Regulation of Cellular and Cancer Stem Cell-Related Putative Gene Expression of Parental and CD44+CD24− Sorted MDA-MB-231 Cells by Cisplatin
Abstract
1. Introduction
2. Results
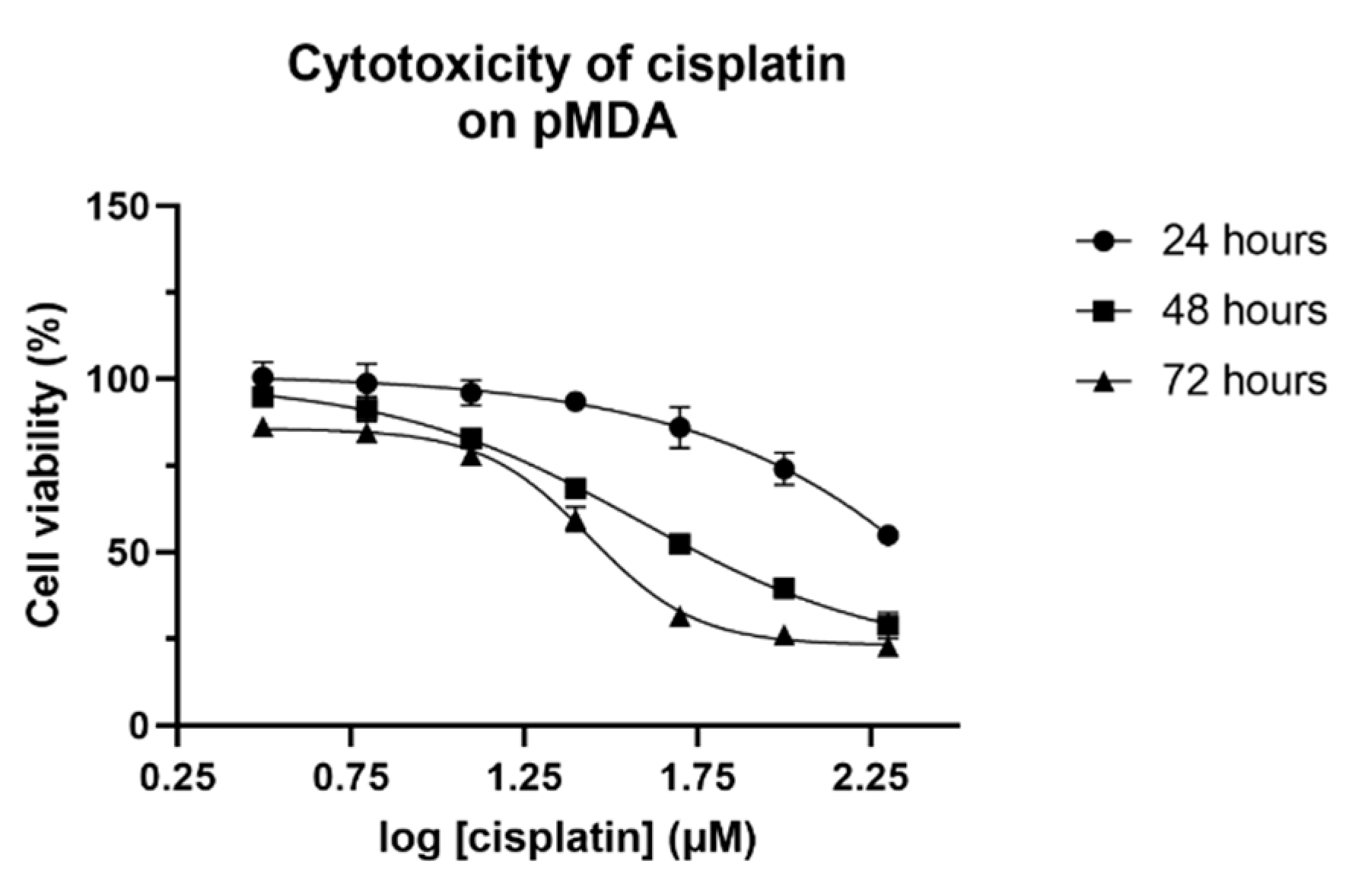
2.1. IC50 Value of Cisplatin on Parental MDA-MB-231
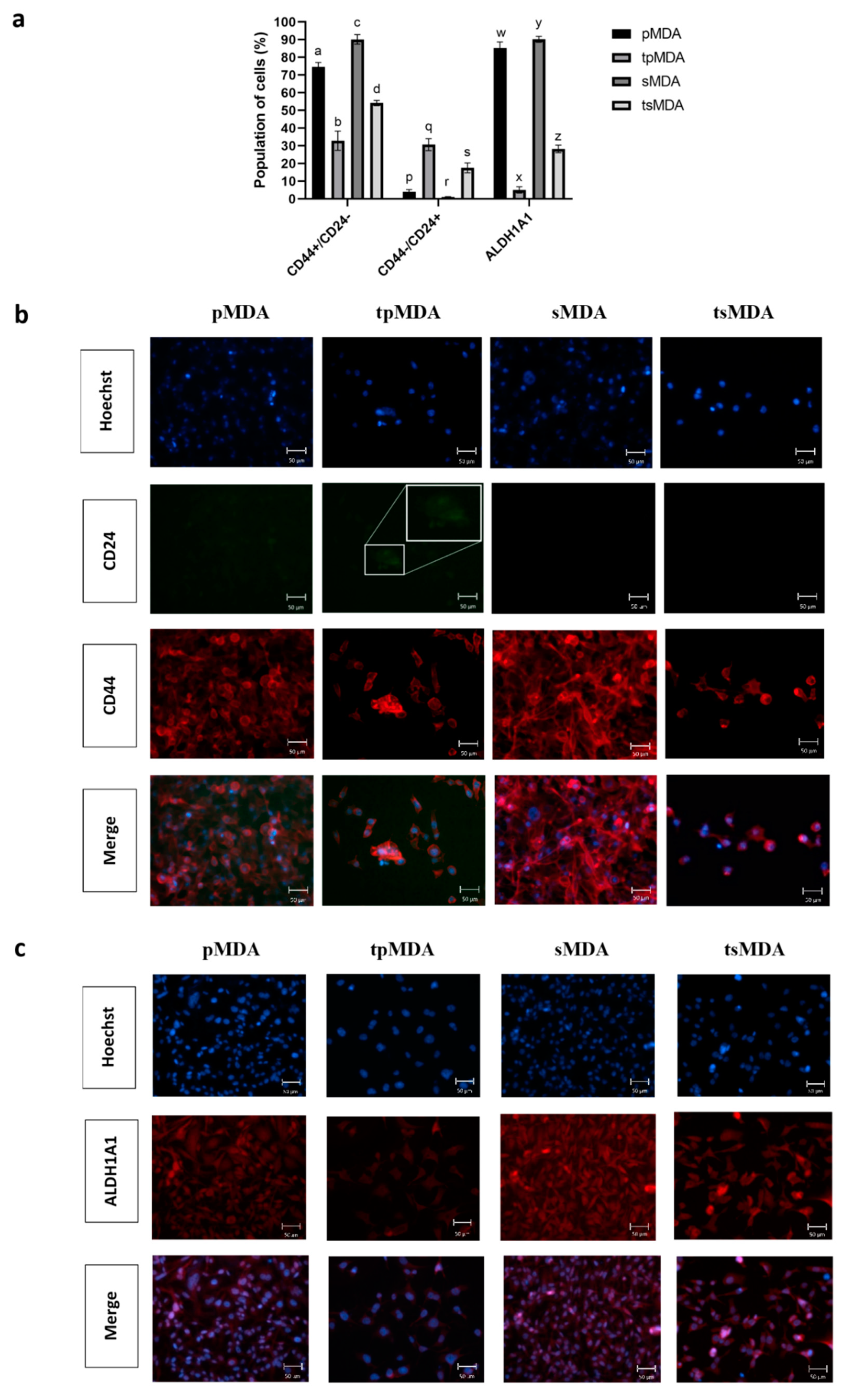
2.2. Sorted MDA-MB-231 Retains Higher Cancer Stem Cell Phenotype Than Parental MDA-MB-231 after Cisplatin Treatment
2.3. Sorted MDA-MB-231 Is More Resistant to Anticancer Drugs
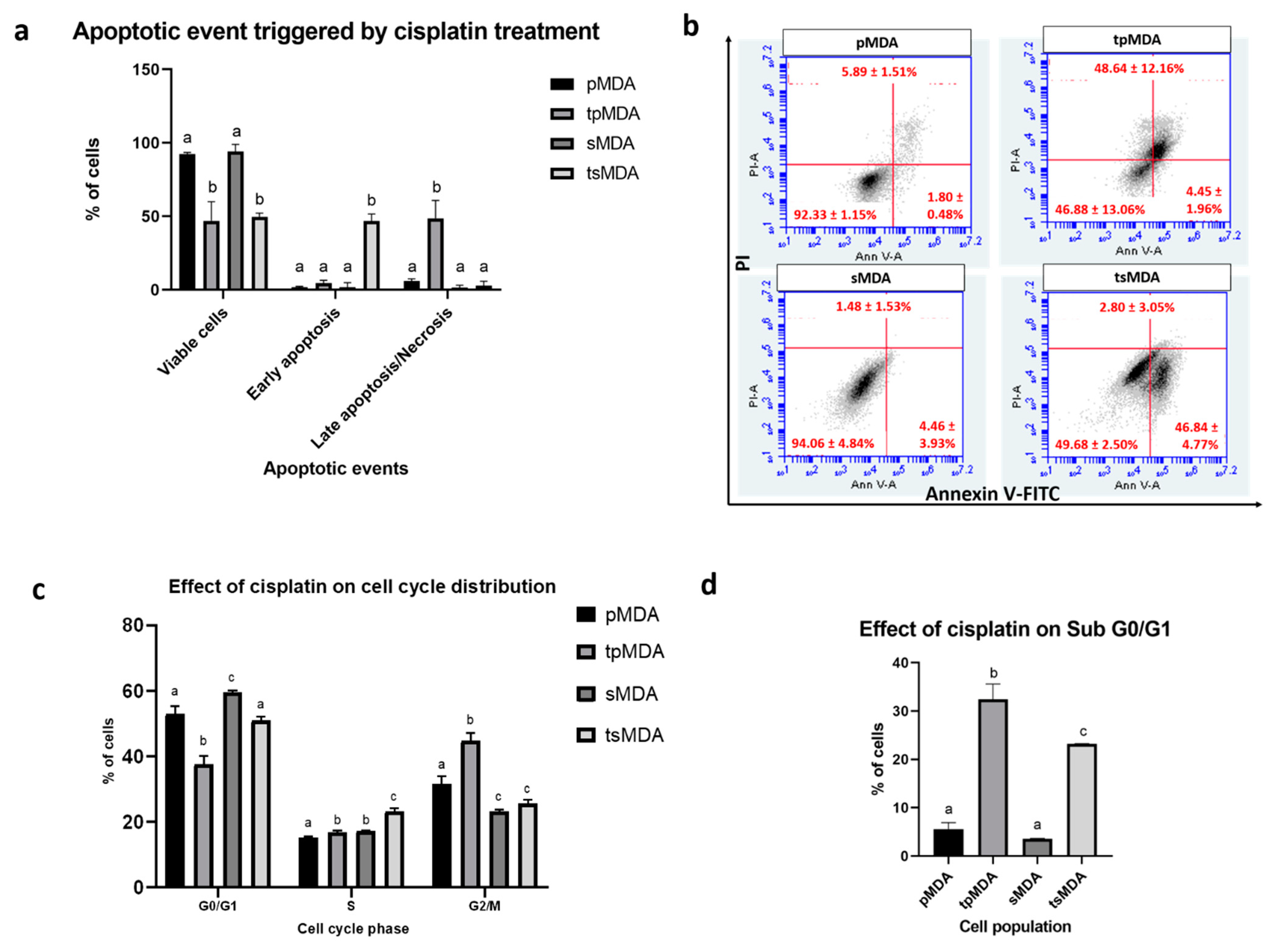
2.4. Sorted MDA-MB-231 Is More Resistant to Cisplatin-Induced Apoptosis
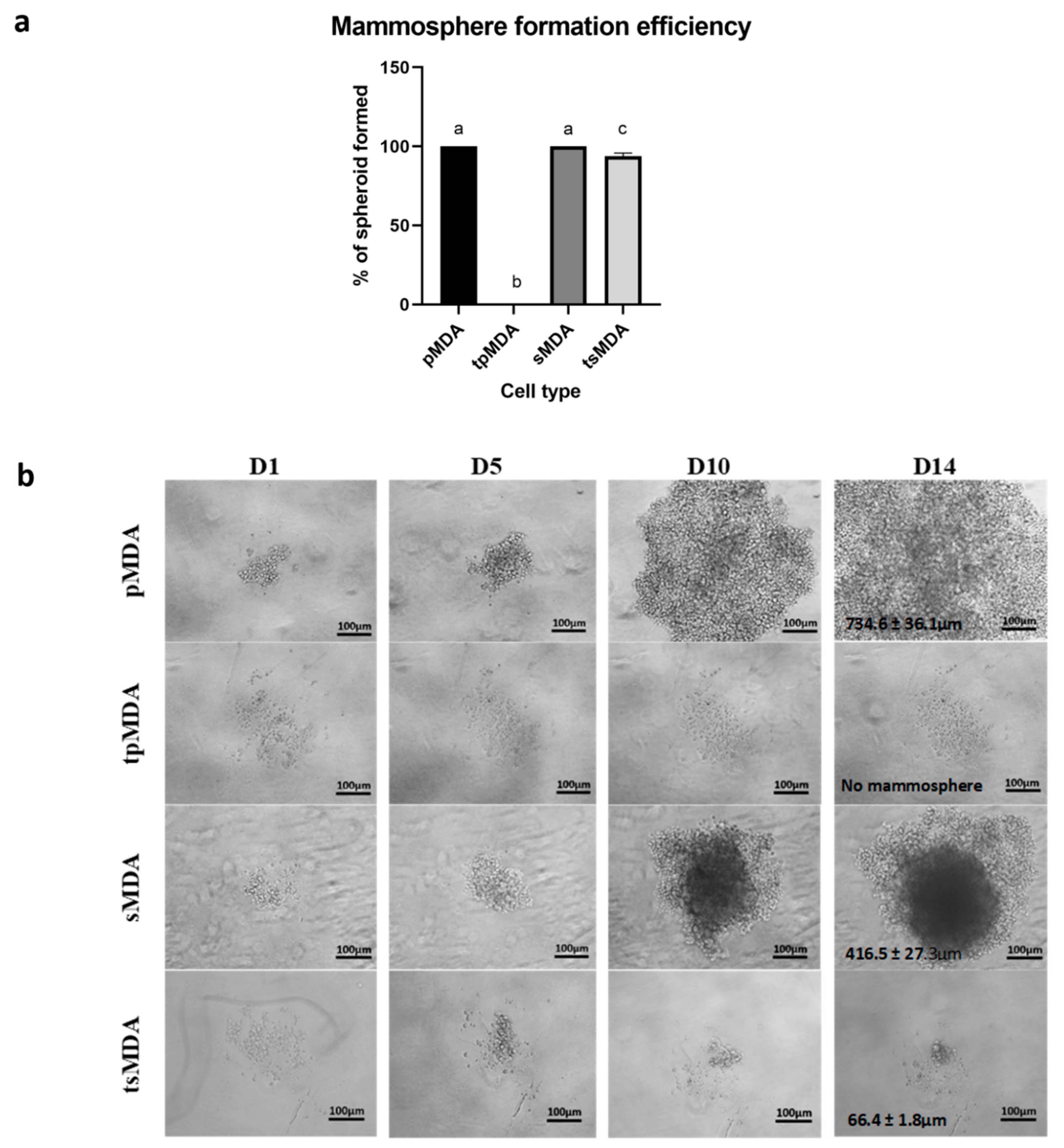
2.5. Sorted MDA-MB-231 Retains Remarkably Higher Self-Renewability after Treatment
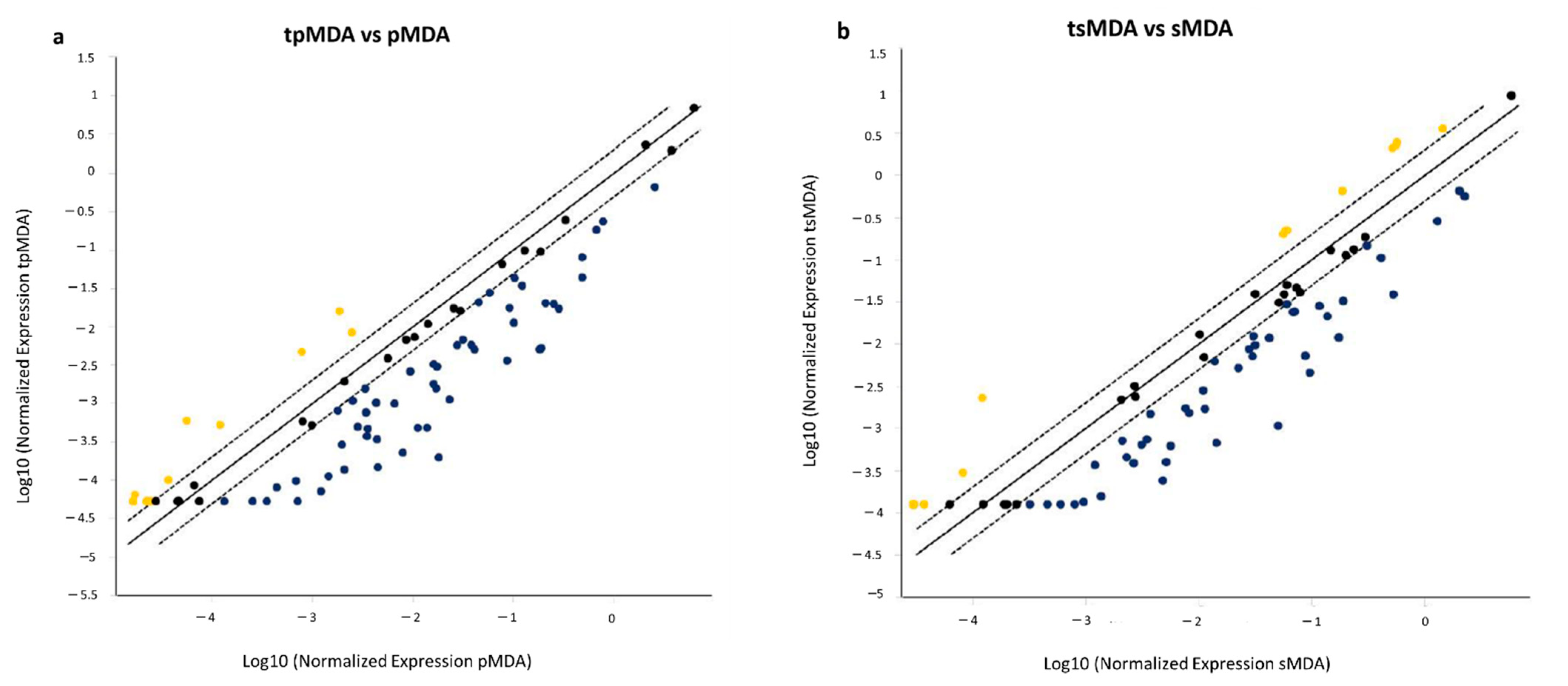
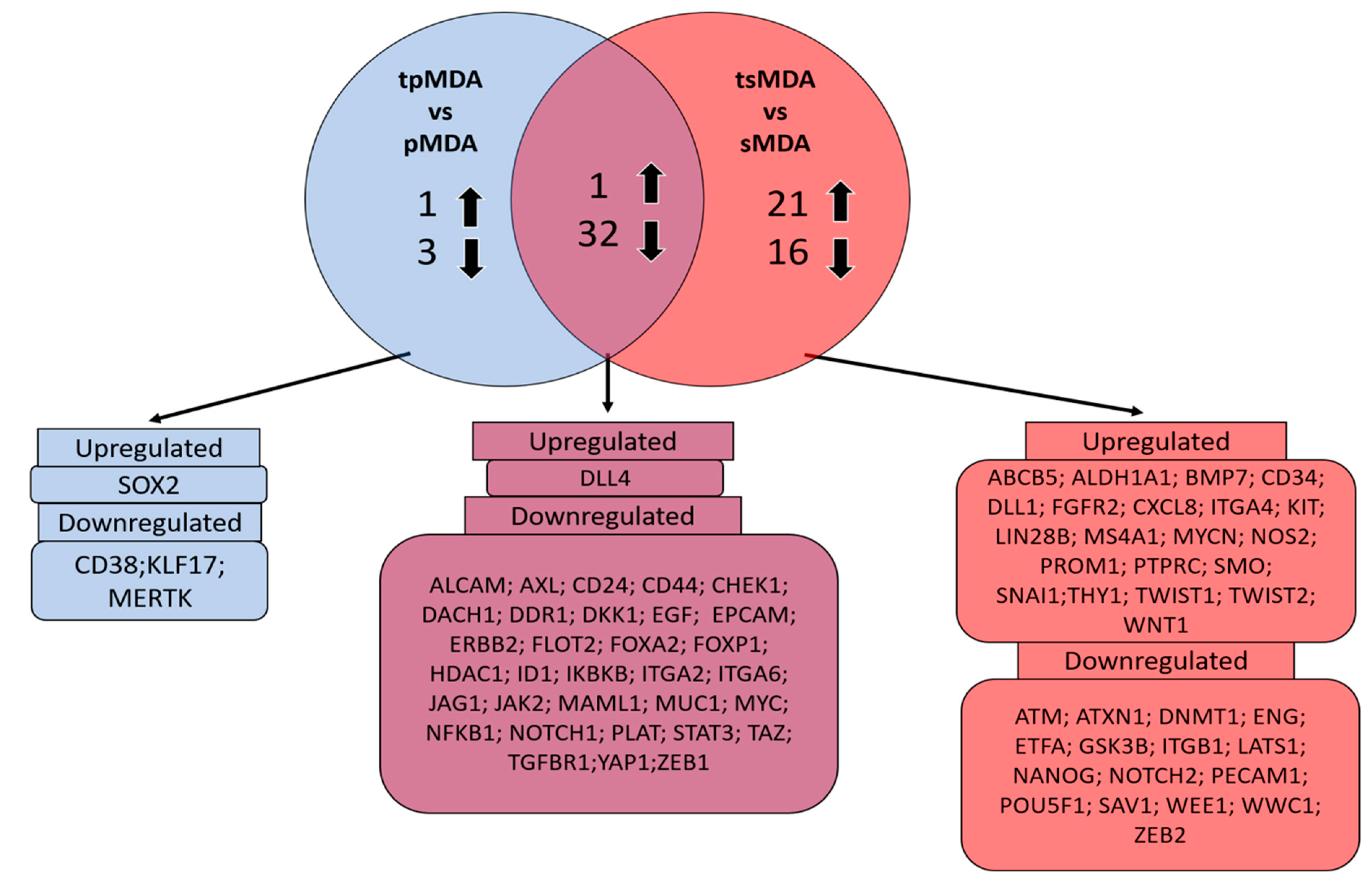
2.6. Dysregulation of Cancer Stem Cell-Correlated Putative Target Genes in MDA-MB-231 Parental and Sorted MDA-MB-231 Induced by Cisplatin Treatment
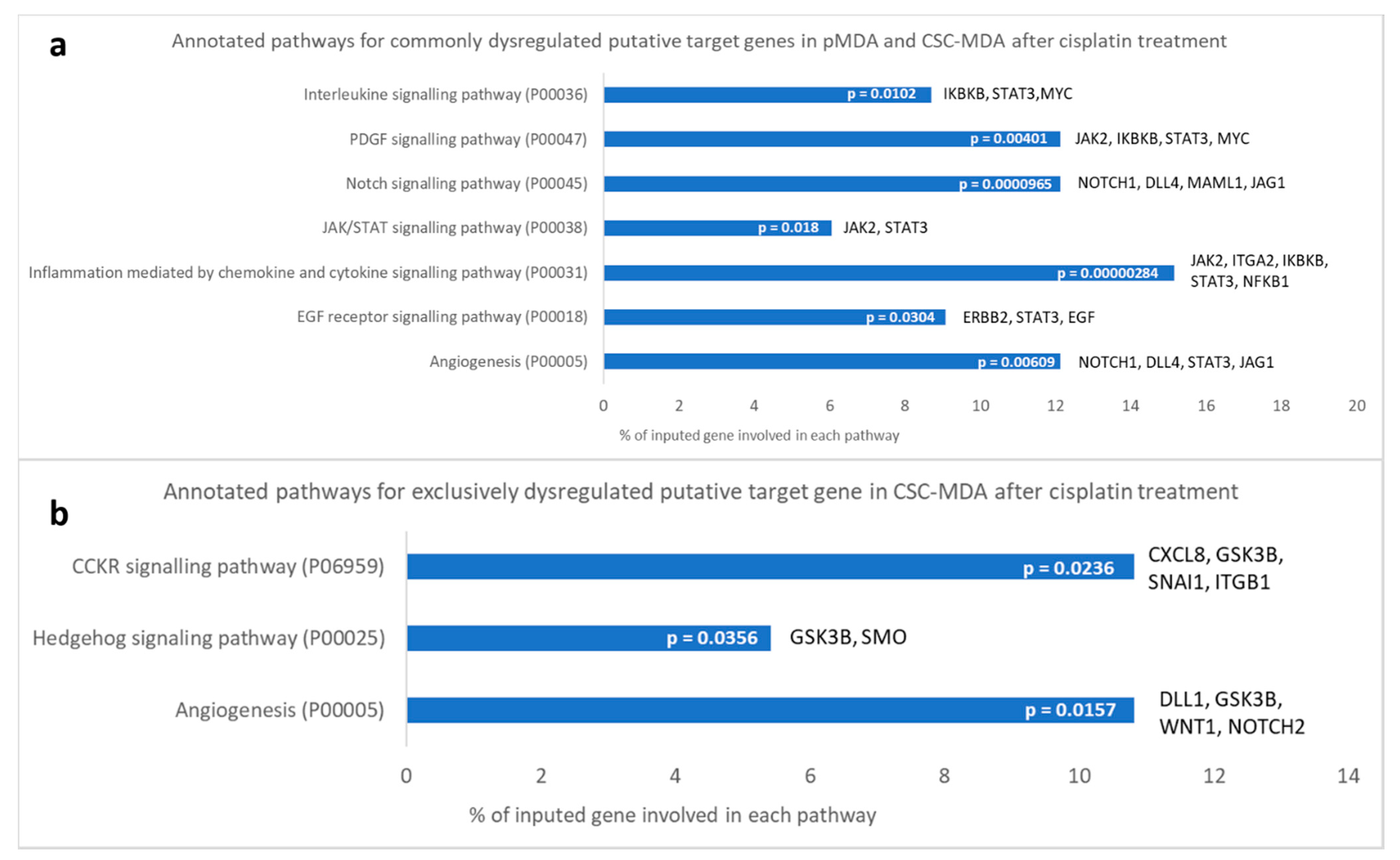
2.7. Dysregulated Cancer Stem Cell-Correlated Putative Target Genes and Corresponding Signaling Pathways Induced by Cisplatin Treatment
3. Discussion
4. Materials and Methods
4.1. Cell Culture for Triple-Negative Breast Cancer Cells
4.2. Isolation of Triple-Negative Breast Cancer Stem Cells via Magnetic-Activated Column Sorting
4.3. Assessment for the Cytotoxicity of Cisplatin
4.4. Acquirement of Cisplatin-Treated Triple-Negative Breast Cancer Subpopulations
- pMDA control
- Treated pMDA (tpMDA)
- sMDA control
- Treated sMDA (tsMDA)
4.5. Immuno-Phenotyping for Expression of Breast Cancer Stem Cell Surface Markers CD44/CD24 via Flow Cytometry
4.6. Immuno-Phenotyping for Expression of Breast Cancer Stem Cell Intracellular Marker ALDH1A1 via Flow Cytometry
4.7. Immunofluorescence Microscopy Detection of Breast Cancer Markers CD44/CD24 and ALDH1A1
4.8. RealTime-Glo™ MT Cell Viability Assay
4.9. FITC-Annexin V/PI Staining Apoptosis Assay
4.10. Cell Cycle Analysis
4.11. Mammosphere Formation Efficiency Test
4.12. Quantitative Real-Time PCR Array for Cancer Stem Cell-Correlated Putative Target Genes
4.13. Gene Ontology Enrichment Pathway Analysis of Dysregulated Putative Target Genes
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liao, H.-Y.; Zhang, W.-W.; Sun, J.-Y.; Li, F.-Y.; He, Z.-Y.; Wu, S.-G. The Clinicopathological Features and Survival Outcomes of Different Histological Subtypes in Triple-negative Breast Cancer. J. Cancer 2018, 9, 296–303. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; Poornima, R.; Challa, V.R.; Goud, Y.G.B. Triple Negative Breast Cancer-Our Experience and Review. Indian J. Surg. Oncol. 2012, 3, 12–16. [Google Scholar] [CrossRef]
- Tan, G.H.; Taib, N.A.; Choo, W.Y.; Teo, S.H.; Yip, C.H. Clinical characteristics of triple-negative breast cancer: Experience in an Asian developing country. Asian Pac. J. Cancer Prev. 2009, 10, 395–398. [Google Scholar]
- Kanapathy Pillai, S.K.; Tay, A.; Nair, S.; Leong, C.-O. Triple-negative breast cancer is associated with EGFR, CK5/6 and c-KIT expression in Malaysian women. BMC Clin. Pathol. 2012, 12, 18. [Google Scholar] [CrossRef]
- Abdul Aziz, A.A.; Md Salleh, M.S.; Ankathil, R. Clinicopathological and Prognostic Characteristics of Malaysian Triple Negative Breast Cancer Patients Undergoing TAC Chemotherapy Regimen. Int. J. Breast Cancer 2020, 2020, 8424365. [Google Scholar] [CrossRef]
- Seyfried, T.N.; Huysentruyt, L.C. On the origin of cancer metastasis. Crit. Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef]
- André, F.; Zielinski, C.C. Optimal strategies for the treatment of metastatic triple-negative breast cancer with currently approved agents. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2012, 23, vi46–vi51. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.D.; Tung, N.M.; Isakoff, S.J.; Golshan, M.; Richardson, A.; Corben, A.D.; Smith, B.L.; Gelman, R.; Winer, E.P.; Garber, J.E. Neoadjuvant cisplatin and bevacizumab in triple negative breast cancer (TNBC): Safety and efficacy. J. Clin. Oncol. 2009, 27, 551. [Google Scholar] [CrossRef]
- Goel, A.K.; Nandy, M.; Sharma, G. Cisplatin as neoadjuvant chemotherapy in triple negative breast cancer: Exciting early results. Indian J. Med. Paediatr. Oncol. 2010, 31, 76–78. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, J.; Wang, Z.; Hu, X.; Wang, B.; Wang, L.; Yang, W.; Liu, Y.; Liu, G.; Di, G.; Hu, Z.; et al. Cisplatin and gemcitabine as the first line therapy in metastatic triple negative breast cancer. Int. J. Cancer 2015, 136, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Rabik, C.A.; Dolan, M.E. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat. Rev. 2007, 33, 9–23. [Google Scholar] [CrossRef]
- Wahba, H.A.; El-Hadaad, H.A. Current approaches in treatment of triple-negative breast cancer. Cancer Biol. Med. 2015, 12, 106–116. [Google Scholar]
- Leong, C.-O.; Vidnovic, N.; DeYoung, M.P.; Sgroi, D.; Ellisen, L.W. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J. Clin. Investig. 2007, 117, 1370–1380. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.-C.; Zhang, J.; Xu, B.-H.; Cai, L.; Ragaz, J.; Wang, Z.-H.; Wang, B.-Y.; Teng, Y.-E.; Tong, Z.-S.; Pan, Y.-Y.; et al. Cisplatin plus gemcitabine versus paclitaxel plus gemcitabine as first-line therapy for metastatic triple-negative breast cancer (CBCSG006): A randomised, open-label, multicentre, phase 3 trial. Lancet Oncol. 2015, 16, 436–446. [Google Scholar] [CrossRef]
- Baselga, J.; Stemmer, S.; Pego, A.; Chan, A.; Goeminne, J.-C.; Graas, M.-P.; Kennedy, J.; Ciruelos Gil, E.M.; Zubel, A.; Groos, J.; et al. Abstract PD01-01: Cetuximab + Cisplatin in Estrogen Receptor-Negative, Progesterone Receptor-Negative, HER2-Negative (Triple-Negative) Metastatic Breast Cancer: Results of the Randomized Phase II BALI-1 Trial. Cancer Res. 2010, 70, PD01. [Google Scholar]
- Byrski, T.; Huzarski, T.; Dent, R.; Gronwald, J.; Zuziak, D.; Cybulski, C.; Kladny, J.; Gorski, B.; Lubinski, J.; Narod, S.A. Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res. Treat. 2009, 115, 359–363. [Google Scholar] [CrossRef]
- Petrelli, F.; Barni, S.; Bregni, G.; de Braud, F.; Di Cosimo, S. Platinum salts in advanced breast cancer: A systematic review and meta-analysis of randomized clinical trials. Breast Cancer Res. Treat. 2016, 160, 425–437. [Google Scholar] [CrossRef]
- Cancello, G.; Bagnardi, V.; Sangalli, C.; Montagna, E.; Dellapasqua, S.; Sporchia, A.; Iorfida, M.; Viale, G.; Barberis, M.; Veronesi, P.; et al. Phase II Study with Epirubicin, Cisplatin, and Infusional Fluorouracil Followed by Weekly Paclitaxel with Metronomic Cyclophosphamide as a Preoperative Treatment of Triple-Negative Breast Cancer. Clin. Breast Cancer 2015, 15, 259–265. [Google Scholar] [CrossRef]
- Somali, I.; Alacacioglu, A.; Tarhan, M.O.; Meydan, N.; Erten, C.; Usalp, S.; Yilmaz, U. Cisplatin plus gemcitabine chemotherapy in taxane/anthracycline-resistant metastatic breast cancer. Chemotherapy 2009, 55, 155–160. [Google Scholar] [CrossRef]
- Telli, M.L.; Timms, K.M.; Reid, J.; Hennessy, B.; Mills, G.B.; Jensen, K.C.; Szallasi, Z.; Barry, W.T.; Winer, E.P.; Tung, N.M.; et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin. Cancer Res. 2016, 22, 3764–3773. [Google Scholar] [CrossRef]
- Li, Q.; Li, Q.; Zhang, P.; Yuan, P.; Wang, J.; Ma, F.; Luo, Y.; Fan, Y.; Cai, R.; Xu, B. A phase II study of capecitabine plus cisplatin in metastatic triple-negative breast cancer patients pretreated with anthracyclines and taxanes. Cancer Biol. Ther. 2015, 16, 1746–1753. [Google Scholar] [CrossRef]
- Shafee, N.; Smith, C.R.; Wei, S.; Kim, Y.; Mills, G.B.; Hortobagyi, G.N.; Stanbridge, E.J.; Lee, E.Y.-H.P. Cancer Stem Cells Contribute to Cisplatin Resistance in Brca1/p53–Mediated Mouse Mammary Tumors. Cancer Res. 2008, 68, 3243–3250. [Google Scholar] [CrossRef]
- Jin, F.; Zhao, L.; Zhao, H.-Y.; Guo, S.-G.; Feng, J.; Jiang, X.-B.; Zhang, S.-L.; Wei, Y.-J.; Fu, R.; Zhao, J.-S. Comparison between cells and cancer stem-like cells isolated from glioblastoma and astrocytoma on expression of anti-apoptotic and multidrug resistance–associated protein genes. Neuroscience 2008, 154, 541–550. [Google Scholar] [CrossRef]
- Carter, B.Z.; Qiu, Y.; Huang, X.; Diao, L.; Zhang, N.; Coombes, K.R.; Mak, D.H.; Konopleva, M.; Cortes, J.; Kantarjian, H.M.; et al. Survivin is highly expressed in CD34(+)38(−) leukemic stem/progenitor cells and predicts poor clinical outcomes in AML. Blood 2012, 120, 173–180. [Google Scholar] [CrossRef]
- Liu, G.; Yuan, X.; Zeng, Z.; Tunici, P.; Ng, H.; Abdulkadir, I.R.; Lu, L.; Irvin, D.; Black, K.L.; Yu, J.S. Analysis of gene expression and chemoresistance of CD133(+) cancer stem cells in glioblastoma. Mol. Cancer 2006, 5, 67. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, L.N.; Chow, E.K.-H. Mechanisms of chemoresistance in cancer stem cells. Clin. Transl. Med. 2013, 2, 1–9. [Google Scholar] [CrossRef]
- Velasco-Velázquez, M.A.; Popov, V.M.; Lisanti, M.P.; Pestell, R.G. The Role of Breast Cancer Stem Cells in Metastasis and Therapeutic Implications. Am. J. Pathol. 2011, 179, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Abraham, B.K.; Fritz, P.; McClellan, M.; Hauptvogel, P.; Athelogou, M.; Brauch, H. Prevalence of CD44+/CD24-/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin. Cancer Res. 2005, 11, 1154–1159. [Google Scholar] [PubMed]
- Liu, H.; Patel, M.R.; Prescher, J.A.; Patsialou, A.; Qian, D.; Lin, J.; Wen, S.; Chang, Y.-F.; Bachmann, M.H.; Shimono, Y.; et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc. Natl. Acad. Sci. USA 2010, 107, 18115–18120. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Tang, H.; Xiao, Q.; He, M.; Zhao, L.; Fu, Y.; Wu, H.; Yu, Z.; Jiang, Q.; Yan, Y.; et al. The Hedgehog signaling pathway is associated with poor prognosis in breast cancer patients with the CD44+/CD24 phenotype. Mol. Med. Rep. 2016, 14, 5261–5270. [Google Scholar] [CrossRef] [PubMed]
- Ghebeh, H.; Sleiman, G.M.; Manogaran, P.S.; Al-Mazrou, A.; Barhoush, E.; Al-Mohanna, F.H.; Tulbah, A.; Al-Faqeeh, K.; Adra, C.N. Profiling of normal and malignant breast tissue show CD44high/CD24lowphenotype as a predominant stem/progenitor marker when used in combination with Ep-CAM/CD49f markers. BMC Cancer 2013, 13, 289. [Google Scholar] [CrossRef]
- Ciccone, V.; Terzuoli, E.; Donnini, S.; Giachetti, A.; Morbidelli, L.; Ziche, M. Stemness marker ALDH1A1 promotes tumor angiogenesis via retinoic acid/HIF-1α/VEGF signalling in MCF-7 breast cancer cells. J. Exp. Clin. Cancer Res. 2018, 37, 311. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Guo, F.; Albers, A.E.; Sehouli, J.; Kaufmann, A.M. Disulfiram modulates ROS accumulation and overcomes synergistically cisplatin resistance in breast cancer cell lines. Biomed. Pharm. 2019, 113, 108727. [Google Scholar] [CrossRef]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Krishnamurthy, S. Cellular responses to Cisplatin-induced DNA damage. J. Nucleic Acids 2010, 2010, 201367. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef]
- Peitsch, M.C.; Müller, C.; Tschopp, J. DNA fragmentation during apoptosis is caused by frequent single-strand cuts. Nucleic Acids Res. 1993, 21, 4206–4209. [Google Scholar] [CrossRef]
- Demarcq, C.; Bunch, R.T.; Creswell, D.; Eastman, A. The role of cell cycle progression in cisplatin-induced apoptosis in Chinese hamster ovary cells. Cell Growth Differ. Publ. Am. Assoc. Cancer Res. 1994, 5, 983–993. [Google Scholar]
- Sorenson, C.M.; Eastman, A. Mechanism of Cisdiamminedichloroplatinum(II)-induced Cytotoxicity: Role of G2 Arrest and DNA Double-Strand Breaks. Cancer Res. 1988, 48, 4484–4488. [Google Scholar]
- Zheng, H.; Shao, F.; Martin, S.; Xu, X.; Deng, C.-X. WEE1 inhibition targets cell cycle checkpoints for triple negative breast cancers to overcome cisplatin resistance. Sci. Rep. 2017, 7, 43517. [Google Scholar] [CrossRef]
- Croker, A.K.; Allan, A.L. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44(+) human breast cancer cells. Breast Cancer Res. Treat. 2012, 133, 75–87. [Google Scholar] [CrossRef]
- Kida, K.; Ishikawa, T.; Yamada, A.; Shimada, K.; Narui, K.; Sugae, S.; Shimizu, D.; Tanabe, M.; Sasaki, T.; Ichikawa, Y.; et al. Effect of ALDH1 on prognosis and chemoresistance by breast cancer subtype. Breast Cancer Res. Treat. 2016, 156, 261–269. [Google Scholar] [CrossRef]
- Mukherjee, P.; Gupta, A.; Chattopadhyay, D.; Chatterji, U. Modulation of SOX2 expression delineates an end-point for paclitaxel-effectiveness in breast cancer stem cells. Sci. Rep. 2017, 7, 9170. [Google Scholar] [CrossRef] [PubMed]
- Sládek, N.E.; Kollander, R.; Sreerama, L.; Kiang, D.T. Cellular levels of aldehyde dehydrogenases (ALDH1A1 and ALDH3A1) as predictors of therapeutic responses to cyclophosphamide-based chemotherapy of breast cancer: A retrospective study. Rational individualization of oxazaphosphorine-based cancer chemothera. Cancer Chemother. Pharmacol. 2002, 49, 309–321. [Google Scholar] [PubMed]
- Tanei, T.; Morimoto, K.; Shimazu, K.; Kim, S.J.; Tanji, Y.; Taguchi, T.; Tamaki, Y.; Noguchi, S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin. Cancer Res. 2009, 15, 4234–4241. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.-K.; Wang, Z.; Fong, C.-C.; Liu, D.; Yip, T.-C.; Au, S.-K.; Zhu, G.; Yang, M. Chemoresistant lung cancer stem cells display high DNA repair capability to remove cisplatin-induced DNA damage. Br. J. Pharmacol. 2017, 174, 302–313. [Google Scholar] [CrossRef]
- Mueller, S.; Schittenhelm, M.; Honecker, F.; Malenke, E.; Lauber, K.; Wesselborg, S.; Hartmann, J.T.; Bokemeyer, C.; Mayer, F. Cell-cycle progression and response of germ cell tumors to cisplatin in vitro. Int. J. Oncol. 2006, 29, 471–479. [Google Scholar] [CrossRef]
- Prabhakaran, P.; Hassiotou, F.; Blancafort, P.; Filgueira, L. Cisplatin Induces Differentiation of Breast Cancer Cells. Front. Oncol. 2013, 3, 134. [Google Scholar] [CrossRef]
- Deng, X.; Apple, S.; Zhao, H.; Song, J.; Lee, M.; Luo, W.; Wu, X.; Chung, D.; Pietras, R.J.; Chang, H.R. CD24 Expression and differential resistance to chemotherapy in triple-negative breast cancer. Oncotarget 2017, 8, 38294. [Google Scholar] [CrossRef]
- Croker, A.K.; Rodriguez-Torres, M.; Xia, Y.; Pardhan, S.; Leong, H.S.; Lewis, J.D.; Allan, A.L. Differential Functional Roles of ALDH1A1 and ALDH1A3 in Mediating Metastatic Behavior and Therapy Resistance of Human Breast Cancer Cells. Int. J. Mol. Sci. 2017, 18, 2039. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ma, H.; Zhang, J.; Zhu, L.; Wang, C.; Yang, Y. Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci. Rep. 2017, 7, 13856. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Lee, H.E.; Li, H.; Shipitsin, M.; Gelman, R.; Polyak, K. Heterogeneity for Stem Cell–Related Markers According to Tumor Subtype and Histologic Stage in Breast Cancer. Clin. Cancer Res. 2010, 16, 876–887. [Google Scholar] [CrossRef] [PubMed]
- Calcagno, A.M.; Salcido, C.D.; Gillet, J.-P.; Wu, C.-P.; Fostel, J.M.; Mumau, M.D.; Gottesman, M.M.; Varticovski, L.; Ambudkar, S. V Prolonged drug selection of breast cancer cells and enrichment of cancer stem cell characteristics. J. Natl. Cancer Inst. 2010, 102, 1637–1652. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Shen, S.; Zhou, Y.; Mao, F.; Lin, Y.; Guan, J.; Xu, Y.; Zhang, S.; Liu, X.; Sun, Q. NOTCH1 is a poor prognostic factor for breast cancer and is associated with breast cancer stem cells. Onco. Targets Ther. 2016, 9, 6865. [Google Scholar] [CrossRef]
- Simoes, B.M.; O’Brien, C.S.; Eyre, R.; Silva, A.; Yu, L.; Sarmiento-Castro, A.; Alferez, D.G.; Spence, K.; Santiago-Gomez, A.; Chemi, F.; et al. Anti-estrogen Resistance in Human Breast Tumors Is Driven by JAG1-NOTCH4-Dependent Cancer Stem Cell Activity. Cell Rep. 2015, 12, 1968–1977. [Google Scholar] [CrossRef]
- Zetter, B.R. Angiogenesis and tumor metastasis. Annu. Rev. Med. 1998, 49, 407–424. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Li, S.; Chepeha, D.B.; Giordano, T.J.; Li, J.; Zhang, H.; Polverini, P.J.; Nor, J.; Kitajewski, J.; Wang, C.-Y. Crosstalk between tumor and endothelial cells promotes tumor angiogenesis by MAPK activation of Notch signaling. Cancer Cell 2005, 8, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Benedito, R.; Roca, C.; Sörensen, I.; Adams, S.; Gossler, A.; Fruttiger, M.; Adams, R.H. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell 2009, 137, 1124–1135. [Google Scholar] [CrossRef]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.I.; He, X.; Perou, C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010, 12, 1–18. [Google Scholar] [CrossRef]
- Casciano, J.C.; Perry, C.; Cohen-Nowak, A.J.; Miller, K.D.; Vande Voorde, J.; Zhang, Q.; Chalmers, S.; Sandison, M.E.; Liu, Q.; Hedley, A.; et al. MYC regulates fatty acid metabolism through a multigenic program in claudin-low triple negative breast cancer. Br. J. Cancer 2020, 122, 868–884. [Google Scholar] [CrossRef]
- Wang, P.-S.; Chou, C.-H.; Lin, C.-H.; Yao, Y.-C.; Cheng, H.-C.; Li, H.-Y.; Chuang, Y.-C.; Yang, C.-N.; Ger, L.-P.; Chen, Y.-C.; et al. A novel long non-coding RNA linc-ZNF469-3 promotes lung metastasis through miR-574-5p-ZEB1 axis in triple negative breast cancer. Oncogene 2018, 37, 4662–4678. [Google Scholar] [CrossRef] [PubMed]
- Ariad, S.; Seymour, L.; Bezwoda, W.R. Platelet-derived growth factor (PDGF) in plasma of breast cancer patients: Orrelation with stage and rate of progression. Breast Cancer Res. Treat. 1991, 20, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Seymour, L.; Dajee, D.; Bezwoda, W.R. Tissue platelet derived-growth factor (PDGF) predicts for shortened survival and treatment failure in advanced breast cancer. Breast Cancer Res. Treat. 1993, 26, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Jansson, S.; Aaltonen, K.; Bendahl, P.-O.; Falck, A.-K.; Karlsson, M.; Pietras, K.; Rydén, L. The PDGF pathway in breast cancer is linked to tumour aggressiveness, triple-negative subtype and early recurrence. Breast Cancer Res. Treat. 2018, 169, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Wang, W.; Zhou, J.; Chang, M.; Peng, W.; Zhang, G.; Li, J.; Li, H.; Bai, C. Knockdown of PLAT enhances the anticancer effect of gefitinib in non-small cell lung cancer. J. Thorac. Dis. 2020, 12, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Duan, W.; Kang, L.; Mao, J.; Yu, X.; Fan, S.; Li, L.; Tao, Y. Smoothened activates breast cancer stem-like cell and promotes tumorigenesis and metastasis of breast cancer. Biomed. Pharm. 2014, 68, 1099–1104. [Google Scholar] [CrossRef]
- Wang, Y.; Lam, J.B.; Lam, K.S.L.; Liu, J.; Lam, M.C.; Hoo, R.L.C.; Wu, D.; Cooper, G.J.S.; Xu, A. Adiponectin modulates the glycogen synthase kinase-3beta/beta-catenin signaling pathway and attenuates mammary tumorigenesis of MDA-MB-231 cells in nude mice. Cancer Res. 2006, 66, 11462–11470. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Xu, L.; Bandyopadhyay, S.; Sethi, S.; Reddy, K.B. Cisplatin and TRAIL enhance breast cancer stem cell death. Int. J. Oncol. 2011, 39, 891–898. [Google Scholar] [PubMed]
- Jang, G.-B.; Kim, J.-Y.; Cho, S.-D.; Park, K.-S.; Jung, J.-Y.; Lee, H.-Y.; Hong, I.-S.; Nam, J.S. Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci. Rep. 2015, 5, 12465. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, C.F.; Urs, S.; Cinelli, C.; Lincoln, A.; Nadeau, R.J.; León, R.; Toher, J.; Mouta-Bellum, C.; Friesel, R.E.; Liaw, L. Notch2 signaling induces apoptosis and inhibits human MDA-MB-231 xenograft growth. Am. J. Pathol. 2007, 171, 1023–1036. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koh, M.Z.; Ho, W.Y.; Yeap, S.K.; Ali, N.M.; Boo, L.; Alitheen, N.B. Regulation of Cellular and Cancer Stem Cell-Related Putative Gene Expression of Parental and CD44+CD24− Sorted MDA-MB-231 Cells by Cisplatin. Pharmaceuticals 2021, 14, 391. https://doi.org/10.3390/ph14050391
Koh MZ, Ho WY, Yeap SK, Ali NM, Boo L, Alitheen NB. Regulation of Cellular and Cancer Stem Cell-Related Putative Gene Expression of Parental and CD44+CD24− Sorted MDA-MB-231 Cells by Cisplatin. Pharmaceuticals. 2021; 14(5):391. https://doi.org/10.3390/ph14050391
Chicago/Turabian StyleKoh, May Zie, Wan Yong Ho, Swee Keong Yeap, Norlaily Mohd Ali, Lily Boo, and Noorjahan Banu Alitheen. 2021. "Regulation of Cellular and Cancer Stem Cell-Related Putative Gene Expression of Parental and CD44+CD24− Sorted MDA-MB-231 Cells by Cisplatin" Pharmaceuticals 14, no. 5: 391. https://doi.org/10.3390/ph14050391
APA StyleKoh, M. Z., Ho, W. Y., Yeap, S. K., Ali, N. M., Boo, L., & Alitheen, N. B. (2021). Regulation of Cellular and Cancer Stem Cell-Related Putative Gene Expression of Parental and CD44+CD24− Sorted MDA-MB-231 Cells by Cisplatin. Pharmaceuticals, 14(5), 391. https://doi.org/10.3390/ph14050391





