Sample Preparation Strategies for Antibody-Free Quantitative Analysis of High Mobility Group Box 1 Protein
Abstract
1. Introduction
2. Results and Discussion
2.1. Detection of HMGB1 Peptides
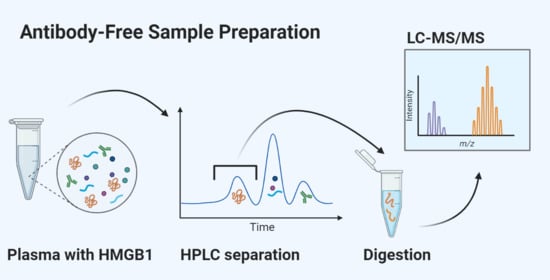
2.2. Sample Preparation
2.2.1. Challenges and Alternative Strategies
2.2.2. Targeted Extraction (A)
2.2.3. Precipitation of HAPs (B)
2.2.4. Processing Whole Samples (C)
2.3. Future Perspectives
2.3.1. Method Improvement
2.3.2. Choice of Sample Preparation
3. Materials and Methods
3.1. Materials
3.1.1. Chemicals
3.1.2. Plasma Samples
3.2. Methods
3.2.1. Separation of Proteins Using HPLC
3.2.2. Reduction, Alkylation, and Trypsin Digestion
3.2.3. HRMS
3.2.4. Protein Identification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sessa, L.; Bianchi, M.E. The evolution of High Mobility Group Box (HMGB) chromatin proteins in multicellular animals. Gene 2007, 387, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Gardella, S.; Andrei, C.; Ferrera, D.; Lotti, L.V.; Torrisi, M.R.; Bianchi, M.E.; Rubartelli, A. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep. 2002, 3, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Scott, M.J.; Fan, J.; Billiar, T.R. Location is the key to function: HMGB1 in sepsis and trauma-induced inflammation. J. Leukoc. Biol. 2019, 106, 161–169. [Google Scholar] [CrossRef]
- Hreggvidsdóttir, H.S.; Lundberg, A.M.; Aveberger, A.-C.; Klevenvall, L.; Andersson, U.; Harris, H.E. High Mobility Group Box Protein 1 (HMGB1)-Partner Molecule Complexes Enhance Cytokine Production by Signaling Through the Partner Molecule Receptor. Mol. Med. 2011, 18, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Hreggvidsdottir, H.S.; Palmblad, K.; Wang, H.; Ochani, M.; Li, J.; Lu, B.; Chavan, S.; Rosas-Ballina, M.; Al-Abed, Y.; et al. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc. Natl. Acad. Sci. USA 2010, 107, 11942–11947. [Google Scholar] [CrossRef]
- Smith, D.E.; Lipsky, B.P.; Russell, C.; Ketchem, R.R.; Kirchner, J.; Hensley, K.; Huang, Y.; Friedman, W.J.; Boissonneault, V.; Plante, M.-M.; et al. A Central Nervous System-Restricted Isoform of the Interleukin-1 Receptor Accessory Protein Modulates Neuronal Responses to Interleukin-1. Immunity 2009, 30, 817–831. [Google Scholar] [CrossRef]
- Dantzer, R. Cytokine, Sickness Behavior, and Depression. Immunol. Allergy Clin. N. Am. 2009, 29, 247–264. [Google Scholar] [CrossRef]
- Bårdsen, K.; Nilsen, M.M.; Kvaløy, J.T.; Norheim, K.B.; Jonsson, G.; Omdal, R. Heat shock proteins and chronic fatigue in primary Sjögren’s syndrome. Innate Immun. 2016, 22, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Norheim, K.B.; Imgenberg-Kreuz, J.; Jonsdottir, K.; Janssen, E.A.M.; Syvänen, A.-C.; Sandling, J.; Nordmark, G.; Omdal, R. Epigenome-wide DNA methylation patterns associated with fatigue in primary Sjögren’s syndrome. Rheumatology 2016, 55, 1074–1082. [Google Scholar] [CrossRef][Green Version]
- Bårdsen, K.; Brede, C.; Kvivik, I.; Kvaløy, J.T.; Jonsdottir, K.; Tjensvoll, A.B.; Ruoff, P.; Omdal, R. Interleukin-1-related activity and hypocretin-1 in cerebrospinal fluid contribute to fatigue in primary Sjögren’s syndrome. J. Neuroinflamm. 2019, 16, 102. [Google Scholar] [CrossRef]
- Grimstad, T.; Kvivik, I.; Kvaløy, J.T.; Aabakken, L.; Omdal, R. Heat-shock protein 90α in plasma reflects severity of fatigue in patients with Crohn’s disease. Innate Immun. 2019, 26, 146–151. [Google Scholar] [CrossRef]
- Urbonaviciute, V.; Fürnrohr, B.G.; Weber, C.; Haslbeck, M.; Wilhelm, S.; Herrmann, M.; Voll, R.E. Factors masking HMGB1 in human serum and plasma. J. Leukoc. Biol. 2006, 81, 67–74. [Google Scholar] [CrossRef]
- Baker, M. Reproducibility crisis: Blame it on the antibodies. Nat. Cell Biol. 2015, 521, 274–276. [Google Scholar] [CrossRef]
- Ottestad, W.; Rognes, I.N.; Skaga, E.; Frisvoll, C.; Haraldsen, G.; Eken, T.; Lundbäck, P. HMGB1 concentration measurements in trauma patients: Assessment of pre-analytical conditions and sample material. Mol. Med. 2019, 26, 1–8. [Google Scholar] [CrossRef]
- Seger, C.; Salzmann, L. After another decade: LC–MS/MS became routine in clinical diagnostics. Clin. Biochem. 2020, 82, 2–11. [Google Scholar] [CrossRef]
- Chen, D.Z.; Spivia, W.R.; Gao, Y.; Van Eyk, J.; Kanodia, S. Abstract 2216: A novel MRM-based mass spectrometry assay to quantify HMGB1. Cancer Chem. 2017, 77, 2216. [Google Scholar] [CrossRef]
- Weng, L.; Guo, L.; Vachani, A.; Mesaros, C.; Blair, I.A. Quantification of Serum High Mobility Group Box 1 by Liquid Chromatography/High-Resolution Mass Spectrometry: Implications for Its Role in Immunity, Inflammation, and Cancer. Anal. Chem. 2018, 90, 7552–7560. [Google Scholar] [CrossRef]
- Wong, S.L.; To, J.; Santos, J.; Allam, V.S.R.R.; Dalton, J.P.; Djordjevic, S.P.; Donnelly, S.; Padula, M.P.; Sukkar, M.B. Proteomic Analysis of Extracellular HMGB1 Identifies Binding Partners and Exposes Its Potential Role in Airway Epithelial Cell Homeostasis. J. Proteome Res. 2017, 17, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Anselm, V.; Steinhilber, A.; Sommersdorf, C.; Poetz, O. Immunoaffinity-Based Liquid Chromatography Mass Spectrometric Assay to Accurately Quantify the Protein Concentration of HMGB1 in EDTA Plasma. Methods Mol. Biol. 2021, 2261, 277–289. [Google Scholar] [CrossRef]
- Halvorsen, T.G.; Reubsaet, L. Antibody based affinity capture LC-MS/MS in quantitative determination of proteins in biological matrices. TrAC Trends Anal. Chem. 2017, 95, 132–139. [Google Scholar] [CrossRef]
- Schilling, B.; Yoo, C.B.; Collins, C.J.; Gibson, B.W. Determining cysteine oxidation status using differential alkylation. Int. J. Mass Spectrom. 2004, 236, 117–127. [Google Scholar] [CrossRef]
- Gianazza, E.; Miller, I.; Palazzolo, L.; Parravicini, C.; Eberini, I. With or without you—Proteomics with or without major plasma/serum proteins. J. Proteom. 2016, 140, 62–80. [Google Scholar] [CrossRef]
- Anderson, N.L. The Human Plasma Proteome. Mol. Cell. Proteom. 2002, 1, 845–867. [Google Scholar] [CrossRef] [PubMed]
- Pichon, V. Immunoaffinity Extraction. In Reference Module in Chemistry, Molecular Sciences and Chemical Engineering; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Neubert, H.; Shuford, C.M.; Olah, T.V.; Garofolo, F.; Schultz, G.A.; Jones, B.R.; Amaravadi, L.; Laterza, O.F.; Xu, K.; Ackermann, B.L. Protein Biomarker Quantification by Immunoaffinity Liquid Chromatography–Tandem Mass Spectrometry: Current State and Future Vision. Clin. Chem. 2020, 66, 282–301. [Google Scholar] [CrossRef]
- Thomas, S.L.; Thacker, J.B.; Schug, K.A.; Maráková, K. Sample preparation and fractionation techniques for intact proteins for mass spectrometric analysis. J. Sep. Sci. 2021, 44, 211–246. [Google Scholar] [CrossRef]
- Levernæs, M.C.S.; Broughton, M.N.; Reubsaet, L.; Halvorsen, T.G. To elute or not to elute in immunocapture bottom-up LC–MS. J. Chromatogr. B 2017, 1055–1056, 51–60. [Google Scholar] [CrossRef][Green Version]
- GPM v. 3.0. Available online: http://ftp.thegpm.org/projects/gpm/gpm-xe-installer/ (accessed on 11 May 2021).
- Craig, R.; Cortens, J.P.; Beavis, R.C. Open Source System for Analyzing, Validating, and Storing Protein Identification Data. J. Proteome Res. 2004, 3, 1234–1242. [Google Scholar] [CrossRef]
- Kvivik, I.; Brede, C.; Jonsson, G.; Omdal, R. Sample Preparation Strategies for HMGB1. Mendeley Data, Version 1; Available online: https://data.mendeley.com/datasets/p4wfz6hhf4/1 (accessed on 18 May 2021).
- Yang, H.; Wang, H.; Levine, Y.A.; Gunasekaran, M.K.; Wang, Y.; Addorisio, M.; Zhu, S.; Li, W.; Li, J.; De Kleijn, D.P.; et al. Identification of CD163 as an antiinflammatory receptor for HMGB1-haptoglobin complexes. JCI Insight 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Spagnuolo, M.S.; Cigliano, L.; D’Andrea, L.D.; Pedone, C.; Abrescia, P. Assignment of the Binding Site for Haptoglobin on Apolipoprotein A-I. J. Biol. Chem. 2005, 280, 1193–1198. [Google Scholar] [CrossRef]
- Kay, R.; Barton, C.; Ratcliffe, L.; Matharoo-Ball, B.; Brown, P.; Roberts, J.; Teale, P.; Creaser, C. Enrichment of low molecular weight serum proteins using acetonitrile precipitation for mass spectrometry based proteomic analysis. Rapid Commun. Mass Spectrom. 2008, 22, 3255–3260. [Google Scholar] [CrossRef] [PubMed]
- Larssen, E.; Brede, C.; Hjelle, A.B.; Øysaed, K.B.; Tjensvoll, A.B.; Omdal, R.; Ruoff, P.; Øysæd, K.B. A rapid method for preparation of the cerebrospinal fluid proteome. Proteomics 2014, 15, 10–15. [Google Scholar] [CrossRef]
- Bustin, M.; Lehn, D.A.; Landsman, D. Structural features of the HMG chromosomal proteins and their genes. Biochim. Biophys. Acta (BBA) Gene Struct. Expr. 1990, 1049, 231–243. [Google Scholar] [CrossRef]
- Gaillard, C.; Borde, C.; Gozlan, J.; Maréchal, V.; Strauss, F. A High-Sensitivity Method for Detection and Measurement of HMGB1 Protein Concentration by High-Affinity Binding to DNA Hemicatenanes. PLoS ONE 2008, 3, e2855. [Google Scholar] [CrossRef] [PubMed]
- Barnay-Verdier, S.; Gaillard, C.; Messmer, M.; Borde, C.; Gibot, S.; Maréchal, V. PCA-ELISA: A sensitive method to quantify free and masked forms of HMGB1. Cytokine 2011, 55, 4–7. [Google Scholar] [CrossRef]
- Wiśniewski, J. Filter-Aided Sample Preparation: The Versatile and Efficient Method for Proteomic Analysis. Methods Enzymol. 2017, 585, 15–27. [Google Scholar] [CrossRef]
- Johns, E.W. Studies on histones. 7. Preparative methods for histone fractions from calf thymus. Biochem. J. 1964, 92, 55–59. [Google Scholar] [CrossRef]
- Andersson, U.; Erlandsson-Harris, H.; Yang, H.; Tracey, K.J. HMGB1 as a DNA-binding cytokine. J. Leukoc. Biol. 2002, 72, 1084–1091. [Google Scholar]
- Li, A.; Sowder, I.R.C.; Henderson, L.E.; Moore, S.P.; Garfinkel, A.D.J.; Fisher, R.J. Chemical Cleavage at Aspartyl Residues for Protein Identification. Anal. Chem. 2001, 73, 5395–5402. [Google Scholar] [CrossRef]
- Kwon, J.; Lee, T. Protein cleavage at ASPARTIC Acid Using Chemical Reagents. International Patent WO2006031063A1, 23 March 2006. [Google Scholar]
- Mazrimas, J.A.; Laskaris, M.; Corzett, M.; Balhorn, R. Separation of HMG Proteins by Reverse-Phase HPLC. J. Liq. Chromatogr. 1984, 7, 907–916. [Google Scholar] [CrossRef]
- Zetterström, C.K.; Bergman, T.; Rynnel-Dagoo, B.; Harris, H.E.; Söder, O.; Andersson, U.; Boman, H.G. High Mobility Group Box Chromosomal Protein 1 (HMGB1) Is an Antibacterial Factor Produced by the Human Adenoid. Pediatr. Res. 2002, 52, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.L.; Haines, L.R.; Hardie, D.B.; Olafson, R.W.; Pearson, T.W. Mass Spectrometric Quantitation of Peptides and Proteins Using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J. Proteome Res. 2004, 3, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.S.; Kim, H.S.; Lee, B.; Kim, Y.H.; Son, M.; Shin, J.-S. Immunological Significance of HMGB1 Post-Translational Modification and Redox Biology. Front. Immunol. 2020, 11, 1189. [Google Scholar] [CrossRef]
- Barnay-Verdier, S.; Fattoum, L.; Borde, C.; Kaveri, S.; Gibot, S.; Marechal, V. Emergence of autoantibodies to HMGB1 is associated with survival in patients with septic shock. Intensiv. Care Med. 2011, 37, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Kvivik, I.; Grimstad, T.; Jonsson, G.; Kvaløy, J.T.; Omdal, R. Anti-HMGB1 auto-Abs influence fatigue in patients with Crohn’s disease. Innate Immun. 2021. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lundbäck, P.; Ottosson, L.; Erlandsson-Harris, H.; Venereau, E.; Bianchi, M.E.; Al-Abed, Y.; Andersson, U.; Tracey, K.J.; Antoine, D.J. Retraction Note to: Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1). Mol. Med. 2020, 26, 1–2. [Google Scholar] [CrossRef]




| Sample Preparation Strategy | Recovery (%) | Results from Search in the GPM Database | File Name (.mgf) | |||
|---|---|---|---|---|---|---|
| Number of Specific HMGB1-Peptides | Total Proteins Identified | Rank# for HMGB1 | ||||
| A | Targeted extraction using anti-HMGB1 Ab | 7 | None | 5 | N/A | HMGB1_1 |
| Targeted extraction using anti-haptoglobin Ab | 16 | 2 | 5 a | 3 | HMGB1_2 | |
| B | Precipitation of HAPs using PCA | 35 | 6 | 61 | 28 | HMGB1_3 |
| C | Acid digestion of whole plasma | 12 b | 2 c | 68 c | 51 c | HMGB1_4 |
| Separation of proteins using HPLC | 33 | 14 | 19 | 3 | HMGB1_5 | |
| Pure recombinant HMGB1 | 100 | 17 | 1 | 1 | HMGB1_6 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kvivik, I.; Jonsson, G.; Omdal, R.; Brede, C. Sample Preparation Strategies for Antibody-Free Quantitative Analysis of High Mobility Group Box 1 Protein. Pharmaceuticals 2021, 14, 537. https://doi.org/10.3390/ph14060537
Kvivik I, Jonsson G, Omdal R, Brede C. Sample Preparation Strategies for Antibody-Free Quantitative Analysis of High Mobility Group Box 1 Protein. Pharmaceuticals. 2021; 14(6):537. https://doi.org/10.3390/ph14060537
Chicago/Turabian StyleKvivik, Ingeborg, Grete Jonsson, Roald Omdal, and Cato Brede. 2021. "Sample Preparation Strategies for Antibody-Free Quantitative Analysis of High Mobility Group Box 1 Protein" Pharmaceuticals 14, no. 6: 537. https://doi.org/10.3390/ph14060537
APA StyleKvivik, I., Jonsson, G., Omdal, R., & Brede, C. (2021). Sample Preparation Strategies for Antibody-Free Quantitative Analysis of High Mobility Group Box 1 Protein. Pharmaceuticals, 14(6), 537. https://doi.org/10.3390/ph14060537