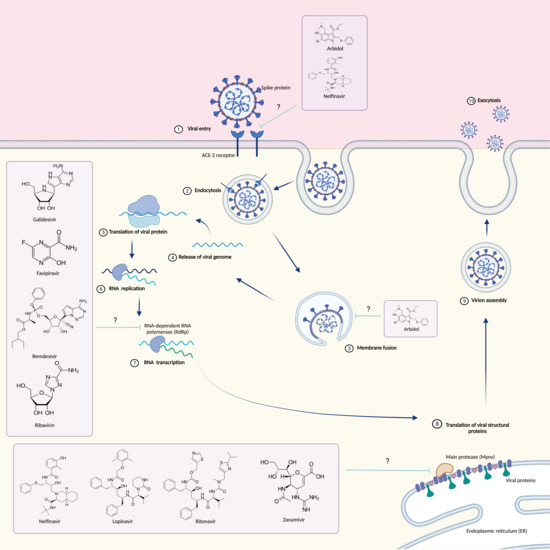
New Approaches and Repurposed Antiviral Drugs for the Treatment of the SARS-CoV-2 Infection
Abstract
1. Introduction
2. Monoclonal Antibodies
3. Antiviral Drugs
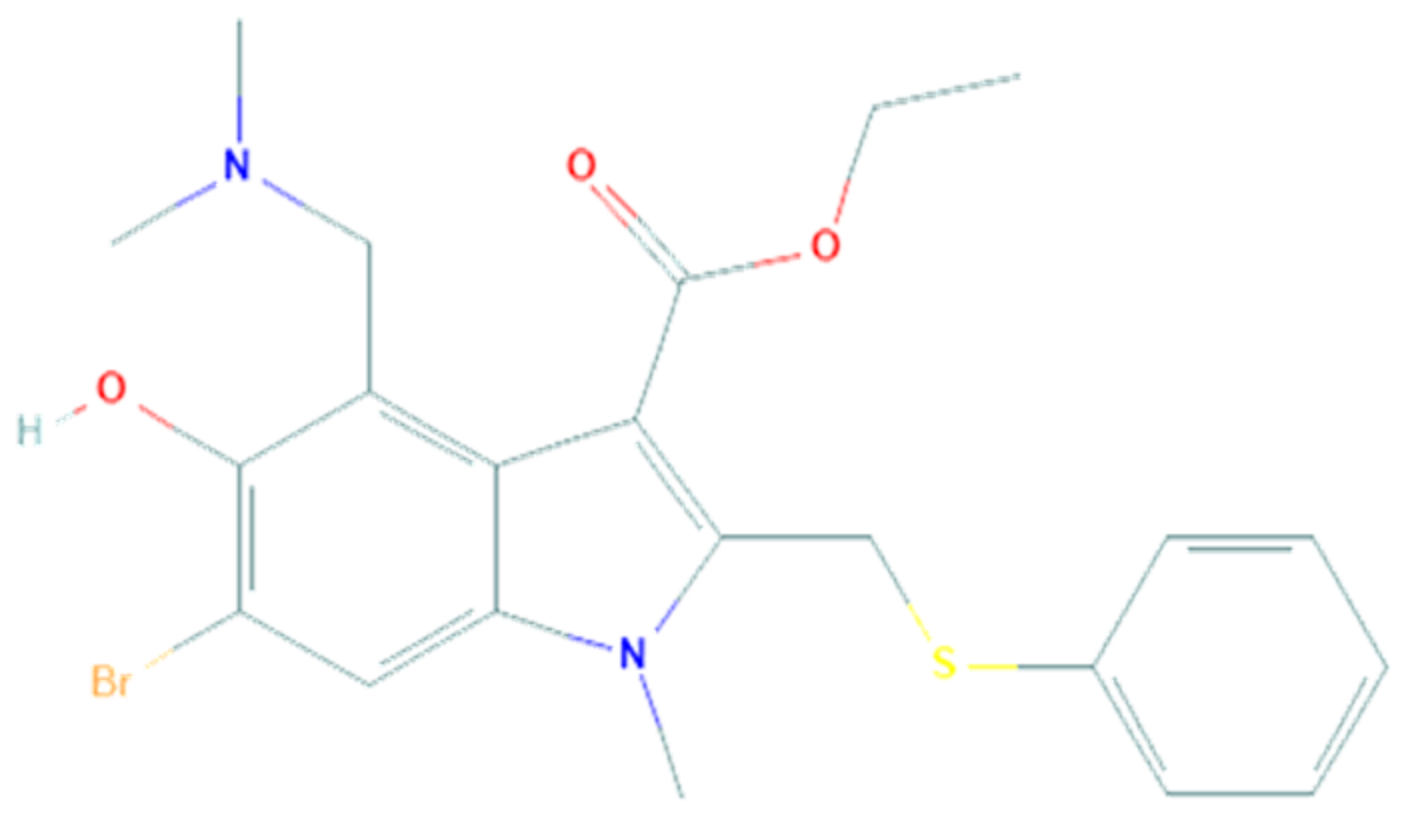
3.1. Arbidol
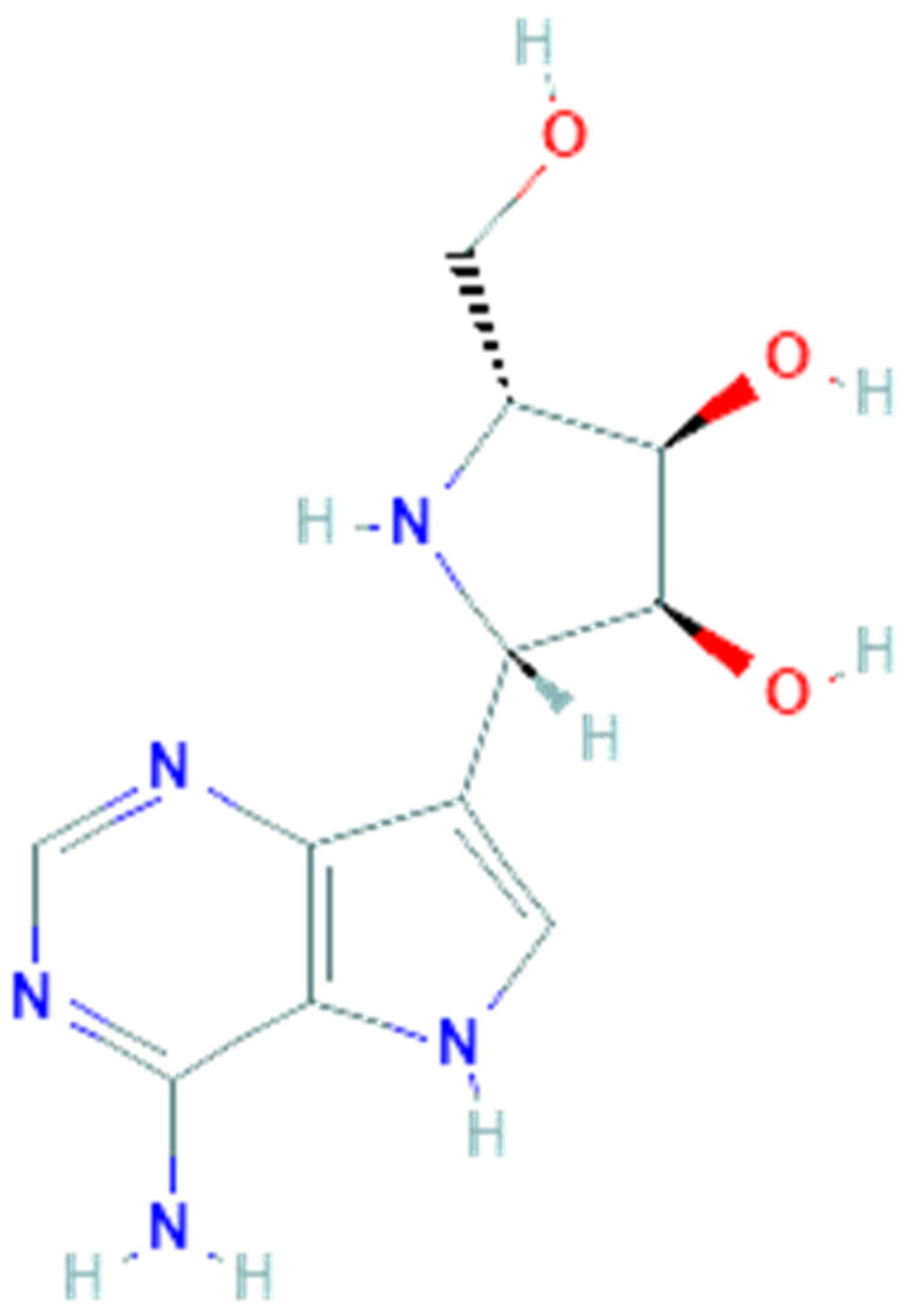
3.2. Galidesivir
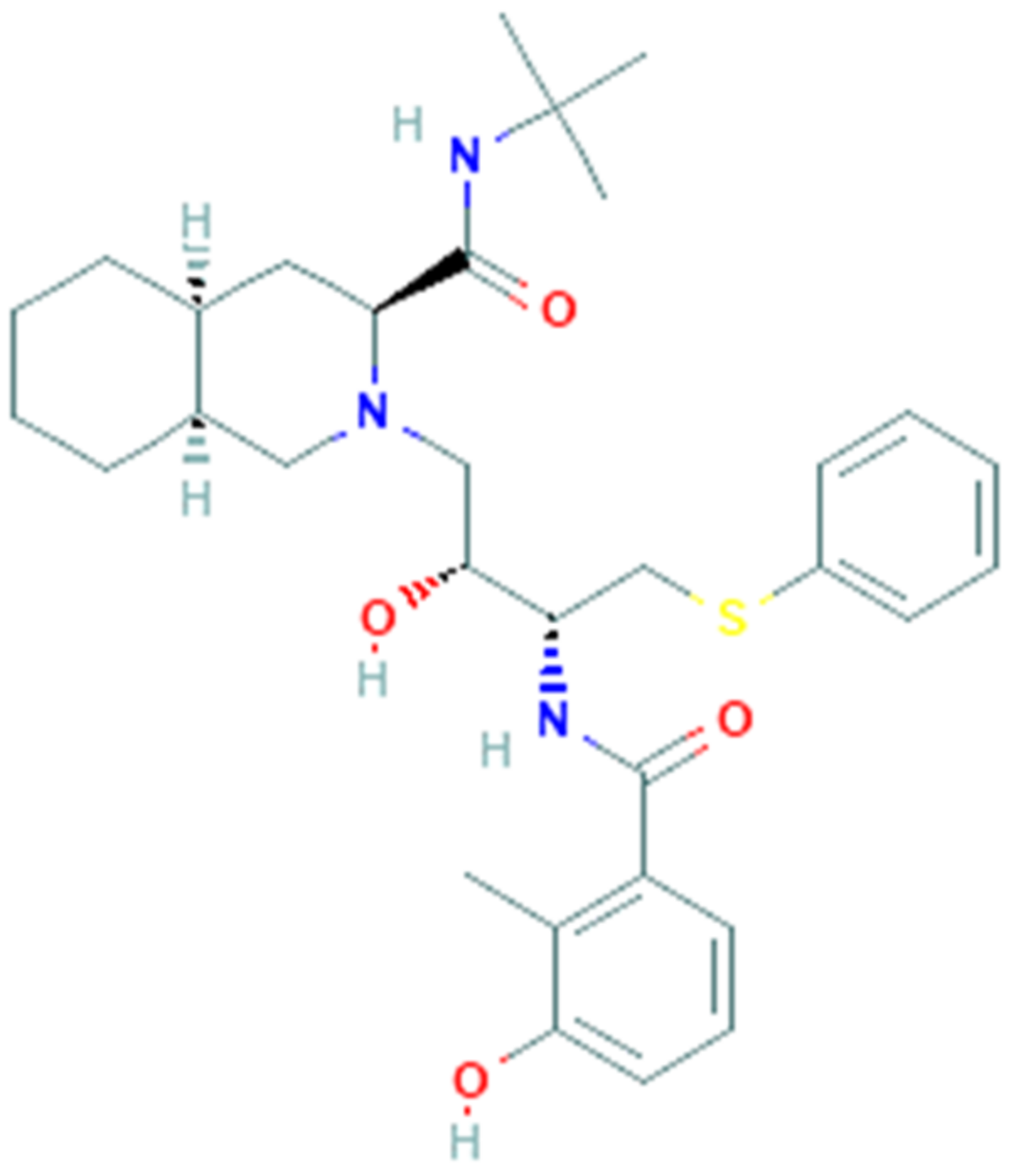
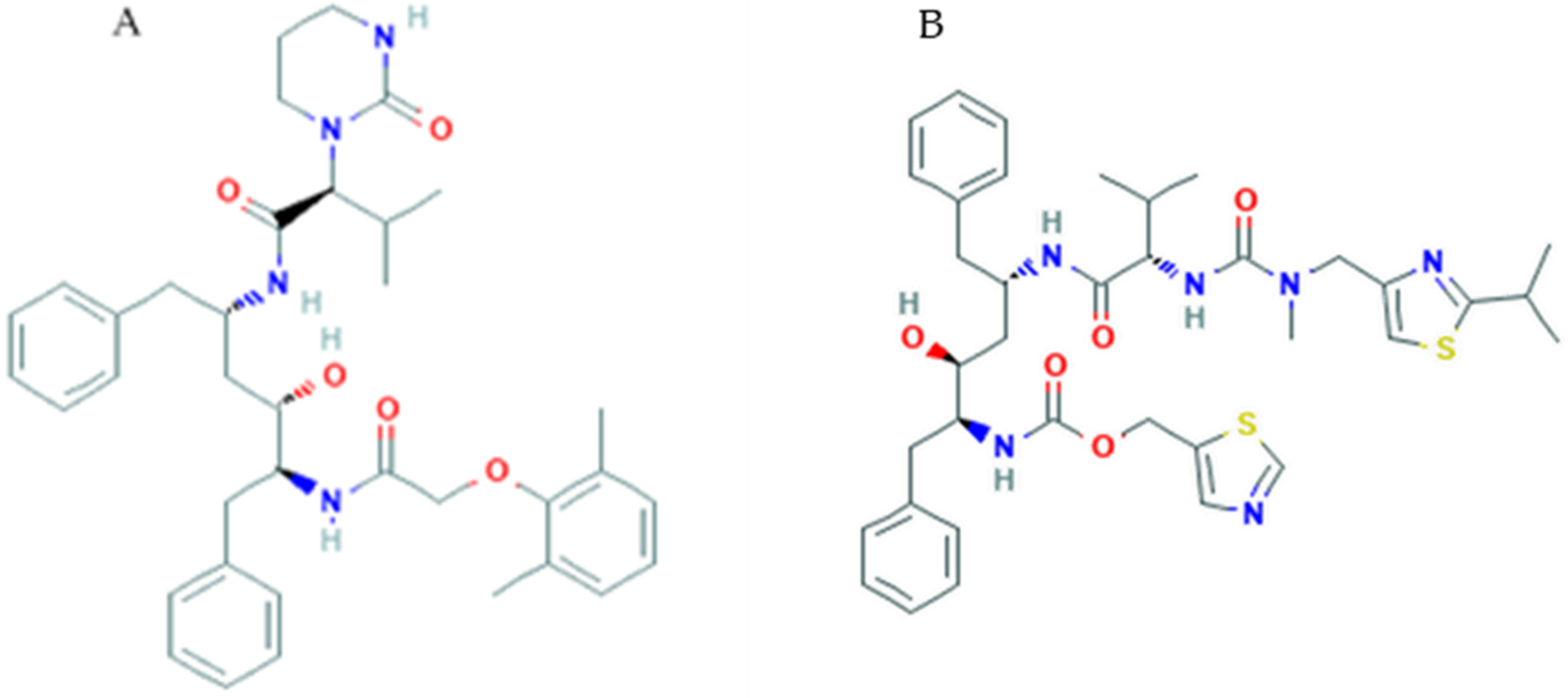
3.3. Nelfinavir
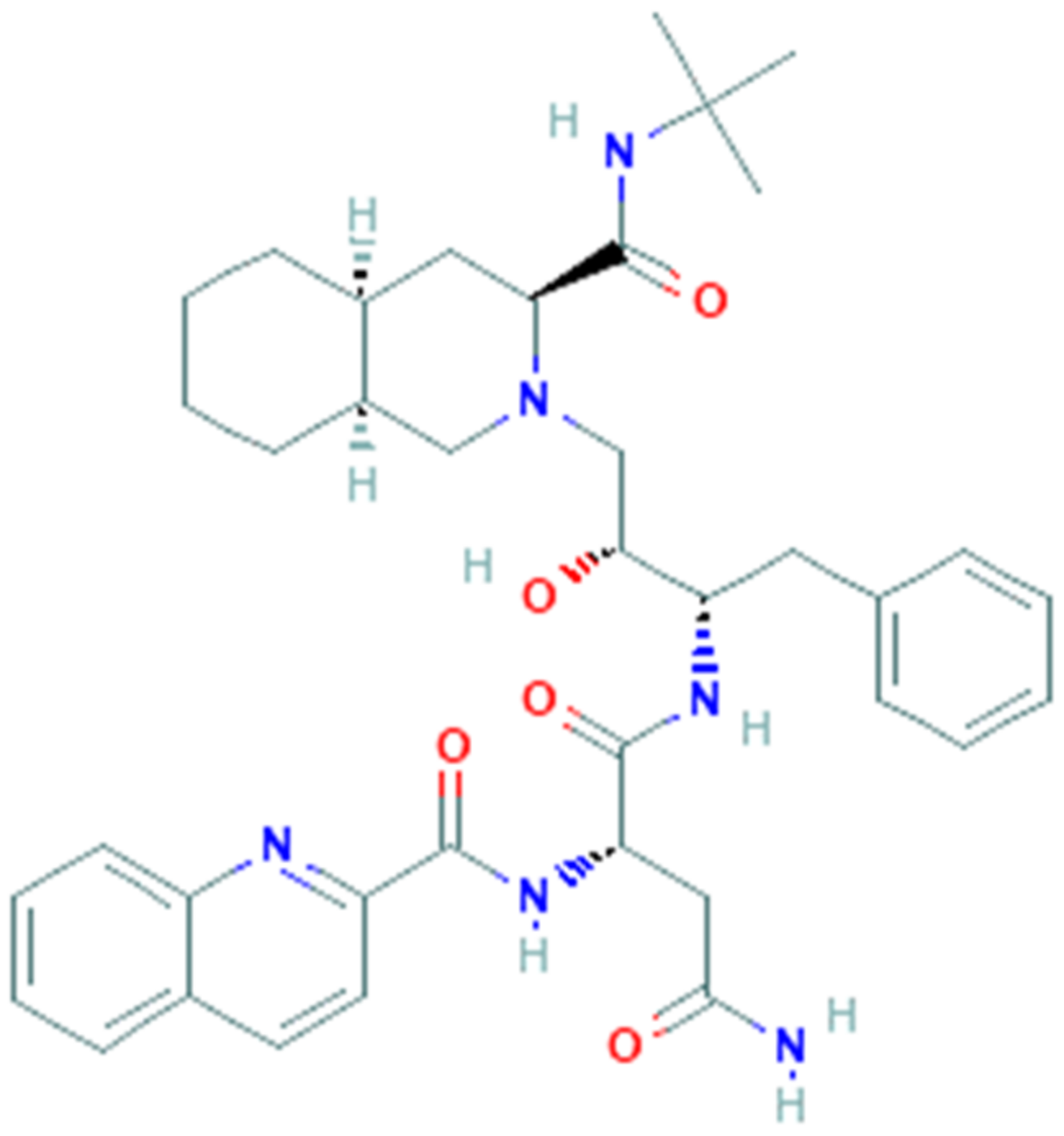
3.4. Saquinavir
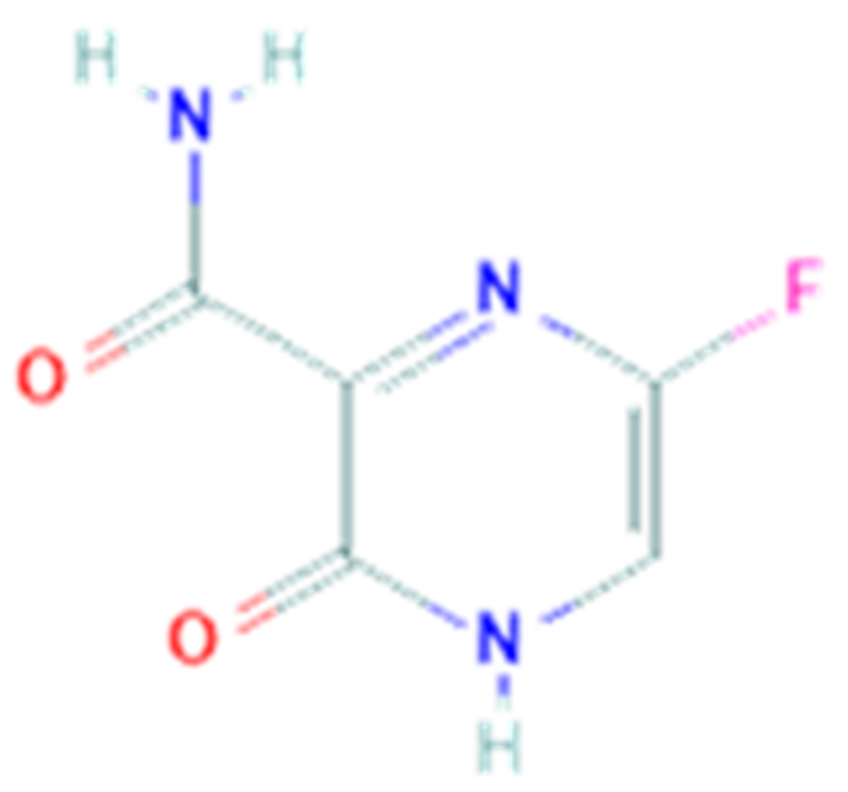
3.5. Favipiravir
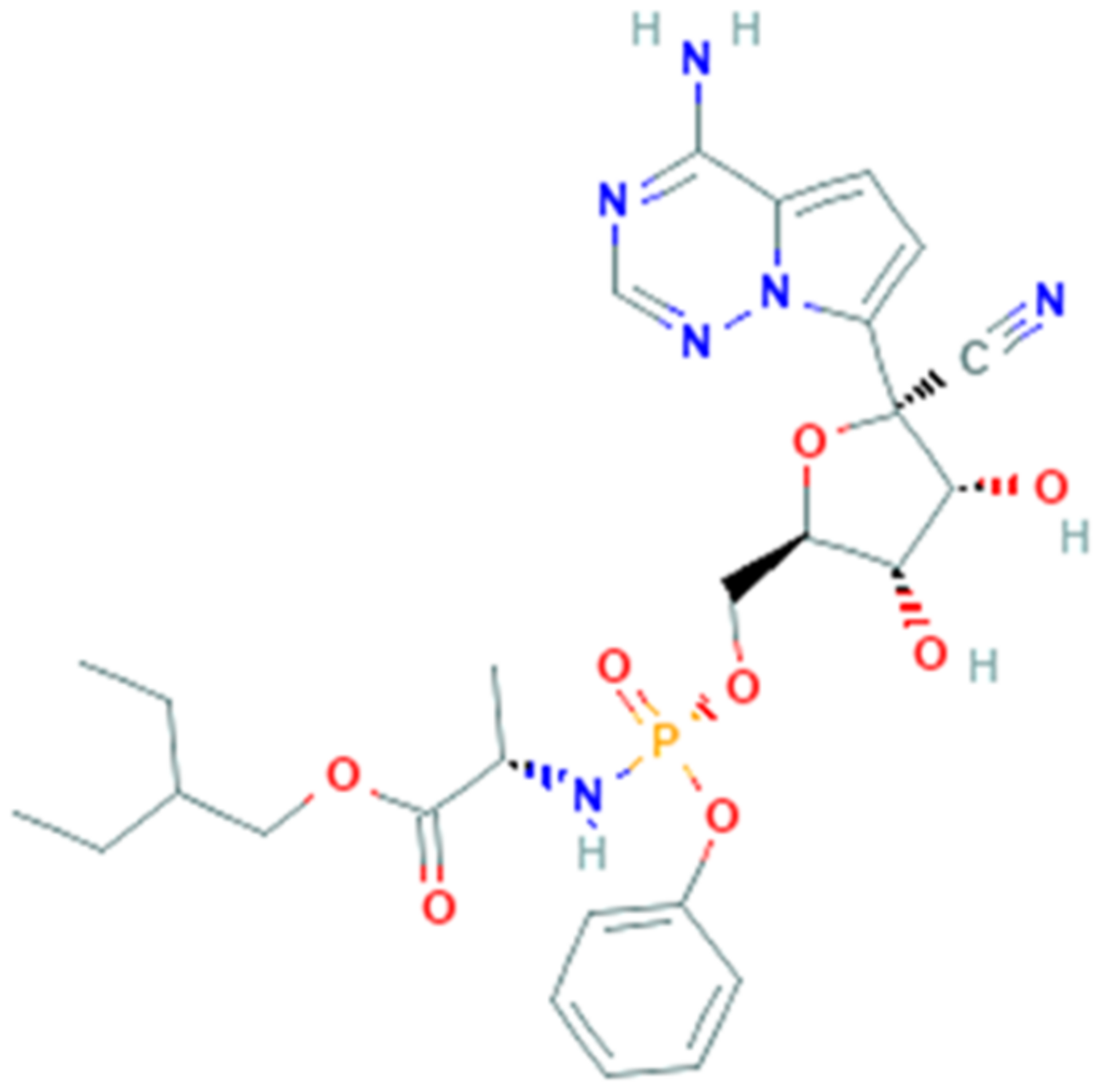
3.6. Remdesivir
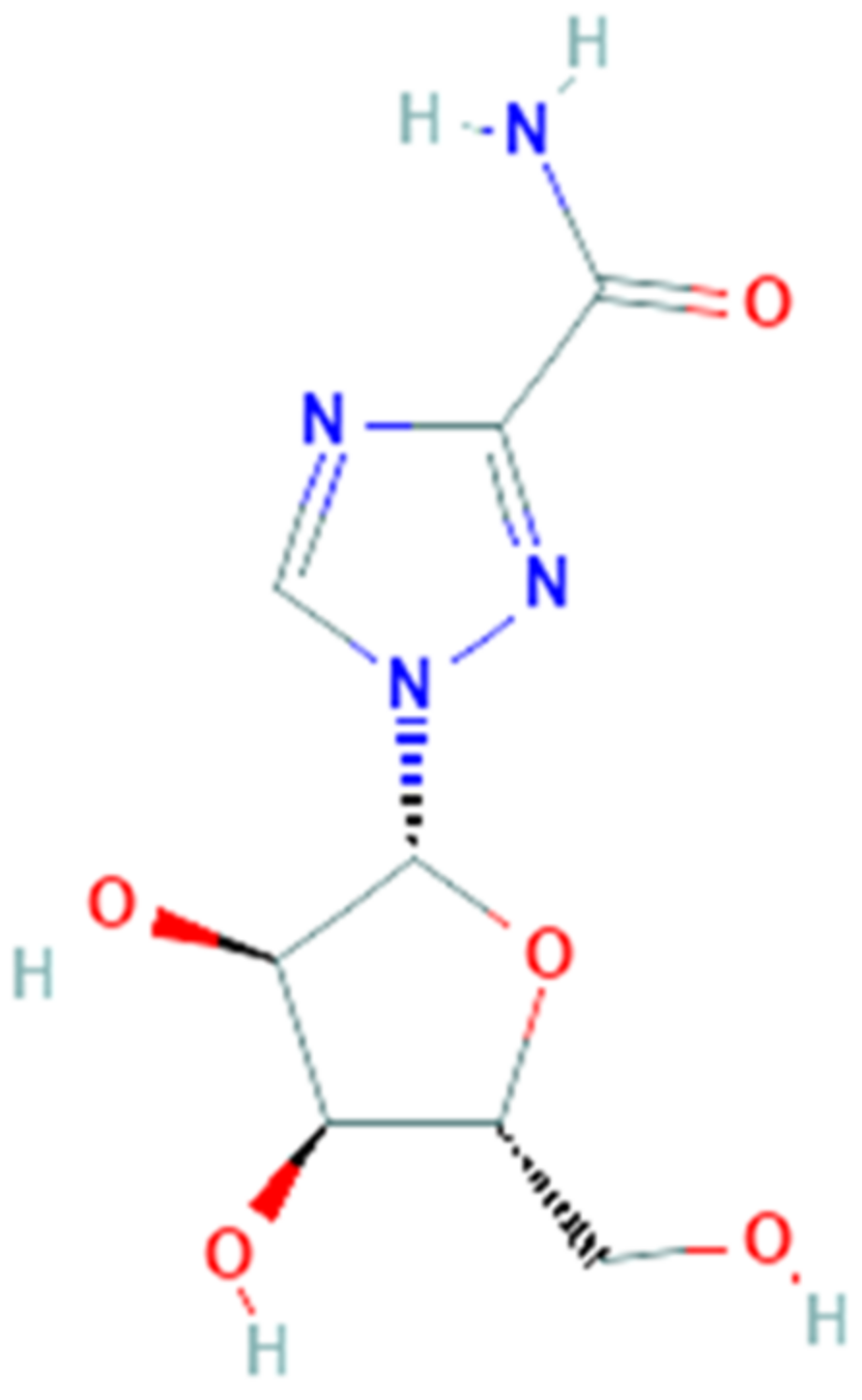
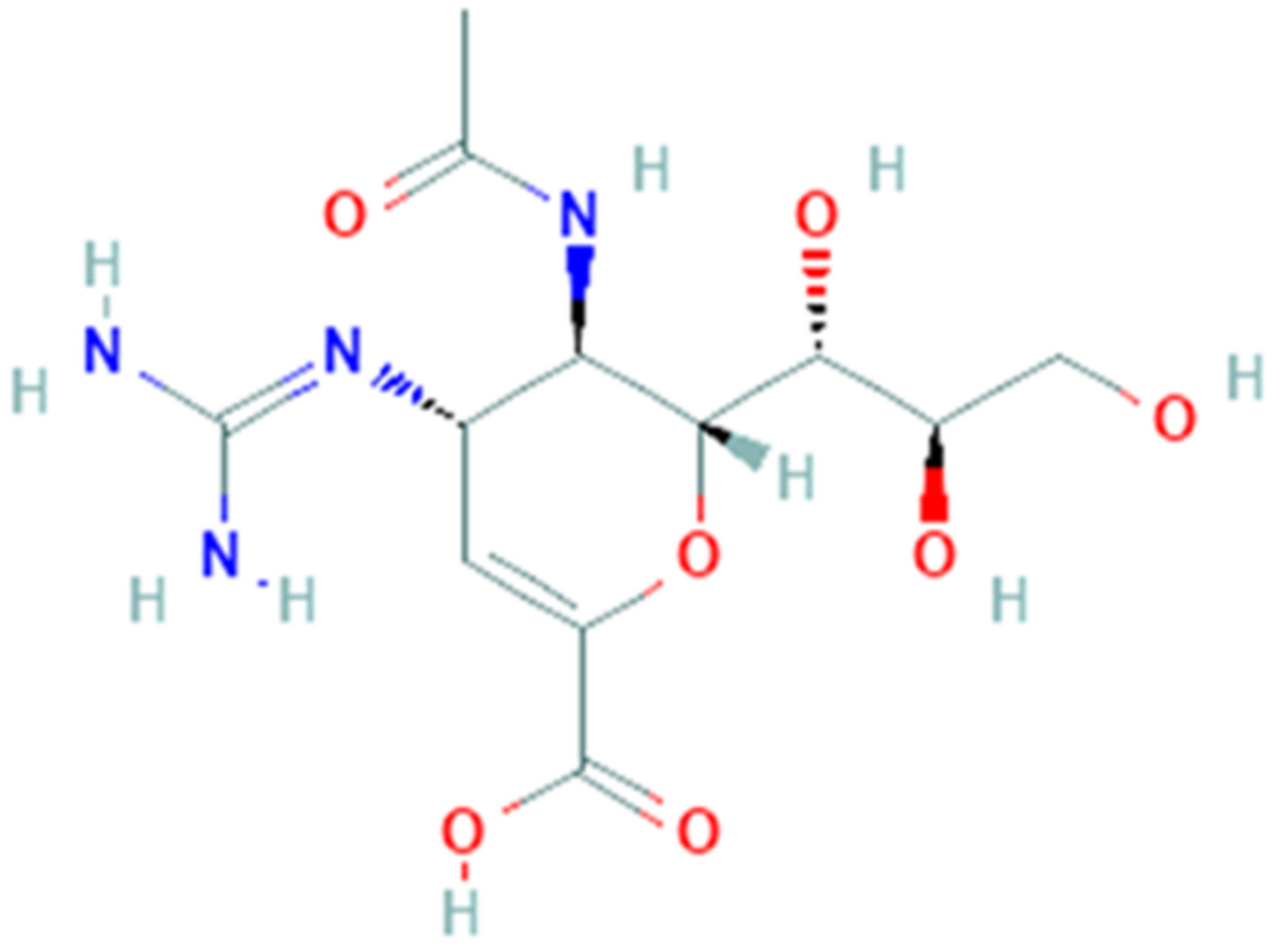
3.7. Ribavirin
3.8. Lopinavir/Ritonavir
3.9. Zanamivir
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kahn, J.; McIntosh, K. History and recent advances in coronavirus discovery. Pediatr. Infect. Dis. J. 2005, 24, S223–S227. [Google Scholar] [CrossRef] [PubMed]
- Sexton, N.R.; Smith, E.C.; Blanc, H.; Vignuzzi, M.; Peersen, O.B.; Denison, M.R. Homology-Based identification of a mutation in the coronavirus RNA- dependent RNA polymerase that confers resistance to multiple mutagens. J. Virol. 2016, 90, 7415–7428. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, C.S.; Tatti, K.M.; Ksiazek, T.G.; Rollin, P.E.; Comer, J.A.; Lee, W.W.; Rota, P.A.; Bankamp, B.; Bellini, W.J.; Zaki, S.R. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004, 10, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Pal, M.; Berhanu, G.; Desalegn, C.; Kandi, V. Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An Update. Cureus 2020, 12, e7423. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Chen, L.; Liu, W.; Zhang, Q.; Xu, K.; Ye, G.; Wu, W.; Sun, Z.; Liu, F.; Wu, K.; Zhong, B.; et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microb. Infect. 2020, 9, 313–319. [Google Scholar] [CrossRef]
- Paules, C.I.; Marston, H.D.; Fauci, A.S. Coronavirus infections more than just the common cold. JAMA 2020, 323, 703–704. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Paraskevis, D.; Kostaki, E.G.; Magiorkinis, G.; Panayiotakopoulos, G.; Sourvinos, G.; Tsiodras, S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020, 79, 104212. [Google Scholar] [CrossRef]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020, 3, 325–328. [Google Scholar] [CrossRef]
- Van Boheemen, S.; de Graaf, M.; Lauber, C.; Bestebroer, T.M.; Raj, V.S.; Zaki, A.M.; Osterhaus, A.D.; Haagmans, B.L.; Gorbalenya, A.E.; Snijder, E.J.; et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio 2012, 3, e00473-12. [Google Scholar] [CrossRef]
- Czub, M.; Weingartl, H.; Czub, S.; He, R.; Cao, J. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine 2005, 23, 2273–2279. [Google Scholar] [CrossRef]
- De Haan, C.A.M.; Kuo, L.; Masters, P.S.; Vennema, H.; Rottier, P.J.M. Coronavirus particle assembly: Primary structure requirements of the membrane protein. J. Virol. 1998, 72, 6838–6850. [Google Scholar] [CrossRef]
- Tortorici, M.A.; Veesler, D. Structural insights into coronavirus entry. Virus Res. 2019, 105, 93–116. [Google Scholar]
- Zhang, N.; Jiang, S.; Du, L. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev. Vaccines 2014, 13, 761–767. [Google Scholar] [CrossRef]
- Xia, S.; Zhu, Y.; Liu, M.; Lan, Q.; Xu, W.; Wu, Y.; Ying, T.; Liu, S.; Shi, Z.; Jiang, S.; et al. Fusion mechanism of 2019nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol. Immunol 2020, 17, 765–767. [Google Scholar] [CrossRef]
- Yu, F.; Du, L.; Ojcius, D.M.; Pan, C.; Jiang, S. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020, 22, 74–79. [Google Scholar] [CrossRef]
- Sola, I.; Almazán, F.; Zúñiga, S.; Enjuanes, L. Continuous and discontinuous RNA synthesis in coronaviruses I. Ann. Rev. Virol. 2015, 2, 265–288. [Google Scholar] [CrossRef]
- Ziebuhr, J. The coronavirus replicase. In Coronavirus Replication and Reverse Genetics; Springer: Berlin/Heidelberg, Germany, 2005; Volume 287, pp. 57–94. [Google Scholar]
- Almazán, F.; De Diego, M.L.; Galán, C.; Escors, D.; Alvarez, E.; Ortego, J.; Sola, I.; Zuñiga, S.; Alonso, S.; Moreno, J.L.; et al. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 2006, 80, 10900–10906. [Google Scholar] [CrossRef]
- Peiris, J.S.M. Coronaviruses. In Medical Microbiology, 18th ed.; Greenwood, D., Barer, M., Slack, R., Irving, W., Eds.; Churchill Livingstone: London, UK, 2012; pp. 587–593. [Google Scholar]
- Wang, Z.; Schmidt, F.; Weisblum, Y.; Muecksch, F.; Barnes, C.O.; Finkin, S.; Schaefer-Babajew, D.; Cipolla, M.; Gaebler, C.; Lieberman, J.A.; et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021, 592, 616–622. [Google Scholar] [CrossRef]
- Greaney, A.J.; Starr, T.N.; Gilchuk, P.; Zost, S.J.; Binshtein, E.; Loes, A.N.; Hilton, S.K.; Huddleston, J.; Eguia, R.; Crawford, K.; et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe 2021, 29, 44–57.e9. [Google Scholar] [CrossRef]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.H.; Michailidis, E.; et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 2020, 9, e61312. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Xia, H.; Zhang, X.; Fontes-Garfias, C.R.; Swanson, K.A.; Cai, H.; Sarkar, R.; Chen, W.; Cutler, M.; et al. Neutralizing activity of BNT162b2-elicited serum-preliminary report. N. Engl. J. Med. 2021, 384, 1466–1468. [Google Scholar] [CrossRef]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; de Oliveira, T.; Vermeulen, M.; van der Berg, K.; et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; VanBlargan, L.A.; Bloyet, L.M.; Rothlauf, P.W.; Chen, R.E.; Stumpf, S.; Zhao, H.; Errico, J.M.; Theel, E.S.; Liebeskind, M.J.; et al. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv 2020, in press. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Fontanet, A.; Autran, B.; Lina, B.; Kieny, M.P.; Karim, S.; Sridhar, D. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet 2021, 397, 952–954. [Google Scholar] [CrossRef]
- Cascella, M.; Rajnik, M.; Cuomo, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation and Treatment Coronavirus (COVID-19). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Jans, D.A.; Wagstaff, K.M. Ivermectin as a Broad-Spectrum Host-Directed Antiviral: The Real Deal? Cells 2020, 9, 2100. [Google Scholar] [CrossRef] [PubMed]
- Wulan, W.N.; Heydet, D.; Walker, E.J.; Gahan, M.E.; Ghildyal, R. Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Front. Microbiol. 2015, 6, 553. [Google Scholar] [CrossRef] [PubMed]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020, 178, 104787. [Google Scholar] [CrossRef]
- Cortegiani, A.; Ingoglia, G.; Ippolito, M.; Giarratano, A.; Einav, S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care 2020, 57, 279–283. [Google Scholar] [CrossRef]
- Li, G.; De Clercq, E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 2020, 19, 149–150. [Google Scholar] [CrossRef]
- Liu, J.; Zheng, X.; Tong, Q.; Li, W.; Wang, B.; Sutter, K.; Lu, M.; Dittmer, U.; Yang, D. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J. Med. Virol. 2020, 92, 491–494. [Google Scholar] [CrossRef]
- Deb, P.; Molla, M.M.A.; Rahman, K.M.S. An update to monoclonal antibody as therapeutic option against COVID-19. Biosaf. Health 2021, in press. [Google Scholar] [CrossRef]
- Weinreich, D.M.; Sivapalasingam, S.; Norton, T.; Ali, S.; Gao, H.; Bhore, R.; Musser, B.J.; Soo, Y.; Rofail, D.; Im, J.; et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021, 384, 238–251. [Google Scholar] [CrossRef]
- Jones, B.E.; Brown-Augsburger, P.L.; Corbett, K.S.; Westendorf, K.; Davies, J.; Cujec, T.P.; Wiethoff, C.M.; Blackbourne, J.L.; Heinz, B.A.; Foster, D.; et al. LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mahase, E. Covid-19: FDA authorises neutralising antibody bamlanivimab for non-admitted patients. BMJ 2020, 371, m4362. [Google Scholar]
- Gottlieb, R.L.; Nirula, A.; Chen, P.; Boscia, J.; Heller, B.; Morris, J.; Huhn, G.; Cardona, J.; Mocherla, B.; Stosor, V.; et al. Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA 2021, 325, 632–644. [Google Scholar] [CrossRef]
- Kaplon, H.; Reichert, J.M. Antibodies to watch in 2021. MAbs 2021, 13, 1860476. [Google Scholar] [CrossRef]
- Liu, H.; Wei, P.; Zhang, Q.; Chen, Z.; Aviszus, K.; Downing, W.; Peterson, S.; Reynoso, L.; Downey, G.P.; Frankel, S.K.; et al. 501Y.V2 and 501Y.V3 variants of SARS-CoV-2 lose binding to Bamlanivimab in vitro. bioRxiv 2021. [Google Scholar] [CrossRef]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef]
- Blaising, J.; Polyak, S.J.; Pécheur, E.I. Arbidol as a broad-spectrum antiviral: An update. Antivir. Res. 2014, 107, 84–94. [Google Scholar] [CrossRef]
- Leneva, I.A.; Fediakina, I.T.; Gus’kova, T.A.; Glushkov, R.G. Sensitivity of Various Influenza Virus Strains to Arbidol. Influence of Arbidol Combination With Different Antiviral Drugs on Reproduction of Influenza Virus A. Ter. Arkh. 2005, 77, 84–88. [Google Scholar]
- Zhao, C.; Zhao, Y.; Chai, H.; Gong, P. Synthesis and in vitro anti-hepatitis B virus activities of some ethyl 5-hydroxy-1H-indole-3-carboxylates. Bioorg. Med. Chem. 2006, 14, 2552–2558. [Google Scholar] [CrossRef]
- Boriskin, Y.S.; Pécheur, E.I.; Polyak, S.J. Arbidol: A broad-spectrum antiviral that inhibits acute and chronic HCV infection. Virol. J. 2006, 3, 56. [Google Scholar] [CrossRef]
- Brooks, M.J.; Sasadeusz, J.J.; Tannock, G.A. Antiviral chemotherapeutic agents against respiratory viruses: Where are we now and what’s in the pipeline? Curr. Opin. Pulm. Med. 2004, 10, 197–203. [Google Scholar] [CrossRef]
- White, J.; Kartenbeck, J.; Helenius, A. Membrane fusion activity of influenza virus. EMBO J. 1982, 1, 217–222. [Google Scholar] [CrossRef]
- Brooks, M.J.; Burtseva, E.I.; Ellery, P.J.; Marsh, G.A.; Lew, A.M.; Slepushkin, A.N.; Crowe, S.M.; Tannock, G.A. Antiviral activity of arbidol, a broad-spectrum drug for use against respiratory viruses, varies according to test conditions. J. Med. Virol. 2012, 84, 170–181. [Google Scholar] [CrossRef]
- Pécheur, E.I.; Borisevich, V.; Halfmann, P.; Morrey, J.D.; Smee, D.F.; Prichard, M.; Mire, C.E.; Kawaoka, Y.; Geisbert, T.W.; Polyak, S.J. The Synthetic Antiviral Drug Arbidol Inhibits Globally Prevalent Pathogenic Viruses. J. Virol. 2016, 90, 3086–3092. [Google Scholar] [CrossRef]
- Wang, H.; Yang, P.; Liu, K.; Guo, F.; Zhang, Y.; Zhang, G.; Jiang, C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008, 18, 290–301. [Google Scholar] [CrossRef]
- Inoue, Y.; Tanaka, N.; Tanaka, Y.; Inoue, S.; Morita, K.; Zhuang, M.; Hattori, T.; Sugamura, K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007, 81, 8722–8729. [Google Scholar] [CrossRef]
- Guo, Y.R.; Cao, Q.D.; Hong, Z.S.; Tan, Y.Y.; Chen, S.D.; Jin, H.J.; Tan, K.S.; Wang, D.Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak-an update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef]
- Vankadari, N. Arbidol: A potential antiviral drug for the treatment of SARS- CoV-2 by blocking trimerization of the spike glycoprotein. Int. J. Antimicrob. Agents 2020, 56, 105998. [Google Scholar] [CrossRef]
- Wang, M.Z.; Cai, B.Q.; Li, L.Y.; Lin, J.T.; Su, N.; Yu, H.X.; Gao, H.; Zhao, J.Z.; Liu, L. Efficacy and safety of arbidol in treatment of naturally acquired influenza. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 2004, 26, 289–293. [Google Scholar] [PubMed]
- Deng, L.; Li, C.; Zeng, Q.; Liu, X.; Li, X.; Zhang, H.; Hong, Z.; Xia, J. Arbidol combined with LPV/r versus LPV/r alone against Corona Virus Disease 2019: A retrospective cohort study. J. Infect. 2020, 81, e1–e5. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Lu, Z.; Xu, T.; Chen, C.; Yang, G.; Zha, T.; Lu, J.; Xue, Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 2020, 81, e21–e23. [Google Scholar] [CrossRef] [PubMed]
- Warren, T.K.; Wells, J.; Panchal, R.G.; Stuthman, K.S.; Garza, N.L.; Van Tongeren, S.A.; Dong, L.; Retterer, C.J.; Eaton, B.P.; Pegoraro, G.; et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature 2014, 508, 402–405. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.; Kotian, P.; Warren, T.; Panchal, R.; Bavari, S.; Julander, J.; Dobo, S.; Rose, A.; El-Kattan, Y.; Taubenheim, B.; et al. BCX4430-A broad-spectrum antiviral adenosine nucleoside analog under development for the treatment of Ebola virus disease. J. Infect. Public Health 2016, 9, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Julander, J.G.; Bantia, S.; Taubenheim, B.R.; Minning, D.M.; Kotian, P.; Morrey, J.D.; Smee, D.F.; Sheridan, W.P.; Babu, Y.S. BCX4430, a novel nucleoside analog, effectively treats yellow fever in a Hamster model. Antimicrob. Agents Chemother. 2014, 58, 6607–6614. [Google Scholar] [CrossRef] [PubMed]
- Julander, J.G.; Siddharthan, V.; Evans, J.; Taylor, R.; Tolbert, K.; Apuli, C.; Stewart, J.; Collins, P.; Gebre, M.; Neilson, S.; et al. Efficacy of the broad-spectrum antiviral compound BCX4430 against Zika virus in cell culture and in a mouse model. Antivir. Res. 2017, 137, 14–22. [Google Scholar] [CrossRef]
- Elfiky, A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020, 253, 117592. [Google Scholar] [CrossRef]
- Jarvis, B.; Faulds, D. Nelfinavir. A review of its therapeutic efficacy in HIV infection. Drugs 1998, 56, 147–167. [Google Scholar] [CrossRef]
- Debouck, C. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res. Hum. Retrovir. 1992, 8, 153–164. [Google Scholar] [CrossRef]
- Pai, V.B.; Nahata, M.C. Nelfinavir mesylate: A protease inhibitor. Ann. Pharm. 1999, 33, 325–339. [Google Scholar] [CrossRef]
- Gantt, S.; Carlsson, J.; Ikoma, M.; Gachelet, E.; Gray, M.; Geballe, A.P.; Corey, L.; Casper, C.; Lagunoff, M.; Vieira, J. The HIV protease inhibitor nelfinavir inhibits Kaposi’s sarcoma-associated herpesvirus replication in vitro. Antimicrob. Agents Chemother. 2011, 55, 2696–2703. [Google Scholar] [CrossRef]
- Kalu, N.N.; Desai, P.J.; Shirley, C.M.; Gibson, W.; Dennis, P.A.; Ambinder, R.F. Nelfinavir inhibits maturation and export of herpes simplex virus 1. J. Virol. 2014, 88, 5455–5461. [Google Scholar] [CrossRef]
- Yamamoto, N.; Yang, R.; Yoshinaka, Y.; Amari, S.; Nakano, T.; Cinatl, J.; Rabenau, H.; Doerr, H.W.; Hunsmann, G.; Otaka, A.; et al. HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem. Biophys. Res. Commun. 2004, 318, 719–725. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Wang, N.; Li, H.; Shi, Y.; Guo, G.; Liu, K.; Zeng, H.; Zou, Q. Therapeutic drugs targeting 2019-nCoV main protease by high-throughput screening. bioRxiv 2020, in press. [Google Scholar]
- Cardoso, W.B.; Mendanha, S.A. Molecular dynamics simulation of docking structures of SARS-CoV-2 main protease and HIV protease inhibitors. J. Mol. Struct. 2021, 1225, 129143. [Google Scholar] [CrossRef]
- Musarrat, F.; Chouljenko, V.; Dahal, A.; Nabi, R.; Chouljenko, T.; Jois, S.D.; Kousoulas, K.G. The anti-HIV drug nelfinavir mesylate (Viracept) is a potent inhibitor of cell fusion caused by the SARSCoV-2 spike (S) glycoprotein warranting further evaluation as an antiviral against COVID-19. J. Med. Virol. 2020, 92, 2087–2095. [Google Scholar] [CrossRef]
- Moyle, G.J. Saquinavir-soft gel: Establishing saquinavir in HAART regimens. Expert Opin. Investig. Drug 1998, 7, 1313–1322. [Google Scholar] [CrossRef]
- Noble, S.; Faulds, D. Saquinavir. A review of its pharmacology and clinical potential in the management of HIV infection. Drugs 1996, 52, 93–112. [Google Scholar] [CrossRef]
- Vella, S.; Floridia, M. Saquinavir. Clinical pharmacology and efficacy. Clin. Pharm. 1998, 34, 189–201. [Google Scholar] [CrossRef]
- Merry, C.; Barry, M.G.; Mulcahy, F.; Ryan, M.; Heavey, J.; Tjia, J.F.; Gibbons, S.E.; Breckenridge, A.M.; Back, D.J. Saquinavir pharmacokinetics alone and in combination with ritonavir in HIV-infected patients. AIDS 1997, 11, F29–F33. [Google Scholar] [CrossRef]
- Kumar, G.N.; Rodrigues, D.; Buko, A.M.; Denissen, J.F. Cytochrome P450- mediated metabolism of the HIV-1 protease inhibitor ritonavir (ABT-538) in human liver microsomes. J. Pharmacol. Exp. Ther. 1996, 277, 423–431. [Google Scholar]
- Perry, C.M.; Noble, S. Saquinavir soft-gel capsule formulation. A review of its use in patients with HIV infection. Drugs 1998, 55, 461–486. [Google Scholar] [CrossRef]
- Savarino, A. Expanding the frontiers of existing antiviral drugs: Possible effects of HIV-1 protease inhibitors against SARS and avian influenza. J. Clin. Virol. 2005, 34, 170–178. [Google Scholar] [CrossRef]
- Lai, S.T. Treatment of severe acute respiratory syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 583–591. [Google Scholar] [CrossRef]
- Ortega, J.T.; Serrano, M.L.; Pujol, F.H.; Rangel, H.R. Unrevealing sequence and structural features of novel coronavirus using in silico approaches: The main protease as molecular target. EXCLI J. 2020, 19, 400–409. [Google Scholar] [PubMed]
- Hall, D.C., Jr.; Ji, H.F. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med. Infect. Dis. 2020, 35, 101646. [Google Scholar] [CrossRef]
- Khan, S.A.; Zia, K.; Ashraf, S.; Uddin, R.; Ul-Haq, Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J. Biomol. Struct. Dyn. 2021, in press. [Google Scholar] [CrossRef]
- Jin, Z.; Smith, L.K.; Rajwanshi, V.K.; Kim, B.; Deval, J. The ambiguous base-pairing and high substrate efficiency of T-705 (Favipiravir) Ribofuranosyl 5′-triphosphate towards influenza A virus polymerase. PLoS ONE 2013, 8, e68347. [Google Scholar] [CrossRef]
- Oestereich, L.; Lüdtke, A.; Wurr, S.; Rieger, T.; Muñoz-Fontela, C.; Günther, S. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antivir. Res. 2014, 105, 17–21. [Google Scholar] [CrossRef]
- Itoh, Y.; Shinya, K.; Kiso, M.; Watanabe, T.; Sakoda, Y.; Hatta, M.; Muramoto, Y.; Tamura, D.; Sakai-Tagawa, Y.; Noda, T.; et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 2009, 460, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Furuta, Y.; Komeno, T.; Nakamura, T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 449–463. [Google Scholar] [CrossRef]
- Cai, Q.; Yang, M.; Liu, D.; Chen, J.; Shu, D.; Xia, J.; Liao, X.; Gu, Y.; Cai, Q.; Yang, Y.; et al. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering 2020, 6, 1192–1198. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Y.; Huang, J.; Yin, P.; Cheng, Z.; Wu, J.; Chen, S.; Zhang, Y.; Chen, B.; Lu, M.; et al. Favipiravir versus Arbidol for COVID-19: A Randomized Clinical Trial. medRxiv 2020. [Google Scholar] [CrossRef]
- Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Götte, M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses 2019, 11, 326. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS- 5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 2018, 9, e00221-18. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Götte, M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020, 295, 4773–4779. [Google Scholar] [CrossRef]
- Ko, W.C.; Rolain, J.M.; Lee, N.Y.; Chen, P.L.; Huang, C.T.; Lee, P.I.; Hsueh, P.R. Arguments in favour of remdesivir for treating SARS-CoV-2 infections. Int. J. Antimicrob. Agents 2020, 55, 105933. [Google Scholar] [CrossRef]
- Warren, T.K.; Jordan, R.; Lo, M.K.; Ray, A.S.; Mackman, R.L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H.C.; et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016, 531, 381–385. [Google Scholar] [CrossRef]
- Lo, M.K.; Jordan, R.; Arvey, A.; Sudhamsu, J.; Shrivastava-Ranjan, P.; Hotard, A.L.; Flint, M.; McMullan, L.K.; Siegel, D.; Clarke, M.O.; et al. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017, 7, 43395. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Graham, R.L.; Menachery, V.D.; Gralinski, L.E.; Case, J.B.; Leist, S.R.; Pyrc, K.; Feng, J.Y.; Trantcheva, I.; et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017, 9, eaal3653. [Google Scholar] [CrossRef]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef]
- Tu, Y.F.; Chien, C.S.; Yarmishyn, A.A.; Lin, Y.Y.; Luo, Y.H.; Lin, Y.T.; Lai, W.Y.; Yang, D.M.; Chou, S.J.; Yang, Y.P.; et al. A Review of SARS-CoV-2 and the Ongoing Clinical Trials. Int. J. Mol. Sci. 2020, 21, 2657. [Google Scholar] [CrossRef]
- Grein, J.; Ohmagari, N.; Shin, D.; Diaz, G.; Asperges, E.; Castagna, A.; Feldt, T.; Green, G.; Green, M.L.; Lescure, F.X.; et al. Compassionate Use of Remdesivir for Patients with Severe Covid-19. N. Engl. J. Med. 2020, 382, 2327–2336. [Google Scholar] [CrossRef]
- Hillaker, E.; Belfer, J.J.; Bondici, A.; Murad, H.; Dumkow, L.E. Delayed Initiation of Remdesivir in a COVID-19-Positive Patient. Pharmacotherapy 2020, 40, 592–598. [Google Scholar] [CrossRef]
- Graci, J.D.; Cameron, C.E. Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 2016, 16, 37–48. [Google Scholar] [CrossRef]
- Murata, Y. Respiratory syncytial virus infection in adults. Curr. Opin. Pulm. Med. 2020, 14, 235–240. [Google Scholar] [CrossRef]
- Snell, N.J. Ribavirin—Current status of a broad spectrum antiviral agent. Expert Opin. Pharmacother. 2001, 2, 1317–1324. [Google Scholar] [CrossRef] [PubMed]
- Bierman, S.M.; Kirkpatrick, W.; Fernandez, H. Clinical efficacy of ribavirin in the treatment of genital herpes simplex virus infection. Chemotherapy 1981, 27, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Habib, A.M.G.; Ali, M.A.E.; Zouaoui, B.R.; Taha, M.A.H.; Mohammed, B.S.; Saquib, N. Clinical outcomes among hospital patients with Middle East respiratory syndrome coronavirus (MERS-CoV) infection. BMC Infect. Dis. 2019, 19, 870. [Google Scholar] [CrossRef] [PubMed]
- Peiris, J.S.; Chu, C.M.; Cheng, V.C.; Chan, K.S.; Hung, I.F.; Poon, L.L.; Law, K.I.; Tang, B.S.; Hon, T.Y.; Chan, C.S.; et al. Clinical progression and viral load in a community outbreak of coronavirus- associated SARS pneumonia: A prospective study. Lancet 2003, 361, 1767–1772. [Google Scholar] [CrossRef]
- Lee, N.; Hui, D.; Wu, A.; Chan, P.; Cameron, P.; Joynt, G.M.; Ahuja, A.; Yung, M.Y.; Leung, C.B.; To, K.F.; et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003, 348, 1986–1994. [Google Scholar] [CrossRef] [PubMed]
- Poutanen, S.M.; Low, D.E.; Henry, B.; Finkelstein, S.; Rose, D.; Green, K.; Tellier, R.; Draker, R.; Adachi, D.; Ayers, M.; et al. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003, 348, 1995–2005. [Google Scholar] [CrossRef]
- Omrani, A.S.; Saad, M.M.; Baig, K.; Bahloul, A.; Abdul-Matin, M.; Alaidaroos, A.Y.; Almakhlafi, G.A.; Albarrak, M.M.; Memish, Z.A.; Albarrak, A.M. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: A retrospective cohort study. Lancet Infect. Dis. 2015, 14, 1090–1095. [Google Scholar] [CrossRef]
- Hung, I.F.; Lung, K.; Tso, E.Y.; Liu, R.; Chung, T.W.; Chu, M.; Ng, Y.; Lo, J.; Chan, J.; Tam, A.R.; et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: An open-label, randomised, phase 2 trial. Lancet 2020, 395, 1695–1704. [Google Scholar] [CrossRef]
- De Clercq, E. Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int. J. Antimicrob. Agents 2009, 33, 307–320. [Google Scholar] [CrossRef]
- Sundquist, W.I.; Kräusslich, H.G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012, 2, a006924. [Google Scholar] [CrossRef]
- Chu, C.M.; Cheng, V.C.; Hung, I.F.; Wong, M.M.; Chan, K.H.; Chan, K.S.; Kao, R.Y.; Poon, L.L.; Wong, C.L.; Guan, Y.; et al. Role of lopinavir/ritonavir in the treatment of SARS: Initial virological and clinical findings. Thorax 2004, 59, 252–256. [Google Scholar] [CrossRef]
- De Wilde, A.H.; Jochmans, D.; Posthuma, C.C.; Zevenhoven-Dobbe, J.C.; van Nieuwkoop, S.; Bestebroer, T.M.; van den Hoogen, B.G.; Neyts, J.; Snijder, E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014, 58, 4875–4884. [Google Scholar] [CrossRef]
- Choy, K.T.; Wong, A.Y.; Kaewpreedee, P.; Sia, S.F.; Chen, D.; Hui, K.; Chu, D.; Chan, M.; Cheung, P.P.; Huang, X.; et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020, 178, 104786. [Google Scholar] [CrossRef]
- Muralidharan, N.; Sakthivel, R.; Velmurugan, D.; Gromiha, M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J. Biomol. Struct. Dyn. 2021, in press. [Google Scholar] [CrossRef]
- Baden, L.R.; Rubin, E.J. Covid-19-The Search for Effective Therapy. N. Engl. J. Med. 2020, 382, 1851–1852. [Google Scholar] [CrossRef]
- De Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef]
- Ye, X.T.; Luo, Y.L.; Xia, S.C.; Sun, Q.F.; Ding, J.G.; Zhou, Y.; Chen, W.; Wang, X.F.; Zhang, W.W.; Du, W.J.; et al. Clinical efficacy of lopinavir/ritonavir in the treatment of Coronavirus disease 2019. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3390–3396. [Google Scholar]
- Suzuki, Y. Sialobiology of influenza: Molecular mechanism of host range variation of influenza viruses. Biol. Pharm. Bull. 2005, 28, 399–408. [Google Scholar] [CrossRef]
- Nayak, D.P.; Hui, E.K.; Barman, S. Assembly and budding of influenza virus. Virus Res. 2004, 106, 147–165. [Google Scholar] [CrossRef]
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, N.A.; Klenk, H.D. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 2004, 78, 12665–12667. [Google Scholar] [CrossRef]
- Hui, D.S.; I Azhar, E.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health-The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef]
- Anand, K.; Ziebuhr, J.; Wadhwani, P.; Mesters, J.R.; Hilgenfeld, R. Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science 2003, 300, 1763–1767. [Google Scholar] [CrossRef]
- Yousefi, B.; Valizadeh, S.; Ghaffari, H.; Vahedi, A.; Karbalaei, M.; Eslami, M. A global treatments for coronaviruses including COVID-19. J. Cell Physiol. 2020, 235, 9133–9142. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauso, L.V.; Imbesi, C.; Irene, G.; Calì, G.; Bitto, A. New Approaches and Repurposed Antiviral Drugs for the Treatment of the SARS-CoV-2 Infection. Pharmaceuticals 2021, 14, 503. https://doi.org/10.3390/ph14060503
Bauso LV, Imbesi C, Irene G, Calì G, Bitto A. New Approaches and Repurposed Antiviral Drugs for the Treatment of the SARS-CoV-2 Infection. Pharmaceuticals. 2021; 14(6):503. https://doi.org/10.3390/ph14060503
Chicago/Turabian StyleBauso, Luana Vittoria, Chiara Imbesi, Gasparo Irene, Gabriella Calì, and Alessandra Bitto. 2021. "New Approaches and Repurposed Antiviral Drugs for the Treatment of the SARS-CoV-2 Infection" Pharmaceuticals 14, no. 6: 503. https://doi.org/10.3390/ph14060503
APA StyleBauso, L. V., Imbesi, C., Irene, G., Calì, G., & Bitto, A. (2021). New Approaches and Repurposed Antiviral Drugs for the Treatment of the SARS-CoV-2 Infection. Pharmaceuticals, 14(6), 503. https://doi.org/10.3390/ph14060503