Development of Non-Ethoxypropanoic Acid Type Cryptochrome Inhibitors with Circadian Molecular Clock-Enhancing Activity by Bioisosteric Replacement
Abstract
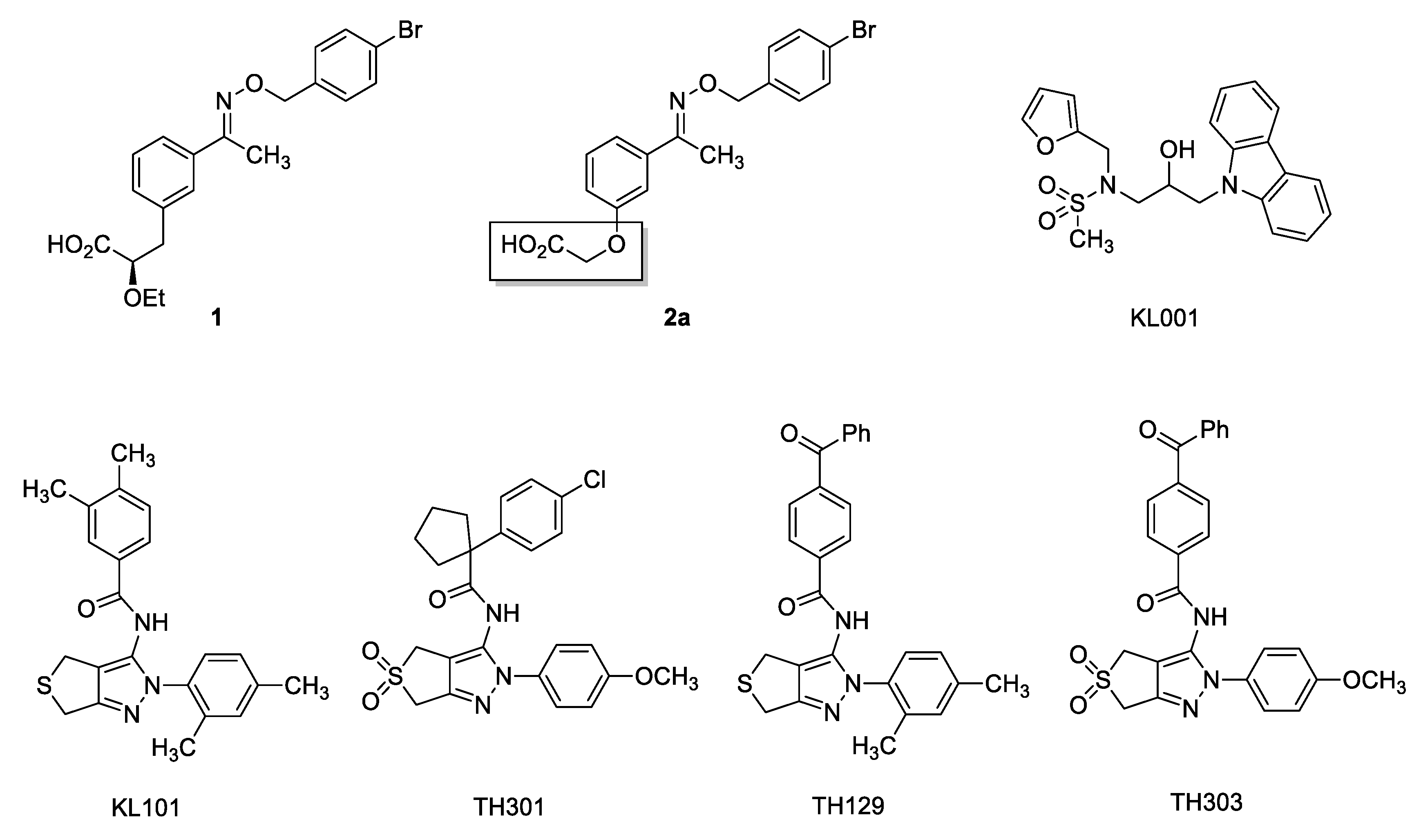
1. Introduction
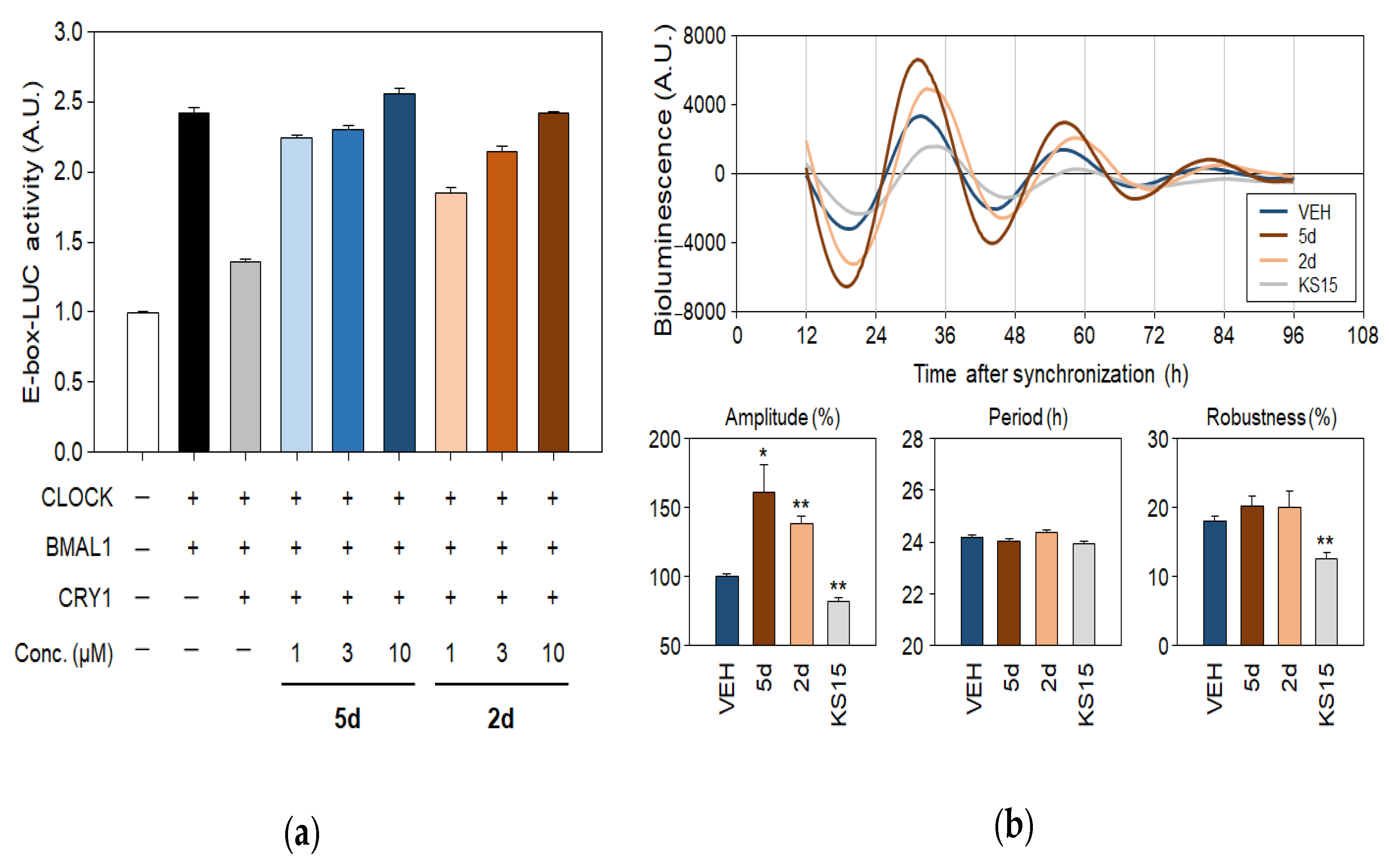
2. Results and Discussion
3. Materials and Methods
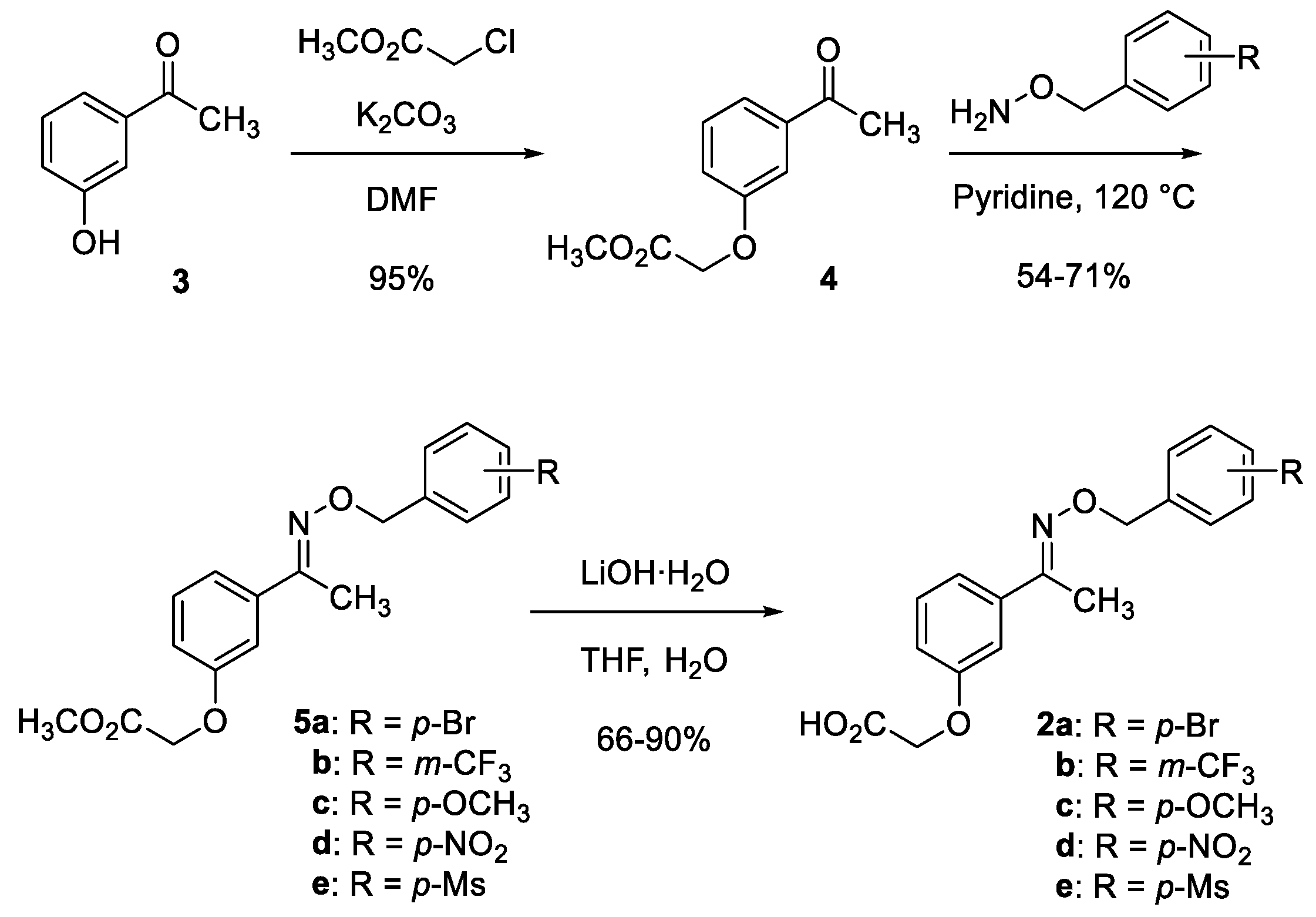
3.1. Chemistry
3.1.1. Methyl 2-(3-acetylphenoxy)acetate (4)
3.1.2. Representative Procedure: Methyl 2-(3-(1-(((4-nitrobenzyl)oxy)imino)ethyl)phenoxy)acetate (5d)
3.1.3. Characterization Data for 5b–e
3.1.4. Representative Procedure: 2-(3-(1-(((4-nitrobenzyl)oxy)imino)ethyl)phenoxy)acetic acid (2d)
3.1.5. Characterization Data for 2b–e
3.2. Cell Cultures and Luciferase Reporter Assay
3.3. Cytochrome P450 Inhibition Assay
3.4. Metabolic Stability Test
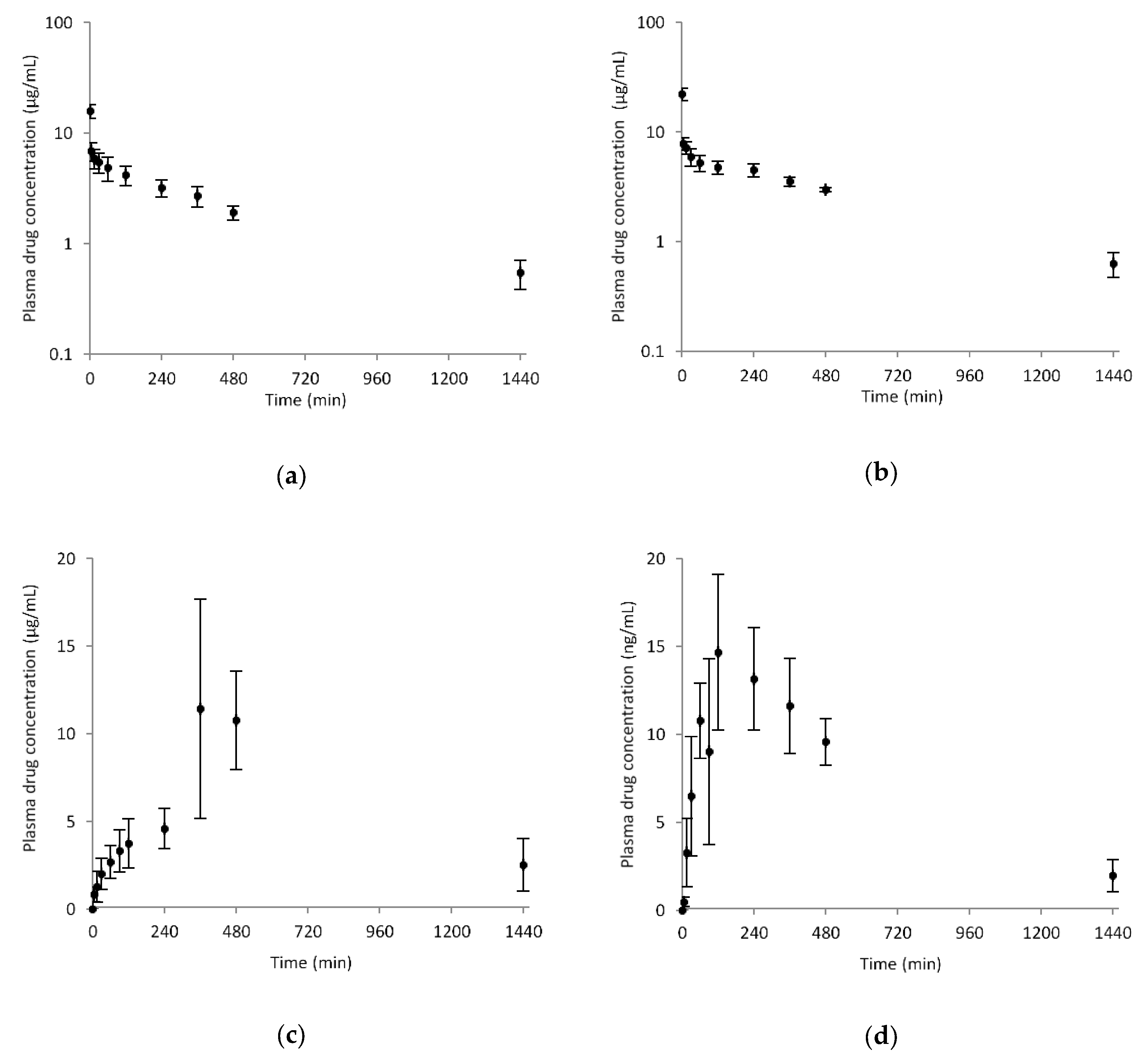
3.5. Pharmacokinetics
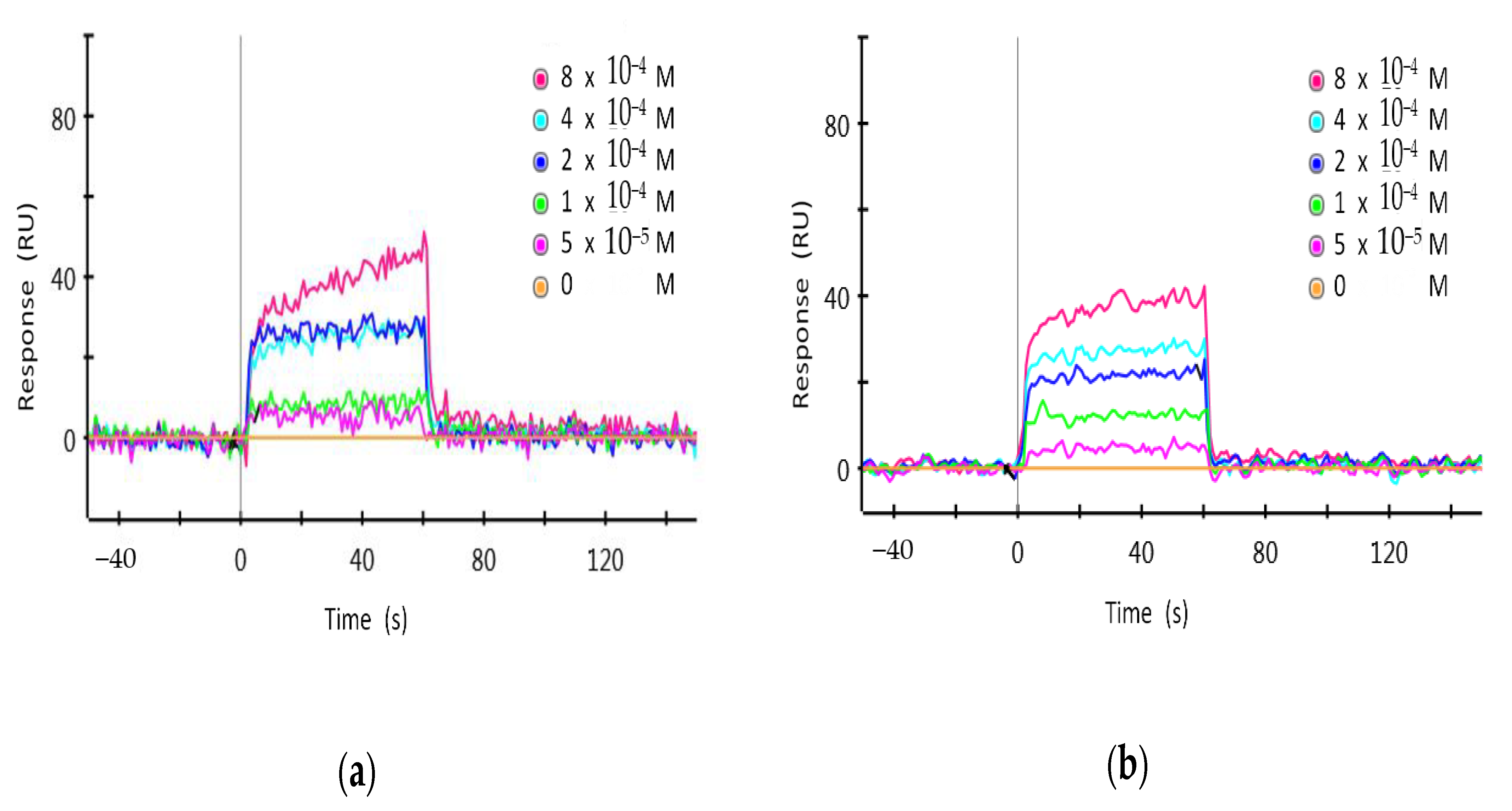
3.6. Surface Plasmon Resonance
3.7. Real-Time Bioluminescence Monitoring
3.8. Statistical Evaluation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patke, A.; Young, M.W.; Axelrod, S. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 2020, 21, 67–84. [Google Scholar] [CrossRef]
- Walker, W.H.; Walton, J.C.; DeVries, A.C.; Nelson, R.J. Circadian rhythm disruption and mental health. Transl. Psychiatry 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yoo, S.-H.; Takahashi, J.S. Development and therapeutic potential of small-molecule modulators of circadian systems. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 231–252. [Google Scholar] [CrossRef] [PubMed]
- Diallo, A.B.; Coiffard, B.; Leone, M.; Mezouar, S.; Mege, J.-L. For whom the clock ticks: Clinical chronobiology for infectious diseases. Front. Immunol. 2020, 11, 1457. [Google Scholar] [CrossRef]
- Ray, S.; Reddy, A.B. COVID-19 management in light of the circadian clock. Nat. Rev. Mol. Cell Biol. 2020, 21, 494–495. [Google Scholar] [CrossRef]
- Hirota, T.; Lee, J.W.; John, P.C.S.; Sawa, M.; Iwaisako, K.; Noguchi, T.; Pongsawakul, P.Y.; Sonntag, T.; Welsh, D.K.; Brenner, D.A.; et al. Identification of small molecule activators of cryptochrome. Science 2012, 337, 1094–1097. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Son, Y.L.; Aikawa, Y.; Makino, E.; Nagai, Y.; Srivastava, A.; Oshima, T.; Sugiyama, A.; Hara, A.; Abe, K.; et al. Isoform-selective regulation of mammalian cryptochromes. Nat. Chem. Biol. 2020, 16, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Kolarski, D.; Miller, M.; Oshima, T.; Nagai, Y.; Aoki, Y.; Kobauri, P.; Srivastava, A.; Sugiyama, A.; Amaike, K.; Sato, A.; et al. Photopharmacological manipulation of mammalian CRY1 for regulation of the circadian clock. J. Am. Chem. Soc. 2021, 143, 2078–2087. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.; Yin, L.; Collins, J.L.; Parks, D.J.; Orband-Miller, L.A.; Wisely, G.B.; Joshi, S.; Lazar, M.A.; Willson, T.M.; Zuercher, W.J. GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor Rev-erbα. ACS Chem. Biol. 2010, 5, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Helleboid, S.; Haug, C.; Lamottke, K.; Zhou, Y.; Wei, J.; Daix, S.; Cambula, L.; Rigou, G.; Hum, D.W.; Walczak, R. The identification of naturally occurring neoruscogenin as a bioavailable, potent, and high-affinity agonist of the nuclear receptor RORα (NR1F1). J. Biomol. Screen. 2014, 19, 399–406. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Nohara, K.; Park, N.; Park, Y.-S.; Guillory, B.; Zhao, Z.; Garcia, J.M.; Koike, N.; Lee, C.C.; Takahashi, J.S.; et al. The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome. Cell Metab. 2016, 23, 610–621. [Google Scholar] [CrossRef]
- Kumar, N.; Solt, L.A.; Conkright, J.J.; Wang, Y.; Istrate, M.A.; Busby, S.A.; Garcia-Ordonez, R.D.; Burris, T.P.; Griffin, P.R. The benzenesulfoamide T0901317 [N-(2, 2, 2-trifluoroethyl)-N-[4-[2, 2, 2-trifluoro-1-hydroxy-1-(trifluoromethyl) ethyl] phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-α/γ inverse agonist. Mol. Pharm. 2010, 77, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Wittine, K.; Sedić, M.; Markova-Car, E.P. Small molecules targeting biological clock; A novel prospective for anti-cancer drugs. Molecules 2020, 25, 4937. [Google Scholar] [CrossRef] [PubMed]
- Amaike, K.; Oshima, T.; Skoulding, N.S.; Toyama, Y.; Hirota, T.; Itami, K. Small molecules modulating mammalian biological clocks: Exciting new opportunities for synthetic chemistry. Chem 2020, 6, 2186–2198. [Google Scholar] [CrossRef]
- Cha, H.K.; Chung, S.; Lim, H.Y.; Jung, J.-W.; Son, G.H. Small molecule modulators of the circadian molecular clock with implications for neuropsychiatric diseases. Front. Mol. Neurosci. 2019, 11, 496. [Google Scholar] [CrossRef] [PubMed]
- Chun, S.K.; Jang, J.; Chung, S.; Yun, H.; Kim, N.-J.; Jung, J.-W.; Son, G.H.; Suh, Y.-G.; Kim, K. Identification and validation of cryptochrome inhibitors that modulate the molecular circadian clock. ACS Chem. Biol. 2014, 9, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Chun, S.K.; Chung, S.; Kim, H.-D.; Lee, J.H.; Jang, J.; Kim, J.; Kim, D.; Son, G.H.; Oh, Y.J.; Suh, Y.-G.; et al. A synthetic cryptochrome inhibitor induces anti-proliferative effects and increases chemosensitivity in human breast cancer cells. Biochem. Biophys. Res. Commun. 2015, 467, 441–446. [Google Scholar] [CrossRef]
- Jang, J.; Chung, S.; Choi, Y.; Lim, H.Y.; Son, Y.; Chun, S.K.; Son, G.H.; Kim, K.; Suh, Y.-G.; Jung, J.-W. The cryptochrome inhibitor KS15 enhances E-box-mediated transcription by disrupting the feedback action of a circadian transcription-repressor complex. Life Sci. 2018, 200, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Nguyen, P.-H.; Kim, G.; Jang, S.-N.; Lee, G.-H.; Phuc, N.M.; Wu, Z.; Liu, K.-H. Strong and selective inhibitory effects of the biflavonoid selamariscina a against CYP2C8 and CYP2C9 enzyme activities in human liver microsomes. Pharmaceutics 2020, 12, 343. [Google Scholar] [CrossRef]
- Song, Y.; Park, S.Y.; Wu, Z.; Liu, K.-H.; Seo, Y.H. Hybrid inhibitors of DNA and HDACs remarkably enhance cytotoxicity in leukaemia cells. J. Enzym. Inhib. Med. Chem. 2020, 35, 1069–1079. [Google Scholar] [CrossRef]
- Yoo, S.-H.; Yamazaki, S.; Lowrey, P.L.; Shimomura, K.; Ko, C.H.; Buhr, E.D.; Siepka, S.M.; Hong, H.-K.; Oh, W.J.; Yoo, O.J.; et al. PERIOD2: LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 2004, 101, 5339–5346. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Carmona, A.V.; Tiwari, A.K.; Trippier, P.C. Amide bond bioisosteres: Strategies, synthesis, and successes. J. Med. Chem. 2020, 63, 12290–12358. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.M.; Barreiro, E.J. Bioisosterism: A useful strategy for molecular modification and drug design. Curr. Med. Chem. 2005, 12, 23–49. [Google Scholar] [CrossRef] [PubMed]
- Dick, A.; Cocklin, S. Bioisosteric replacement as a tool in anti-HIV drug design. Pharmaceuticals 2020, 13, 36. [Google Scholar] [CrossRef]

| Entry | Compound | Max (%) 1 | EC50 (μM) |
|---|---|---|---|
| 1 | 2a | 164.8 | 1.54 |
| 2 | 5a | 149.1 | n/d 2 |
| 3 | 2b | 130.7 | n/d |
| 4 | 5b | 171.5 | n/d |
| 5 | 2c | 182.1 | 9.88 |
| 6 | 5c | 151.8 | 6.32 |
| 7 | 2d | 161.3 | 10.17 |
| 8 | 5d | 193.0 | 6.44 |
| 9 | 2e | 98.7 | n/d |
| 10 | 5e | 104.7 | n/d |
| P450 | Enzyme Activity | IC50 (μM) | |
|---|---|---|---|
| KS15 | 5d | ||
| 1A2 | Phenacetin O-deethylation | 14.9 | >50 |
| 2C9 | Tolbutamide 4-methylhydroxylation | 9.1 | >50 |
| 2C19 | Omeprazole hydroxylation | 20.2 | >50 |
| 2D6 | Dextromethorphan O-demethylation | 23.1 | >50 |
| 3A | Midazolam 1’-hydroxylation | 24.0 | >50 |
| Parameter | Intravenous (2 mg/kg) | Oral (5 mg/kg) | ||
|---|---|---|---|---|
| 5d | 2d | 5d | 2d | |
| AUClast (μg/mL·min) | 2891.2 ± 465.0 | 3885.9 ± 278.5 | 9471.5 ± 1281.7 | 10,204.7 ± 1308.5 |
| AUCinfinite (μg/mL·min) | 3268.2 ± 600.3 | 4290.6 ± 398.7 | n/a | 11,069.2 ± 1130.5 |
| t1/2 (min) | 467.1 ± 61.3 | 433.0 ± 58.0 | n/a | 391.2 ± 47.6 |
| Cmax (μg/mL) | - | - | 13.5 ± 4.7 | 14.6 ± 5.1 |
| tmax (min) | - | - | 420.0 ± 69.3 | 160.0 ± 69.2 |
| CL (mL/min/kg) | 0.6 ± 0.1 | 0.5 ± 0.0 | - | - |
| MRT (min) | 588.5 ± 75.6 | 557.1 ± 73.7 | - | - |
| Vss (mL/kg) | 349.8 ± 34.9 | 259.7 ± 25.8 | - | - |
| F (%) | - | - | > 99 | > 99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, Y.U.; Jin, H.-E.; Lim, H.Y.; Choi, G.; Joo, H.; Kang, B.; Lee, G.-H.; Liu, K.-H.; Maeng, H.-J.; Chung, S.; et al. Development of Non-Ethoxypropanoic Acid Type Cryptochrome Inhibitors with Circadian Molecular Clock-Enhancing Activity by Bioisosteric Replacement. Pharmaceuticals 2021, 14, 496. https://doi.org/10.3390/ph14060496
Jeong YU, Jin H-E, Lim HY, Choi G, Joo H, Kang B, Lee G-H, Liu K-H, Maeng H-J, Chung S, et al. Development of Non-Ethoxypropanoic Acid Type Cryptochrome Inhibitors with Circadian Molecular Clock-Enhancing Activity by Bioisosteric Replacement. Pharmaceuticals. 2021; 14(6):496. https://doi.org/10.3390/ph14060496
Chicago/Turabian StyleJeong, Yong Uk, Hyo-Eon Jin, Hye Young Lim, Goyeong Choi, Hansol Joo, Bohun Kang, Ga-Hyun Lee, Kwang-Hyeon Liu, Han-Joo Maeng, Sooyoung Chung, and et al. 2021. "Development of Non-Ethoxypropanoic Acid Type Cryptochrome Inhibitors with Circadian Molecular Clock-Enhancing Activity by Bioisosteric Replacement" Pharmaceuticals 14, no. 6: 496. https://doi.org/10.3390/ph14060496
APA StyleJeong, Y. U., Jin, H.-E., Lim, H. Y., Choi, G., Joo, H., Kang, B., Lee, G.-H., Liu, K.-H., Maeng, H.-J., Chung, S., Son, G. H., & Jung, J.-W. (2021). Development of Non-Ethoxypropanoic Acid Type Cryptochrome Inhibitors with Circadian Molecular Clock-Enhancing Activity by Bioisosteric Replacement. Pharmaceuticals, 14(6), 496. https://doi.org/10.3390/ph14060496









