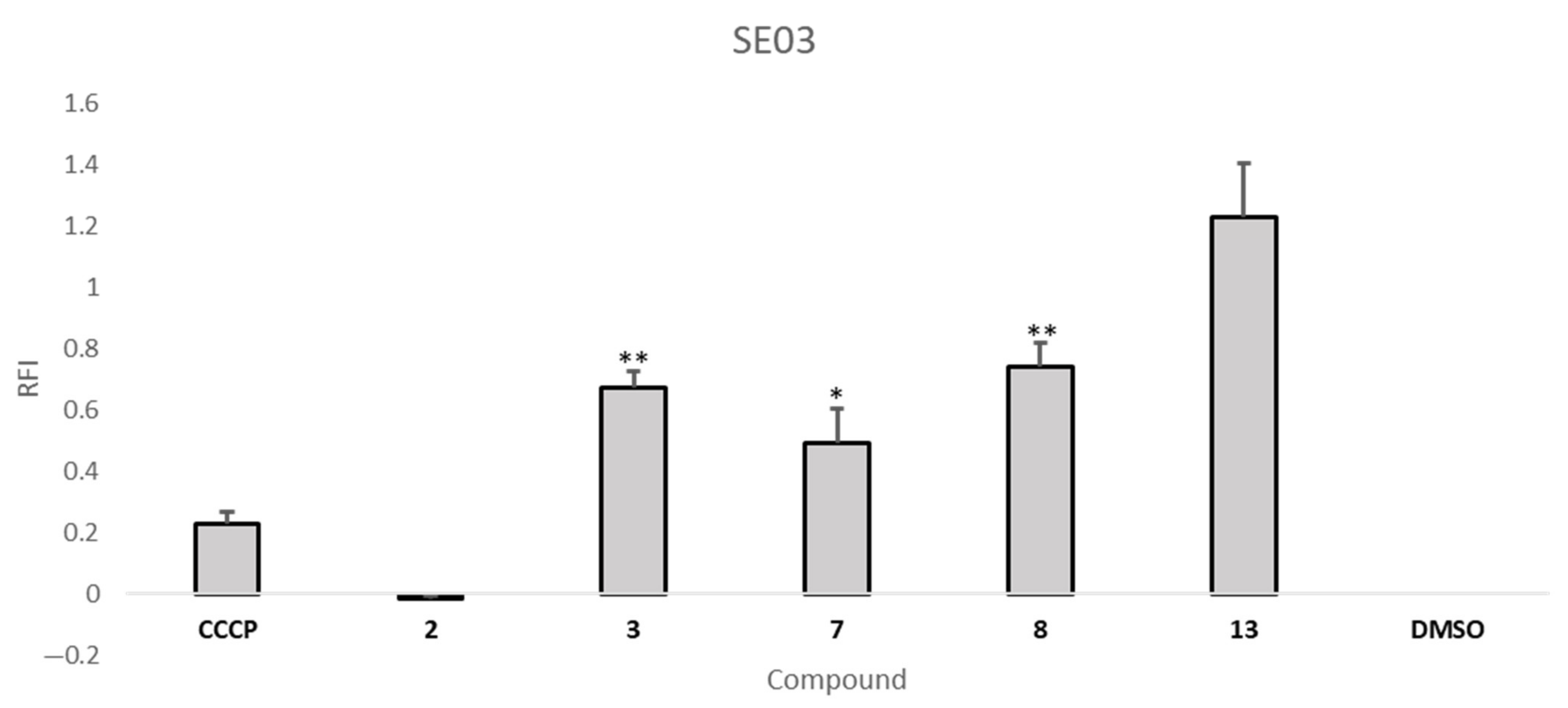
2.5. Efflux Pump Inhibition Assay
2.6. Biofilm Inhibition and Quorum Sensing Inhibition Assays
2.7. In Vitro Studies in Mammalian Cells
2.7.1. Cytotoxicity in Mouse Embryonic Fibroblast Cell Line
2.7.2. Compounds’ Cytotoxicity in Human Cells
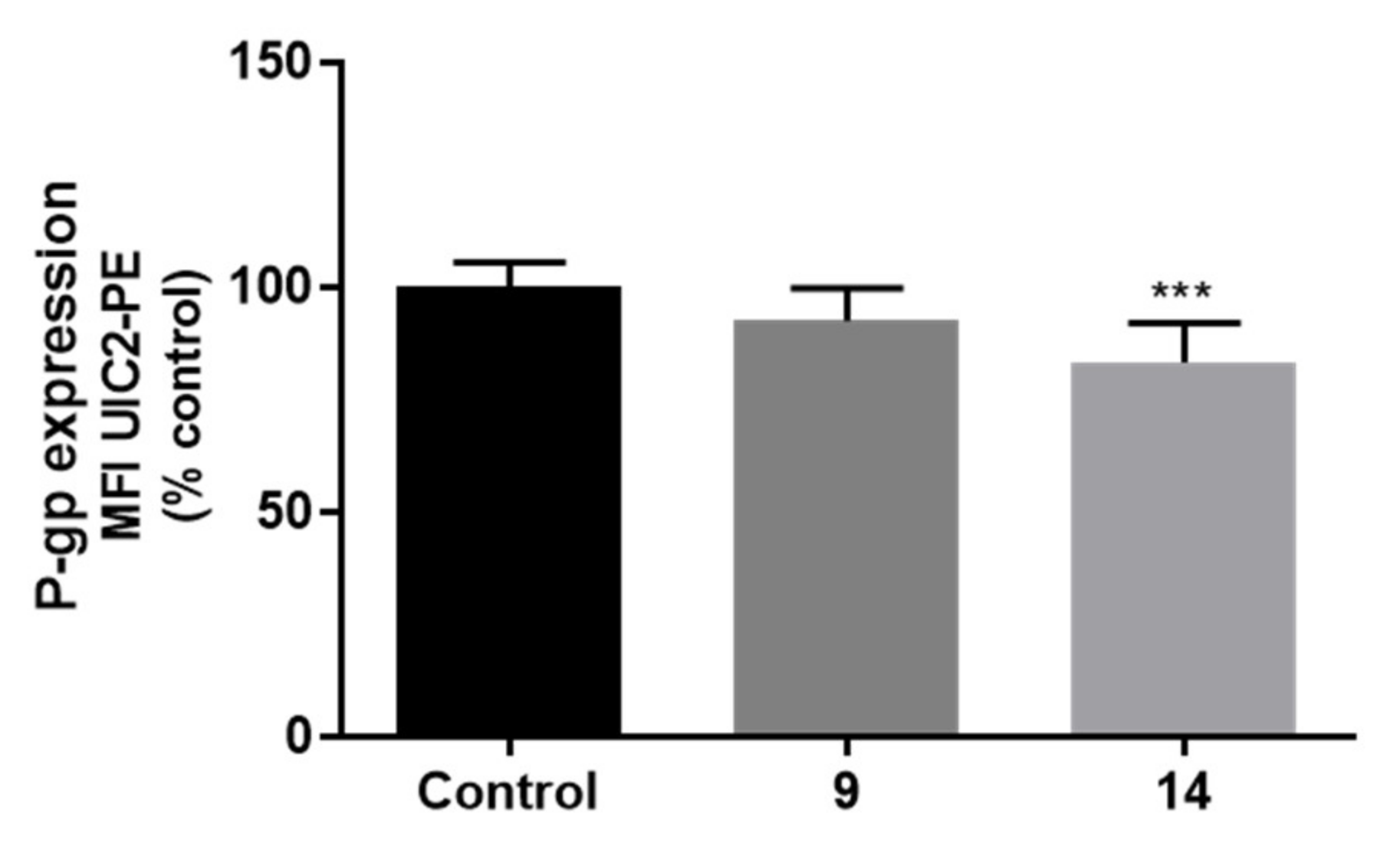
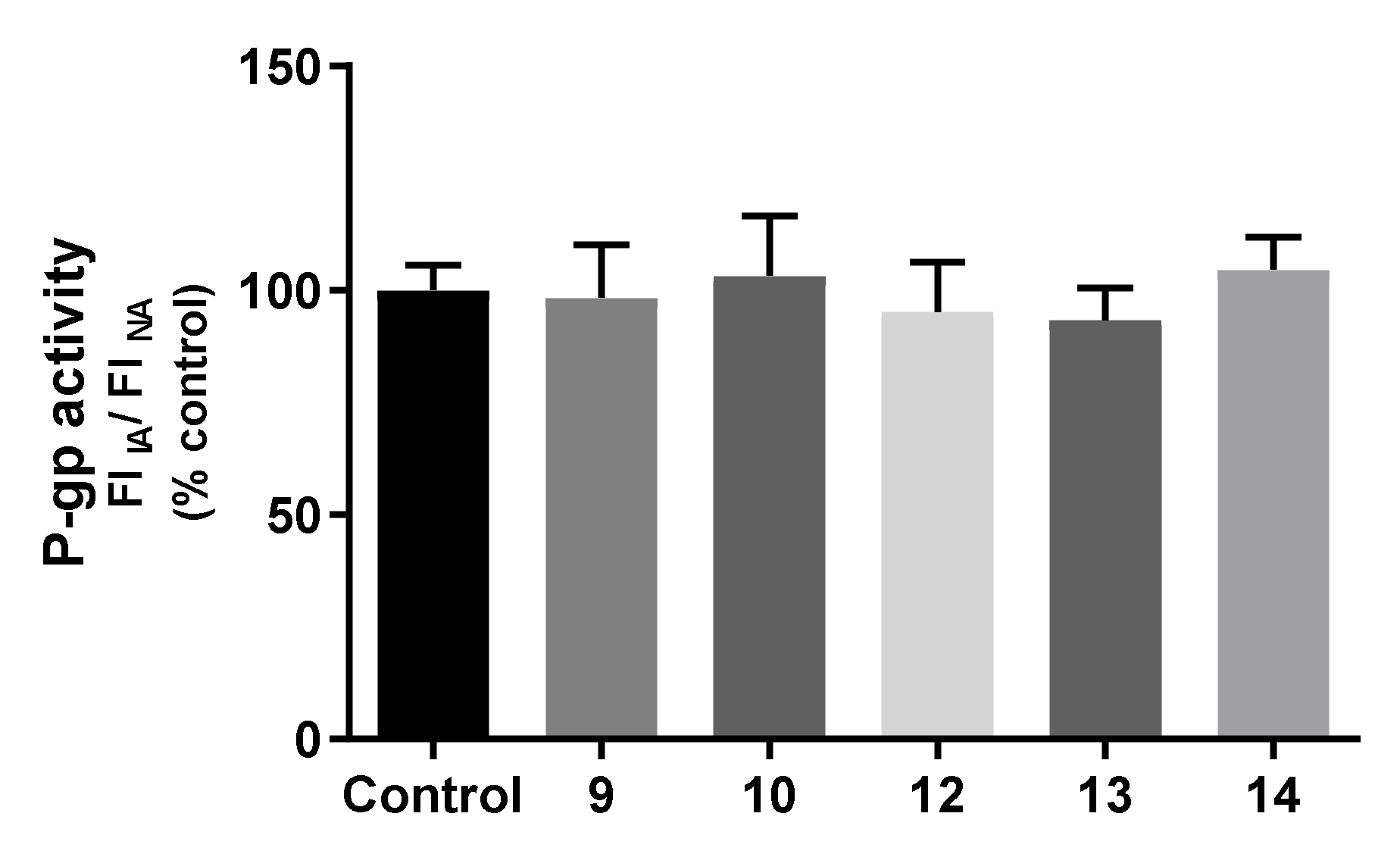
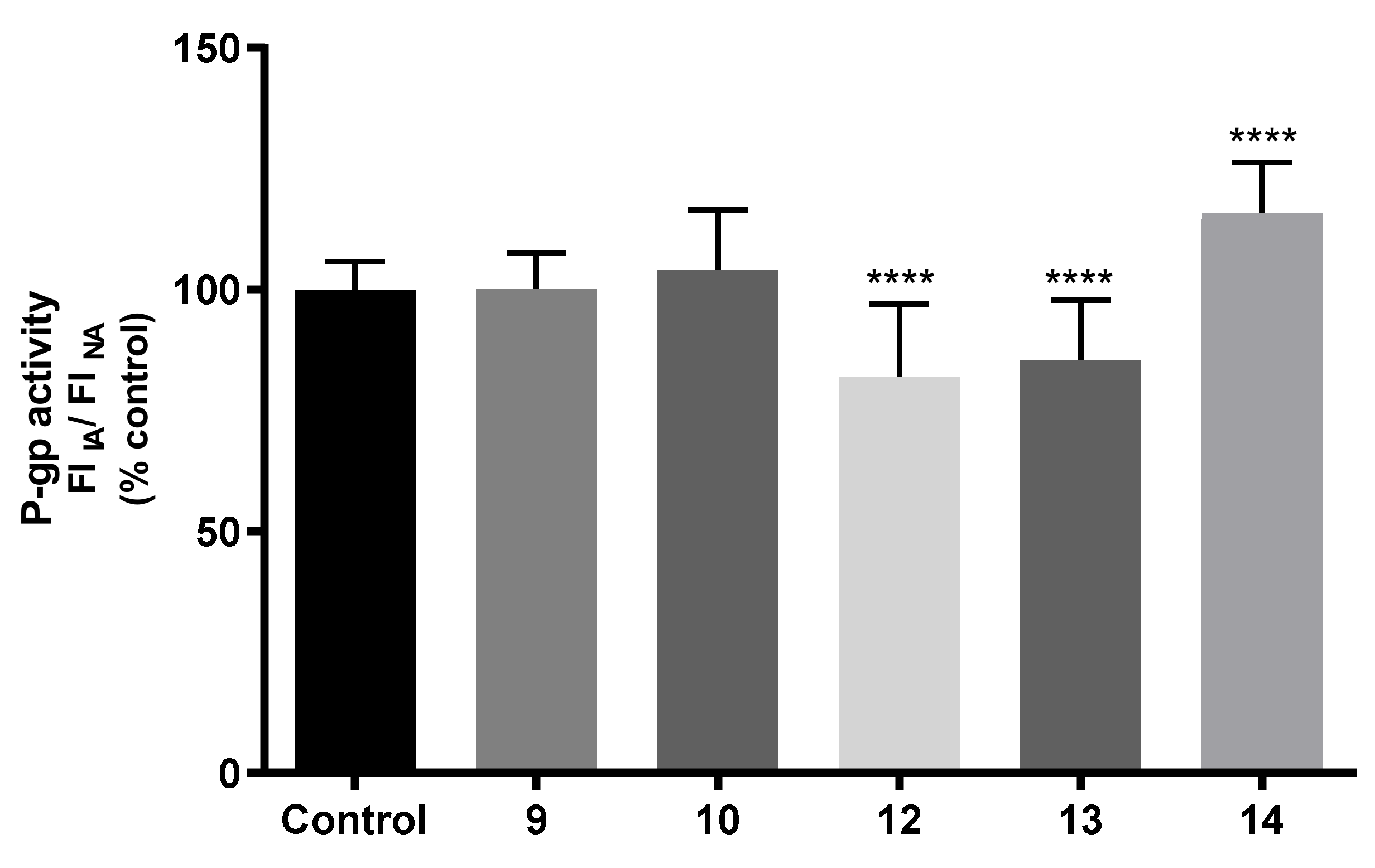
2.7.3. Effects on P-gp Expression and Activity
3. Materials and Methods
3.1. Chemistry
3.1.1. General Procedure for the Synthesis of 1-Nitrogen Substituted Thioxanthones (8–13)
3.1.2. General Procedure for the Synthesis of 1-Bromo-4-Propoxy-9H-Tioxanthen-9-One (14)
3.2. X-ray Crystallography
3.3. Docking Studies
3.4. Bacterial Strains
3.5. Antibacterial Assay and Synergy with Antimicrobials
3.6. Real-Time EB Accumulation Assay
3.7. Inhibition of Biofilm Formation
3.8. Quorum Sensing Assay
3.9. In Vitro Studies in Mammalian Cells
3.9.1. Cytotoxicity in NIH/3T3 Cell Line
3.9.2. Cytotoxicity in Caco-2 Cell Line
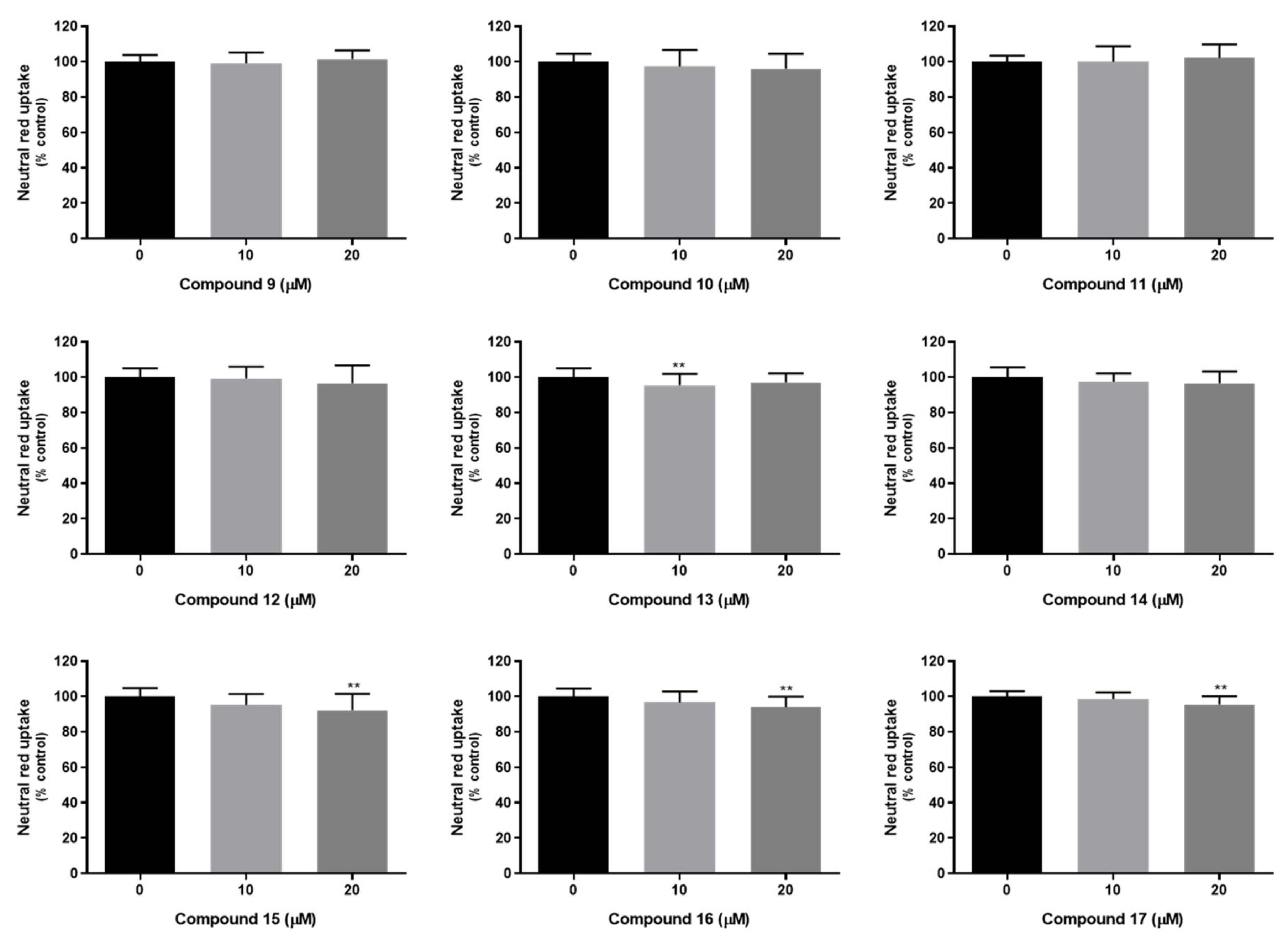
3.9.3. Neutral Red Uptake Assay
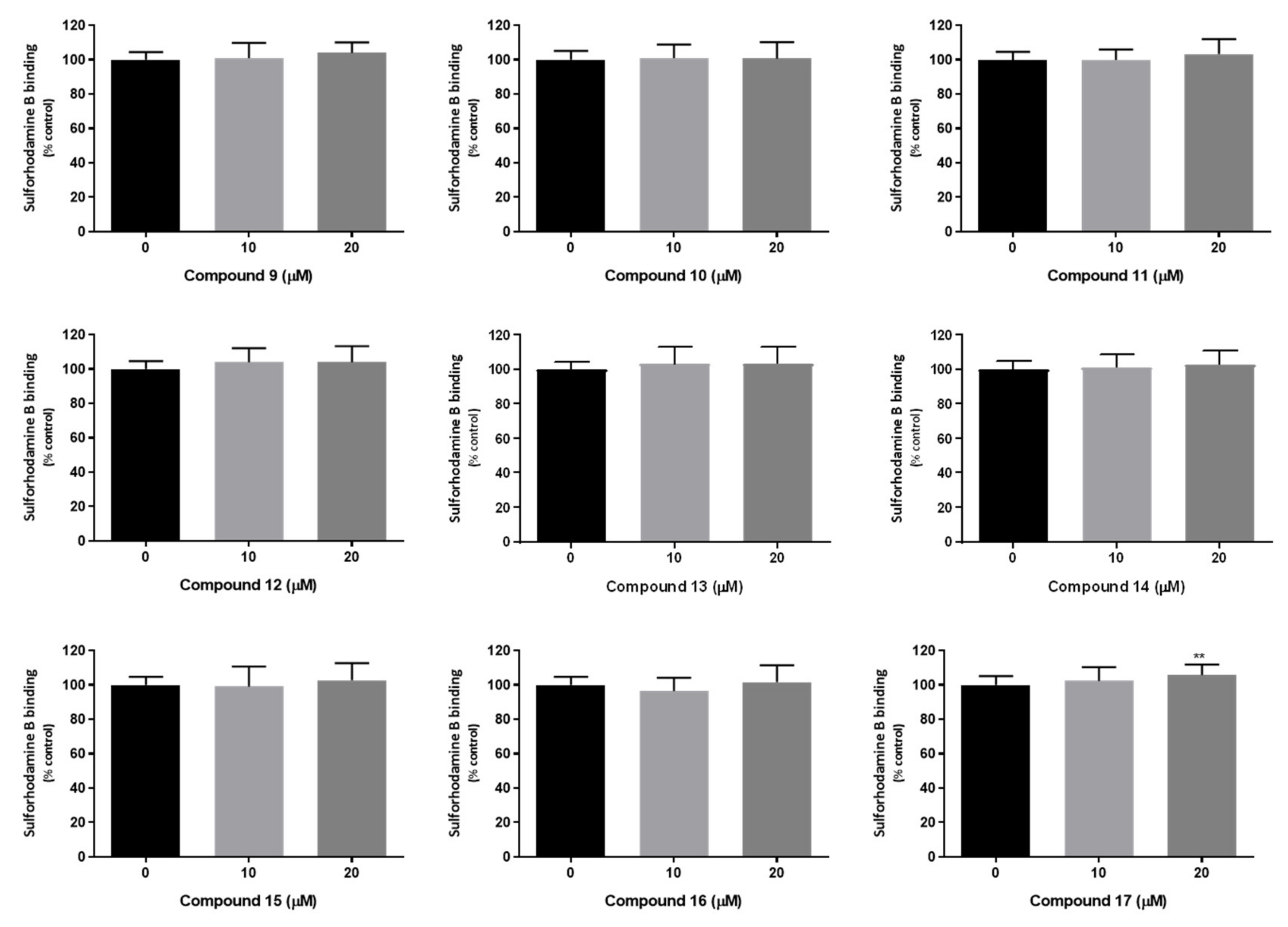
3.9.4. Sulforhodamine B Binding Assay
3.9.5. Flow Cytometry Analysis of P-Glycoprotein Expression
3.9.6. Evaluation of P-Glycoprotein Transport Activity
3.9.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef]
- Palmeira, A.; Sousa, E.; Vasconcelos, M.H.; Pinto, M. Three Decades of P-gp Inhibitors: Skimming Through Several Generations and Scaffolds. Curr. Med. Chem. 2012, 19, 1946–2025. [Google Scholar] [CrossRef]
- Nanayakkara, A.K.; Follit, C.A.; Chen, G.; Williams, N.S.; Vogel, P.D.; Wise, J.G. Targeted inhibitors of P-glycoprotein increase chemotherapeutic-induced mortality of multidrug resistant tumor cells. Sci. Rep. 2018, 8, 967. [Google Scholar] [CrossRef]
- Durães, F.; Pinto, M.; Sousa, E. Medicinal Chemistry Updates on Bacterial Efflux Pump Modulators. Curr. Med. Chem. 2018, 25, 6030–6069. [Google Scholar] [CrossRef]
- Nikaido, H.; Takatsuka, Y. Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta 2009, 1794, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Mukherjee, M.M.; Varela, M.F. Modulation of Bacterial Multidrug Resistance Efflux Pumps of the Major Facilitator Superfamily. Int. J. Bacteriol. 2013, 2013, 204141. [Google Scholar] [CrossRef]
- Alcalde-Rico, M.; Hernando-Amado, S.; Blanco, P.; Martínez, J.L. Multidrug Efflux Pumps at the Crossroad between Antibiotic Resistance and Bacterial Virulence. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef] [PubMed]
- Soto, S.M. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013, 4, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Pinto, M.M.M.; Palmeira, A.; Fernandes, C.; Resende, D.I.S.P.; Sousa, E.; Cidade, H.; Tiritan, M.E.; Correia-da-Silva, M.; Cravo, S. From Natural Products to New Synthetic Small Molecules: A Journey through the World of Xanthones. Molecules 2021, 26, 431. [Google Scholar] [CrossRef]
- Palmeira, A.; Vasconcelos, M.H.; Paiva, A.; Fernandes, M.X.; Pinto, M.; Sousa, E. Dual inhibitors of P-glycoprotein and tumor cell growth: (Re)discovering thioxanthones. Biochem. Pharmacol. 2012, 83, 57–68. [Google Scholar] [CrossRef]
- Bessa, L.J.; Palmeira, A.; Gomes, A.S.; Vasconcelos, V.; Sousa, E.; Pinto, M.; Martins da Costa, P. Synergistic Effects Between Thioxanthones and Oxacillin Against Methicillin-Resistant Staphylococcus aureus. Microb. Drug Resist. 2015, 21, 404–415. [Google Scholar] [CrossRef] [PubMed]
- Durães, F.; Resende, D.I.S.P.; Palmeira, A.; Szemerédi, N.; Pinto, M.M.M.; Spengler, G.; Sousa, E. Xanthones Active against Multidrug Resistance and Virulence Mechanisms of Bacteria. Antibiotics 2021, 10, 600. [Google Scholar] [CrossRef]
- Grossman, O.; Azerraf, C.; Gelman, D. Palladium Complexes Bearing Novel Strongly Bent Trans-Spanning Diphosphine Ligands: Synthesis, Characterization, and Catalytic Activity. Organometallics 2006, 25, 375–381. [Google Scholar] [CrossRef]
- Durães, F.; Silva, P.M.A.; Novais, P.; Amorim, I.; Gales, L.; Esteves, C.I.C.; Guieu, S.; Bousbaa, H.; Pinto, M.; Sousa, E. Tetracyclic Thioxanthene Derivatives: Studies on Fluorescence and Antitumor Activity. Molecules 2021, 26, 3315. [Google Scholar] [CrossRef]
- Gales, L.; Damas, A.M. Xanthones-A Structural Perspective. Curr. Med. Chem. 2005, 12, 2499–2515. [Google Scholar] [CrossRef] [PubMed]
- Freitas, V.L.S.; Gomes, J.R.B.; Gales, L.; Damas, A.M.; Ribeiro da Silva, M.D.M.C. Experimental and Computational Studies on the Structural and Thermodynamic Properties of Two Sulfur Heterocyclic Keto Compounds. J. Chem. Eng. Data 2010, 55, 5009–5017. [Google Scholar] [CrossRef]
- Kaatz, G.W.; Moudgal, V.V.; Seo, S.M.; Kristiansen, J.E. Phenothiazines and thioxanthenes inhibit multidrug efflux pump activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 719–726. [Google Scholar] [CrossRef]
- Shi, X.; Chen, M.; Yu, Z.; Bell, J.M.; Wang, H.; Forrester, I.; Villarreal, H.; Jakana, J.; Du, D.; Luisi, B.F.; et al. In situ structure and assembly of the multidrug efflux pump AcrAB-TolC. Nat. Commun. 2019, 10, 2635. [Google Scholar] [CrossRef]
- Yan, N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 2013, 38, 151–159. [Google Scholar] [CrossRef]
- Aron, Z.; Opperman, T.J. The hydrophobic trap—the Achilles heel of RND efflux pumps. Res. Microbiol. 2018, 169, 393–400. [Google Scholar] [CrossRef]
- Zárate, S.G.; Morales, P.; Świderek, K.; Bolanos-Garcia, V.M.; Bastida, A. A Molecular Modeling Approach to Identify Novel Inhibitors of the Major Facilitator Superfamily of Efflux Pump Transporters. Antibiotics 2019, 8, 25. [Google Scholar] [CrossRef]
- Bessa, L.J.; Barbosa-Vasconcelos, A.; Mendes, A.; Vaz-Pires, P.; Martins da Costa, P. High prevalence of multidrug-resistant Escherichia coli and Enterococcus spp. in river water, upstream and downstream of a wastewater treatment plant. J. Water Health 2014, 12, 426–435. [Google Scholar] [CrossRef]
- Anand, N. Sulfonamides: Structure-Activity Relationships and Mechanism of Action. In Inhibition of Folate Metabolism in Chemotherapy: The Origins and Uses of Co-trimoxazole; Hitchings, G.H., Ed.; Springer: Berlin/Heidelberg, Germany, 1983; pp. 25–54. [Google Scholar]
- Mouwakeh, A.; Kincses, A.; Nové, M.; Mosolygó, T.; Mohácsi-Farkas, C.; Kiskó, G.; Spengler, G. Nigella sativa essential oil and its bioactive compounds as resistance modifiers against Staphylococcus aureus. Phytother. Res. 2019, 33, 1010–1018. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, V.K.; Pathania, R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J. Med. Res. 2019, 149, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Parai, D.; Banerjee, M.; Dey, P.; Mukherjee, S.K. Reserpine attenuates biofilm formation and virulence of Staphylococcus aureus. Microb. Pathog. 2020, 138, 103790. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, S.; Klinger-Strobel, M.; Bohnert, J.A.; Wendler, S.; Rödel, J.; Pletz, M.W.; Löffler, B.; Tuchscherr, L. Clinically Approved Drugs Inhibit the s Multidrug NorA Efflux Pump and Reduce Biofilm Formation. Front. Microbiol. 2019, 10, 2762. [Google Scholar] [CrossRef]
- Gajdács, M.; Spengler, G. The Role of Drug Repurposing in the Development of Novel Antimicrobial Drugs: Non-Antibiotic Pharmacological Agents as Quorum Sensing-Inhibitors. Antibiotics 2019, 8, 270. [Google Scholar] [CrossRef]
- Silva, R.; Palmeira, A.; Carmo, H.; Barbosa, D.; Gameiro, M.; Gomes, A.; Paiva, M.; Sousa, E.; Pinto, M.; Bastos, M.; et al. P-glycoprotein induction in Caco-2 cells by newly synthetized thioxanthones prevents paraquat cytotoxicity. Arch. Toxicol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Takara, K.; Hayashi, R.; Kokufu, M.; Yamamoto, K.; Kitada, N.; Ohnishi, N.; Yokoyama, T. Effects of nonsteroidal anti-inflammatory drugs on the expression and function of P-glycoprotein/MDR1 in Caco-2 cells. Drug Chem. Toxicol. 2009, 32, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Eicher, T.; Cha, H.-j.; Seeger, M.A.; Brandstätter, L.; El-Delik, J.; Bohnert, J.A.; Kern, W.V.; Verrey, F.; Grütter, M.G.; Diederichs, K.; et al. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl. Acad. Sci. USA 2012, 109, 5687. [Google Scholar] [CrossRef]
- Mikolosko, J.; Bobyk, K.; Zgurskaya, H.I.; Ghosh, P. Conformational Flexibility in the Multidrug Efflux System Protein AcrA. Structure 2006, 14, 577–587. [Google Scholar] [CrossRef]
- Koronakis, V.; Sharff, A.; Koronakis, E.; Luisi, B.; Hughes, C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 2000, 405, 914–919. [Google Scholar] [CrossRef]
- Sussman, J.L.; Lin, D.; Jiang, J.; Manning, N.O.; Prilusky, J.; Ritter, O.; Abola, E.E. Protein Data Bank (PDB): Database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. D Biol. Crystallogr. 1998, 54, 1078–1084. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [CrossRef] [PubMed]
- Kim, Y.; Chen, J. Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science 2018, 359, 915. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.B.; Szewczyk, P.; Grimard, V.; Lee, C.W.; Martinez, L.; Doshi, R.; Caya, A.; Villaluz, M.; Pardon, E.; Cregger, C.; et al. Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain. Proc. Natl. Acad. Sci. USA 2013, 110, 13386–13391. [Google Scholar] [CrossRef]
- Li, J.; Jaimes, K.F.; Aller, S.G. Refined structures of mouse P-glycoprotein. Protein Sci. 2014, 23, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Szewczyk, P.; Tao, H.; McGrath, A.P.; Villaluz, M.; Rees, S.D.; Lee, S.C.; Doshi, R.; Urbatsch, I.L.; Zhang, Q.; Chang, G. Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Gajdács, M.; Spengler, G. Standard operating procedure (SOP) for disk diffusion-based quorum sensing inhibition assays. Acta Pharm. Hung. 2020, 89, 117–125. [Google Scholar] [CrossRef][Green Version]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 11th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Ferreira, R.J.; Kincses, A.; Gajdács, M.; Spengler, G.; dos Santos, D.J.V.A.; Molnár, J.; Ferreira, M.-J.U. Terpenoids from Euphorbia pedroi as Multidrug-Resistance Reversers. J. Nat. Prod. 2018, 81, 2032–2040. [Google Scholar] [CrossRef]
- Repetto, G.; del Peso, A.; Zurita, J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 2008, 3, 1125–1131. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Silva, V.; Gil-Martins, E.; Rocha-Pereira, C.; Lemos, A.; Palmeira, A.; Puthongking, P.; Sousa, E.; de Lourdes Bastos, M.; Remião, F.; Silva, R. Oxygenated xanthones as P-glycoprotein modulators at the intestinal barrier: In vitro and docking studies. Med. Chem. Res. 2020, 29, 1041–1057. [Google Scholar] [CrossRef]

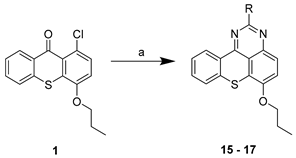
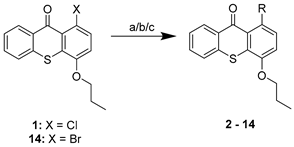 | ||
| Compound | R | Procedure |
| 2 | 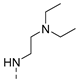 | a |
| 3 | 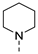 | a |
| 4 | 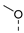 | a |
| 5 | 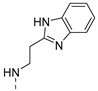 | a |
| 6 | 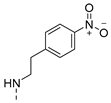 | a |
| 7 | 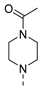 | a |
| 8 | 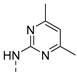 | a |
| 9 | 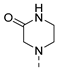 | a |
| 10 | 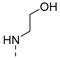 | a |
| 11 | 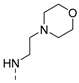 | a |
| 12 | 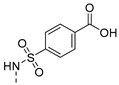 | a, b |
| 13 | 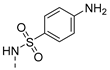 | b |
| 14 | Br | c |
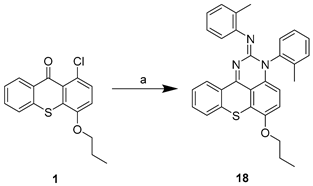
 | ||
| Compound | R | Procedure |
| 15 | H | a |
| 16 | NH2 | a |
| 17 | 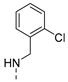 | a |
 | ||
| Compound | Docking Scores | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| AcrB | AcrA | TolC | NorA | P-gp | |||||
| SBS | HT | HH | LD | BCR | CS | TMD | NBD | ||
| 1 | −6.3 | 17.5 | −5.4 | −4.2 | −6.2 | −5.2 | −4.9 | −5.7 | −6.6 |
| 2 | −6.1 | −1.0 | −5.4 | −5.4 | −6.3 | −5.6 | −4.9 | −7.2 | −5.2 |
| 3 | −6.9 | −5.0 | −5.6 | −4.7 | −6.8 | −3.9 | −5.7 | −8.5 | −5.3 |
| 4 | −6.5 | −5.1 | −5.4 | −5.2 | −6.2 | −5.3 | −5.3 | −7.1 | −5.7 |
| 5 | −7.5 | 4.2 | −6.2 | −6.3 | −7.5 | −7.8 | −6.2 | −8.6 | −5.2 |
| 6 | −7.4 | −3.4 | −6.5 | −6.6 | −6.9 | −7.4 | −5.3 | −9.2 | −6.3 |
| 7 | −7.6 | 4.8 | −5.7 | −4.9 | −7.7 | −2.3 | −5.5 | −8.0 | −5.6 |
| 8 | −8.1 | −2.5 | −6.0 | −5.2 | −7.3 | −6.2 | −5.0 | −9.2 | −6.3 |
| 9 | −7.3 | −3.6 | −5.8 | −4.8 | −6.9 | −4.0 | −5.8 | −8.2 | −5.6 |
| 10 | −7.2 | −5.7 | −5.8 | −5.3 | −6.7 | −6.1 | −5.7 | −7.2 | −5.7 |
| 11 | −7.5 | 1.6 | −6.1 | −5.7 | −7.2 | −6.1 | −5.7 | −7.4 | −5.9 |
| 12 | −8.2 | −5.7 | −6.9 | −6.4 | −7.8 | −5.5 | −5.7 | −8.9 | −6.6 |
| 13 | −7.8 | −4.8 | −6.5 | −6.0 | −7.1 | −6.2 | −5.1 | −9.2 | −5.8 |
| 14 | −6.3 | −4.7 | −5.4 | −5.3 | −6.4 | −4.7 | −5.7 | −7.4 | −5.5 |
| 15 | −6.5 | −5.6 | −5.7 | −5.7 | −6.6 | −5.8 | −5.4 | −7.3 | −5.1 |
| 16 | −7.4 | −5.7 | −5.8 | −6.1 | −7.0 | −5.5 | −5.6 | −8.0 | −6.7 |
| 17 | −7.9 | −6.2 | −6.9 | −5.6 | −9.0 | −5.2 | −6.5 | −9.2 | −7.2 |
| 18 | −8.5 | 32.9 | −6.8 | −5.7 | −8.4 | 4.0 | −7.4 | −10.4 | −6.0 |
| D13-9001 | −9.7 | 26.5 | −6.2 | −5.1 | −7.4 | - | - | - | - |
| Doxorubicin | −8.9 | 15.4 | −7.2 | −5.6 | −7.2 | - | - | −8.3 | −7.5 |
| MBX-3132 | −7.9 | 2.9 | −7.9 | −6.2 | −7.7 | - | - | - | - |
| Minocycline | −8.7 | 26.7 | −6.2 | −5.4 | −7.7 | - | - | −6.8 | −6.3 |
| PAβN | −7.1 | −4.7 | −5.8 | −4.9 | −7.1 | −9.4 | −5.3 | - | - |
| Reserpine | −8.7 | 10.9 | 5.6 | 4.6 | −7.5 | 1.0 | −4.6 | - | - |
| trans(E)-flupentixol | - | - | - | - | - | −3.2 | −4.6 | - | - |
| Verapamil | - | - | - | - | - | - | - | −7.1 | −4.1 |
| Compound | Antibacterial Activity | Synergy with Antimicrobials | ||||||
|---|---|---|---|---|---|---|---|---|
| Minimum Inhibitory Concentration (MIC) (µM) | ||||||||
| E. coli ATCC 25922 | P. aeruginosa ATCC 27853 | E. faecalis ATCC 29212 | S. aureus ATCC 29213 | S. aureus 272123 | SE03 | E. coli SA/2 | E. faecalis B3/101 | |
| CTX MIC = 1124 (512 µg/mL) | VAN MIC = 707 (1024 µg/mL) | |||||||
| CTX + Compound 1 | VAN + Compound 2 | |||||||
| 2 | - | - | 83 (32 µg/mL) | 83 (32 µg/mL) | >100 | >100 | - | - |
| 3 | - | - | >100 | >100 | >100 | >100 | - | - |
| 4 | - | - | - | - | >100 | >100 | - | - |
| 5 | - | - | - | - | >100 | >100 | - | - |
| 6 | - | - | - | - | >100 | >100 | - | - |
| 7 | - | - | - | - | >100 | >100 | - | - |
| 8 | >100 | >100 | >100 | >100 | >100 | >100 | 281 (128 µg/mL) | 707 (1024 µg/mL) |
| 9 | >100 | >100 | >100 | >100 | >100 | >100 | 1124 (512 µg/mL) | 44 (64 µg/mL) |
| 10 | >100 | >100 | >100 | >100 | >100 | >100 | 281 (128 µg/mL) | 707 (1024 µg/mL) |
| 11 | >100 | >100 | >100 | >100 | >100 | >100 | 1124 (512 µg/mL) | 707 (1024 µg/mL) |
| 12 | >100 | >100 | 34 (16 µg/mL) | 34 (16 µg/mL) | >100 | 50 | 70 (32 µg/mL) | 707 (1024 µg/mL) |
| 13 | >100 | >100 | >100 | >100 | >100 | >100 | 1124 (512 µg/mL) | 707 (1024 µg/mL) |
| 14 | >100 | >100 | >100 | >100 | >100 | >100 | 1124 (512 µg/mL) | 707 (1024 µg/mL) |
| 15 | >100 | >100 | >100 | >100 | >100 | >100 | 1124 (512 µg/mL) | 707 (1024 µg/mL) |
| 16 | >100 | >100 | >100 | >100 | >100 | >100 | 1124 (512 µg/mL) | 707 (1024 µg/mL) |
| 17 | >100 | >100 | >100 | >100 | >100 | >100 | 1124 (512 µg/mL) | 707 (1024 µg/mL) |
| 18 | >100 | >100 | >100 | >100 | >100 | >100 | 281 (128 µg/mL) | 707 (1024 µg/mL) |
| Compound | RFI ± SD | |
|---|---|---|
| S. aureus 272123 | SE03 | |
| 2 | 0.75 ± 0.08 | −0.02 ± 0.04 |
| 3 | 1.50 ± 0.04 | 0.67 ± 0.01 |
| 4 | 0.11 ± 0.26 | −0.10 ± 0.05 |
| 5 | −0.13 ± 0.04 | −0.20 ± 0.01 |
| 6 | 0.09 ± 0.11 | −0.20 ± 0.01 |
| 7 | −0.10 ± 0.02 | 0.49 ± 0.11 |
| 8 | 0.24 ± 0.07 | 0.74 ± 0.08 |
| 9 | 0.14 ± 0.02 | 1.54 ± 0.09 |
| 10 | 4.71 ± 0.78 | 6.00 ± 0.80 |
| 11 | 0.89 ± 0.06 | 1.37 ± 0.054 |
| 12 | 0.29 ± 0.19 | −0.21 ± 0.07 |
| 13 | −0.18 ± 0.05 | 1.23 ± 0.18 |
| 14 | 1.13 ± 0.13 | 0.87 ± 0.11 |
| 15 | −0.07 ± 0.50 | 0.54 ± 0.11 |
| 16 | 0.96 ± 0.27 | 7.52 ± 0.01 |
| 17 | 9.69 ± 0.95 | 25.51 ± 2.93 |
| 18 | 0.20 ± 0.03 | 0.20 ± 0.02 |
| Reserpine | 0.50 ± 0.04 | --- |
| CCCP | --- | 0.23 ± 0.04 |
| Compound | Biofilm Inhibition (%) ± SD | Quorum Sensing Inhibition (mm) ± SD | |||
|---|---|---|---|---|---|
| S. aureus ATCC 29213 | S. aureus 272123 | S. marcescens | wt85 | EZF + CV026 | |
| 2 | 0.53 ± 1.05 1 | 81.11 ± 1.18 3 | 0 | 0 | 0 |
| 3 | 93.35 ± 0.86 2 | 93.49 ± 0.33 4 | 0 | 0 | 31 ± 0.8 |
| 4 | 0.17 ± 0.83 2 | 47.92 ± 1.08 4 | 0 | 0 | 0 |
| 5 | 0.37 ± 0.47 2 | 0 4 | 0 | 0 | 0 |
| 6 | 0 2 | 0 4 | 0 | 0 | 0 |
| 7 | 0 2 | 47.07 ± 7.54 4 | 0 | 0 | 30 ± 0.5 |
| 8 | 0 2 | 0 5 | 0 | 0 | 32 ± 0.1 |
| 9 | 5.18 ± 1.02 2 | 31.48 ± 8.99 5 | 0 | 0 | 0 |
| 10 | 0 2 | 55.17 ± 4.20 5 | 0 | 0 | 0 |
| 11 | 0 2 | 20.17 ± 4.00 4 | 31 ± 0.8 | 0 | 0 |
| 12 | 53.32 ± 7.81 2 | 96.50 ± 0.67 5 | 0 | 0 | 0 |
| 13 | 0.42 ± 1.36 2 | 72.04 ± 6.66 4 | 0 | 0 | 51 ± 0.1 |
| 14 | 0 2 | 2.46 ± 0.72 4 | 0 | 0 | 0 |
| 15 | 0 2 | 75.95 ± 2.06 4 | 32 ± 0.5 | 0 | 50 ± 0.1 |
| 16 | 0 2 | 73.59 ± 1.80 4 | 0 | 0 | 29 ± 0.8 |
| 17 | 0 2 | 6.32 ± 4.32 4 | 0 | 0 | 49 ± 0.5 |
| 18 | 19.68 ±7.46 2 | 1.62 ± 2.43 5 | 0 | 0 | 0 |
| Reserpine | 1 30.37 ± 7.32 2 22.29 ± 5.10 | 3 90.34 ± 3.35 4 72.10 ± 1.54 5 93.76 ± 2.01 | ND | ND | ND |
| PMZ | ND | ND | 18 ± 0.8 | 40 ± 0.1 | 41 ± 0.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Durães, F.; Palmeira, A.; Cruz, B.; Freitas-Silva, J.; Szemerédi, N.; Gales, L.; da Costa, P.M.; Remião, F.; Silva, R.; Pinto, M.; et al. Antimicrobial Activity of a Library of Thioxanthones and Their Potential as Efflux Pump Inhibitors. Pharmaceuticals 2021, 14, 572. https://doi.org/10.3390/ph14060572
Durães F, Palmeira A, Cruz B, Freitas-Silva J, Szemerédi N, Gales L, da Costa PM, Remião F, Silva R, Pinto M, et al. Antimicrobial Activity of a Library of Thioxanthones and Their Potential as Efflux Pump Inhibitors. Pharmaceuticals. 2021; 14(6):572. https://doi.org/10.3390/ph14060572
Chicago/Turabian StyleDurães, Fernando, Andreia Palmeira, Bárbara Cruz, Joana Freitas-Silva, Nikoletta Szemerédi, Luís Gales, Paulo Martins da Costa, Fernando Remião, Renata Silva, Madalena Pinto, and et al. 2021. "Antimicrobial Activity of a Library of Thioxanthones and Their Potential as Efflux Pump Inhibitors" Pharmaceuticals 14, no. 6: 572. https://doi.org/10.3390/ph14060572
APA StyleDurães, F., Palmeira, A., Cruz, B., Freitas-Silva, J., Szemerédi, N., Gales, L., da Costa, P. M., Remião, F., Silva, R., Pinto, M., Spengler, G., & Sousa, E. (2021). Antimicrobial Activity of a Library of Thioxanthones and Their Potential as Efflux Pump Inhibitors. Pharmaceuticals, 14(6), 572. https://doi.org/10.3390/ph14060572










