A Multi-Objective Approach for Anti-Osteosarcoma Cancer Agents Discovery through Drug Repurposing
Abstract
1. Introduction
2. Results
2.1. Datasets and Molecular Descriptors
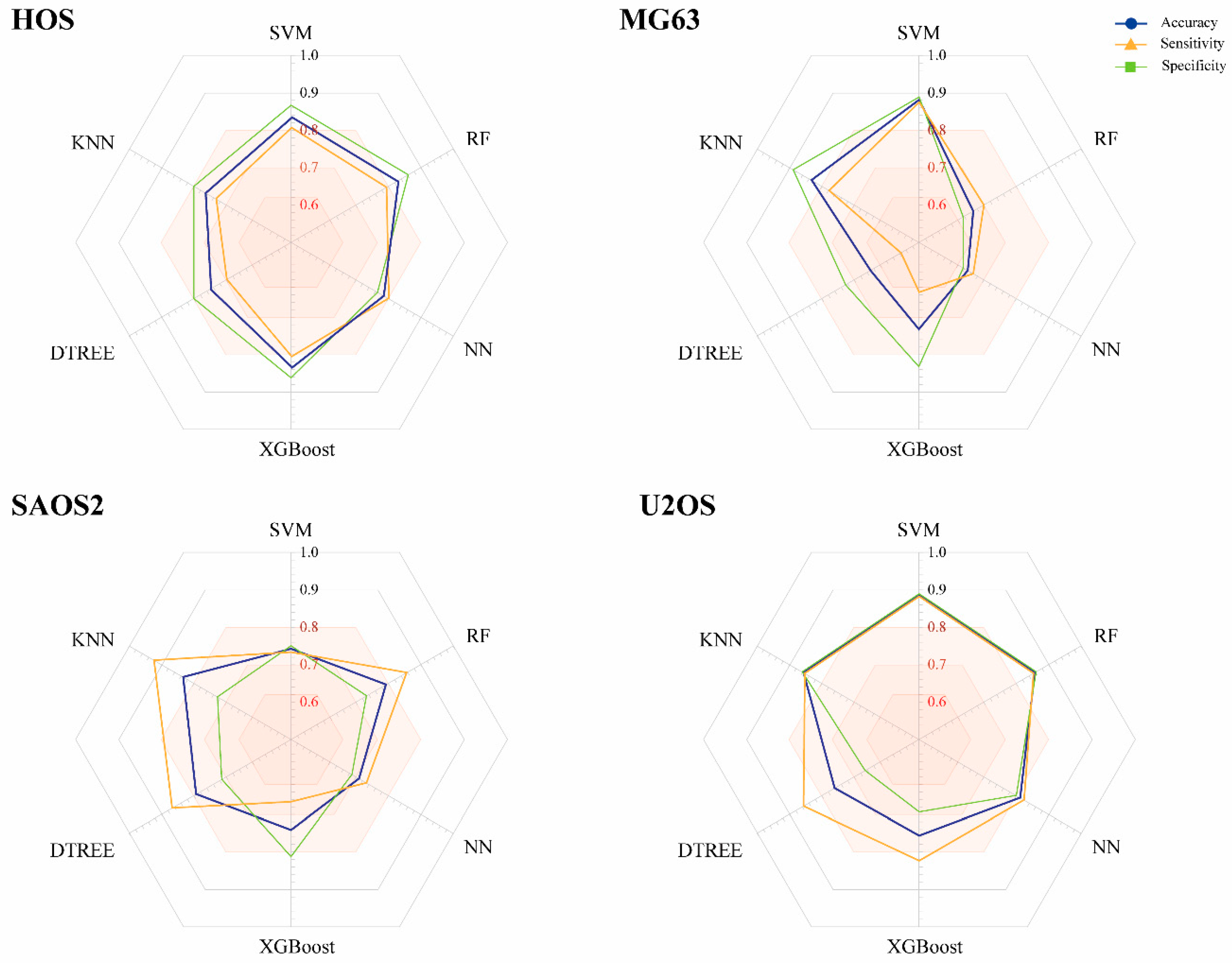
2.2. Construction of Models
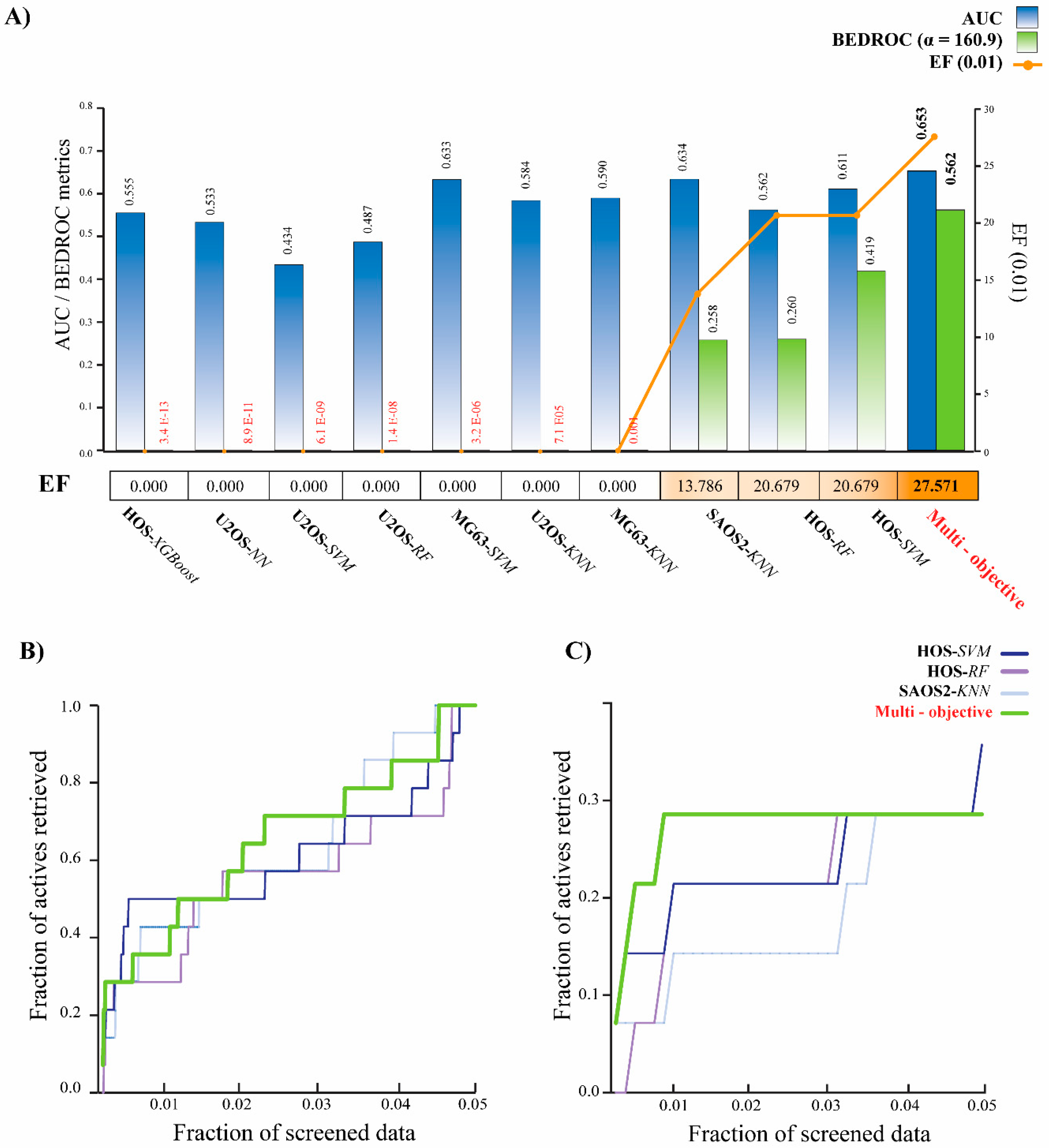
2.3. Multi-Objective Model Assessment and Virtual Screening
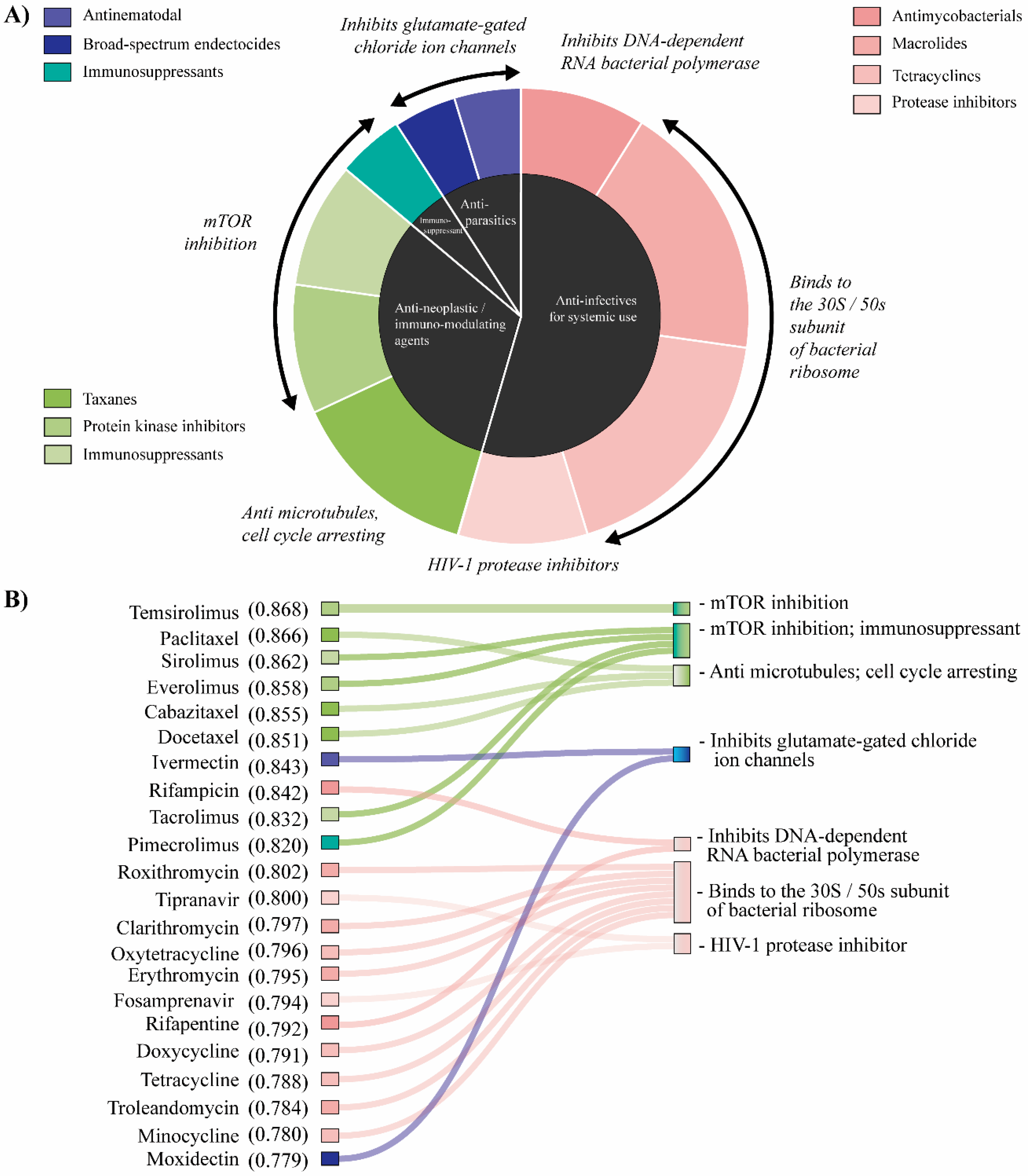
2.4. Analysis of Repurposed Drugs
3. Discussion
4. Materials and Methods
4.1. Preprocessing Datasets and Molecular Descriptors
4.2. Machine Learning Models and Quality Evaluation
4.3. Multi-Objective Model Assembly and Virtual Screening
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ottaviani, G.; Jaffe, N. The epidemiology of osteosarcoma. Cancer Treat. Res. 2009, 152, 3–13. [Google Scholar]
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology (NCC Guidelines)—Bone Cancer. Available online: https://www.nccn.org/ (accessed on 4 March 2020).
- Misaghi, A.; Goldin, A.; Awad, M.; Kulidjian, A.A. Osteosarcoma: A comprehensive review. SICOT J. 2018, 4, 12. [Google Scholar] [CrossRef]
- Biermann, J.S.; Chow, W.; Reed, D.R.; Lucas, D.; Adkins, D.R.; Agulnik, M.; Benjamin, R.S.; Brigman, B.; Budd, G.T.; Curry, W.T.; et al. NCCN Guidelines Insights: Bone Cancer, Version 2.2017. J. Natl. Compr. Cancer Netw. 2017, 15, 155–167. [Google Scholar] [CrossRef]
- de Azevedo, J.W.V.; de Medeiros Fernandes, T.A.A.; Fernandes, J.V., Jr.; de Azevedo, J.C.V.; Lanza, D.C.F.; Bezerra, C.M.; Andrade, V.S.; de Araujo, J.M.G.; Fernandes, J.V. Biology and pathogenesis of human osteosarcoma. Oncol. Lett. 2020, 19, 1099–1116. [Google Scholar]
- Xin, S.; Wei, G. Prognostic factors in osteosarcoma: A study level meta-analysis and systematic review of current practice. J. Bone Oncol. 2020, 21, 100281. [Google Scholar] [CrossRef]
- Marko, T.A.; Diessner, B.J.; Spector, L.G. Prevalence of Metastasis at Diagnosis of Osteosarcoma: An International Comparison. Pediatr. Blood Cancer 2016, 63, 1006–1011. [Google Scholar] [CrossRef]
- Duchman, K.R.; Gao, Y.; Miller, B.J. Prognostic factors for survival in patients with high-grade osteosarcoma using the Surveillance, Epidemiology, and End Results (SEER) Program database. Cancer Epidemiol. 2015, 39, 593–599. [Google Scholar] [CrossRef]
- Song, K.; Song, J.; Lin, K.; Chen, F.; Ma, X.; Jiang, J.; Li, F. Survival analysis of patients with metastatic osteosarcoma: A Surveillance, Epidemiology, and End Results population-based study. Int. Orthop. 2019, 43, 1983–1991. [Google Scholar] [CrossRef]
- Taran, S.J.; Taran, R.; Malipatil, N.B. Pediatric Osteosarcoma: An Updated Review. Indian J. Med. Paediatr. Oncol. 2017, 38, 33–43. [Google Scholar] [CrossRef]
- Vos, H.I.; Coenen, M.J.; Guchelaar, H.J.; Te Loo, D.M. The role of pharmacogenetics in the treatment of osteosarcoma. Drug Discov. Today 2016, 21, 1775–1786. [Google Scholar] [CrossRef]
- Durfee, R.A.; Mohammed, M.; Luu, H.H. Review of Osteosarcoma and Current Management. Rheumatol. Ther. 2016, 3, 221–243. [Google Scholar] [CrossRef]
- Omer, N.; Le Deley, M.C.; Piperno-Neumann, S.; Marec-Berard, P.; Italiano, A.; Corradini, N.; Bellera, C.; Brugieres, L.; Gaspar, N. Phase-II trials in osteosarcoma recurrences: A systematic review of past experience. Eur. J. Cancer 2017, 75, 98–108. [Google Scholar] [CrossRef]
- Harrison, D.J.; Geller, D.S.; Gill, J.D.; Lewis, V.O.; Gorlick, R. Current and future therapeutic approaches for osteosarcoma. Expert Rev. Anticancer Ther. 2018, 18, 39–50. [Google Scholar] [CrossRef]
- Lo, Y.C.; Rensi, S.E.; Torng, W.; Altman, R.B. Machine learning in chemoinformatics and drug discovery. Drug Discov. Today 2018, 23, 1538–1546. [Google Scholar] [CrossRef]
- Brown, A.S.; Patel, C.J. A review of validation strategies for computational drug repositioning. Brief. Bioinform. 2018, 19, 174–177. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Ma, X.H.; Shi, Z.; Tan, C.; Jiang, Y.; Go, M.L.; Low, B.C.; Chen, Y.Z. In-silico approaches to multi-target drug discovery: Computer aided multi-target drug design, multi-target virtual screening. Pharm. Res. 2010, 27, 739–749. [Google Scholar] [CrossRef]
- Huang, G.; Li, J.; Wang, P.; Li, W. A Review of Computational Drug Repositioning Approaches. Comb. Chem. High Throughput Screen 2017, 20, 831–838. [Google Scholar] [CrossRef]
- Park, K. A review of computational drug repurposing. Transl. Clin. Pharmacol. 2019, 27, 59–63. [Google Scholar] [CrossRef]
- Cruz-Monteagudo, M.; Schurer, S.; Tejera, E.; Perez-Castillo, Y.; Medina-Franco, J.L.; Sanchez-Rodriguez, A.; Borges, F. Systemic QSAR and phenotypic virtual screening: Chasing butterflies in drug discovery. Drug Discov. Today 2017, 22, 994–1007. [Google Scholar] [CrossRef]
- Murphy, R.F. An active role for machine learning in drug development. Nat. Chem. Biol. 2011, 7, 327–330. [Google Scholar] [CrossRef]
- Ramsay, R.R.; Popovic-Nikolic, M.R.; Nikolic, K.; Uliassi, E.; Bolognesi, M.L. A perspective on multi-target drug discovery and design for complex diseases. Clin. Transl. Med. 2018, 7, 3. [Google Scholar] [CrossRef]
- Nagamalla, L.; Kumar, J.V.S. In silico screening of FDA approved drugs on AXL kinase and validation for breast cancer cell line. J. Biomol. Struct. Dyn. 2020. [Google Scholar] [CrossRef]
- Kumar, R.; Chaudhary, K.; Singla, D.; Gautam, A.; Raghava, G.P. Designing of promiscuous inhibitors against pancreatic cancer cell lines. Sci. Rep. 2014, 4, 4668. [Google Scholar] [CrossRef]
- Issa, N.T.; Stathias, V.; Schurer, S.; Dakshanamurthy, S. Machine and deep learning approaches for cancer drug repurposing. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Koudijs, K.K.M.; Terwisscha van Scheltinga, A.G.T.; Bohringer, S.; Schimmel, K.J.M.; Guchelaar, H.J. Personalised drug repositioning for Clear Cell Renal Cell Carcinoma using gene expression. Sci. Rep. 2018, 8, 5250. [Google Scholar] [CrossRef]
- Wei, G.G.; Gao, L.; Tang, Z.Y.; Lin, P.; Liang, L.B.; Zeng, J.J.; Chen, G.; Zhang, L.C. Drug repositioning in head and neck squamous cell carcinoma: An integrated pathway analysis based on connectivity map and differential gene expression. Pathol. Res. Pract. 2019, 215, 152378. [Google Scholar] [CrossRef]
- Singh, H.; Kumar, R.; Singh, S.; Chaudhary, K.; Gautam, A.; Raghava, G.P. Prediction of anticancer molecules using hybrid model developed on molecules screened against NCI-60 cancer cell lines. BMC Cancer 2016, 16, 77. [Google Scholar] [CrossRef]
- Speck-Planche, A.; Cordeiro, M. Fragment-based in silico modeling of multi-target inhibitors against breast cancer-related proteins. Mol. Divers. 2017, 21, 511–523. [Google Scholar] [CrossRef]
- Bediaga, H.; Arrasate, S.; Gonzalez-Diaz, H. PTML Combinatorial Model of ChEMBL Compounds Assays for Multiple Types of Cancer. ACS Comb. Sci 2018, 20, 621–632. [Google Scholar] [CrossRef]
- Lopez-Cortes, A.; Paz, Y.M.C.; Guerrero, S.; Cabrera-Andrade, A.; Barigye, S.J.; Munteanu, C.R.; Gonzalez-Diaz, H.; Pazos, A.; Perez-Castillo, Y.; Tejera, E. OncoOmics approaches to reveal essential genes in breast cancer: A panoramic view from pathogenesis to precision medicine. Sci. Rep. 2020, 10, 5285. [Google Scholar] [CrossRef]
- Li, X.; Yan, M.L.; Yu, Q. Identification of candidate drugs for the treatment of metastatic osteosarcoma through a subpathway analysis method. Oncol. Lett. 2017, 13, 4378–4384. [Google Scholar] [CrossRef][Green Version]
- Speck-Planche, A.; Kleandrova, V.V.; Luan, F.; Cordeiro, M.N. Fragment-based QSAR model toward the selection of versatile anti-sarcoma leads. Eur. J. Med. Chem. 2011, 46, 5910–5916. [Google Scholar] [CrossRef]
- Chalise, P.; Koestler, D.C.; Bimali, M.; Yu, Q.; Fridley, B.L. Integrative clustering methods for high-dimensional molecular data. Transl. Cancer Res. 2014, 3, 202–216. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Braga, R.C.; Alves, V.M.; Silva, A.C.; Nascimento, M.N.; Silva, F.C.; Liao, L.M.; Andrade, C.H. Virtual screening strategies in medicinal chemistry: The state of the art and current challenges. Curr. Top. Med. Chem. 2014, 14, 1899–1912. [Google Scholar] [CrossRef]
- Bolognesi, M.L.; Cavalli, A. Multitarget Drug Discovery and Polypharmacology. Chem. Med. Chem. 2016, 11, 1190–1192. [Google Scholar] [CrossRef]
- Perez-Castillo, Y.; Sanchez-Rodriguez, A.; Tejera, E.; Cruz-Monteagudo, M.; Borges, F.; Cordeiro, M.; Le-Thi-Thu, H.; Pham-The, H. A desirability-based multi objective approach for the virtual screening discovery of broad-spectrum anti-gastric cancer agents. PLoS ONE 2018, 13, e0192176. [Google Scholar] [CrossRef]
- Xue, H.; Li, J.; Xie, H.; Wang, Y. Review of Drug Repositioning Approaches and Resources. Int. J. Biol Sci 2018, 14, 1232–1244. [Google Scholar] [CrossRef]
- Langedijk, J.; Mantel-Teeuwisse, A.K.; Slijkerman, D.S.; Schutjens, M.H. Drug repositioning and repurposing: Terminology and definitions in literature. Drug Discov. Today 2015, 20, 1027–1034. [Google Scholar] [CrossRef]
- Parvathaneni, V.; Kulkarni, N.S.; Muth, A.; Gupta, V. Drug repurposing: A promising tool to accelerate the drug discovery process. Drug Discov. Today 2019, 24, 2076–2085. [Google Scholar] [CrossRef]
- Ding, L.; Congwei, L.; Bei, Q.; Tao, Y.; Ruiguo, W.; Heze, Y.; Bo, D.; Zhihong, L. mTOR: An attractive therapeutic target for osteosarcoma? Oncotarget 2016, 7, 50805–50813. [Google Scholar] [CrossRef]
- Bishop, M.W.; Janeway, K.A. Emerging concepts for PI3K/mTOR inhibition as a potential treatment for osteosarcoma. F1000Research 2016, 5, 1590. [Google Scholar] [CrossRef]
- Cabrera-Andrade, A.; Lopez-Cortes, A.; Jaramillo-Koupermann, G.; Paz, Y.M.C.; Perez-Castillo, Y.; Munteanu, C.R.; Gonzalez-Diaz, H.; Pazos, A.; Tejera, E. Gene Prioritization through Consensus Strategy, Enrichment Methodologies Analysis, and Networking for Osteosarcoma Pathogenesis. Int. J. Mol. Sci 2020, 21, 1053. [Google Scholar] [CrossRef]
- de Bono, J.S.; Oudard, S.; Ozguroglu, M.; Hansen, S.; Machiels, J.P.; Kocak, I.; Gravis, G.; Bodrogi, I.; Mackenzie, M.J.; Shen, L.; et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: A randomised open-label trial. Lancet 2010, 376, 1147–1154. [Google Scholar] [CrossRef]
- de Wit, R.; de Bono, J.; Sternberg, C.N.; Fizazi, K.; Tombal, B.; Wulfing, C.; Kramer, G.; Eymard, J.C.; Bamias, A.; Carles, J.; et al. Cabazitaxel versus Abiraterone or Enzalutamide in Metastatic Prostate Cancer. N. Engl. J. Med. 2019, 381, 2506–2518. [Google Scholar] [CrossRef]
- Oudard, S.; Fizazi, K.; Sengelov, L.; Daugaard, G.; Saad, F.; Hansen, S.; Hjalm-Eriksson, M.; Jassem, J.; Thiery-Vuillemin, A.; Caffo, O.; et al. Cabazitaxel Versus Docetaxel As First-Line Therapy for Patients With Metastatic Castration-Resistant Prostate Cancer: A Randomized Phase III Trial-FIRSTANA. J. Clin. Oncol. 2017, 35, 3189–3197. [Google Scholar] [CrossRef]
- Lo, Y.C.; Senese, S.; France, B.; Gholkar, A.A.; Damoiseaux, R.; Torres, J.Z. Computational Cell Cycle Profiling of Cancer Cells for Prioritizing FDA-Approved Drugs with Repurposing Potential. Sci. Rep. 2017, 7, 11261. [Google Scholar] [CrossRef]
- Reynolds, C.P.; Kang, M.H.; Maris, J.M.; Kolb, E.A.; Gorlick, R.; Wu, J.; Kurmasheva, R.T.; Houghton, P.J.; Smith, M.A. Initial testing (stage 1) of the anti-microtubule agents cabazitaxel and docetaxel, by the pediatric preclinical testing program. Pediatr. Blood Cancer 2015, 62, 1897–1905. [Google Scholar] [CrossRef]
- Amoroso, L.; Castel, V.; Bisogno, G.; Casanova, M.; Marquez-Vega, C.; Chisholm, J.C.; Doz, F.; Moreno, L.; Ruggiero, A.; Gerber, N.U.; et al. Phase II results from a phase I/II study to assess the safety and efficacy of weekly nab-paclitaxel in paediatric patients with recurrent or refractory solid tumours: A collaboration with the European Innovative Therapies for Children with Cancer Network. Eur. J. Cancer 2020, 135, 89–97. [Google Scholar] [CrossRef]
- Hussain, A.; Dar, M.S.; Bano, N.; Hossain, M.M.; Basit, R.; Bhat, A.Q.; Aga, M.A.; Ali, S.; Hassan, Q.P.; Dar, M.J. Identification of dinactin, a macrolide antibiotic, as a natural product-based small molecule targeting Wnt/beta-catenin signaling pathway in cancer cells. Cancer Chemother. Pharmacol. 2019, 84, 551–559. [Google Scholar] [CrossRef]
- Gupta, A.; Okesli-Armlovich, A.; Morgens, D.; Bassik, M.C.; Khosla, C. A genome-wide analysis of targets of macrolide antibiotics in mammalian cells. J. Biol. Chem. 2020, 295, 2057–2067. [Google Scholar] [CrossRef]
- Bahrami, F.; Morris, D.L.; Pourgholami, M.H. Tetracyclines: Drugs with huge therapeutic potential. Mini Rev. Med. Chem 2012, 12, 44–52. [Google Scholar] [CrossRef]
- Fiorillo, M.; Toth, F.; Sotgia, F.; Lisanti, M.P. Doxycycline, Azithromycin and Vitamin C (DAV): A potent combination therapy for targeting mitochondria and eradicating cancer stem cells (CSCs). Aging 2019, 11, 2202–2216. [Google Scholar] [CrossRef]
- Lamb, R.; Ozsvari, B.; Lisanti, C.L.; Tanowitz, H.B.; Howell, A.; Martinez-Outschoorn, U.E.; Sotgia, F.; Lisanti, M.P. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: Treating cancer like an infectious disease. Oncotarget 2015, 6, 4569–4584. [Google Scholar] [CrossRef]
- Maksimovic-Ivanic, D.; Fagone, P.; McCubrey, J.; Bendtzen, K.; Mijatovic, S.; Nicoletti, F. HIV-protease inhibitors for the treatment of cancer: Repositioning HIV protease inhibitors while developing more potent NO-hybridized derivatives? Int. J. Cancer 2017, 140, 1713–1726. [Google Scholar] [CrossRef]
- Petroni, G.; Stefanini, M.; Pillozzi, S.; Crociani, O.; Becchetti, A.; Arcangeli, A. Data describing the effects of the Macrolide Antibiotic Clarithromycin on preclinical mouse models of Colorectal Cancer. Data Brief. 2019, 26, 104406. [Google Scholar] [CrossRef]
- Van Nuffel, A.M.; Sukhatme, V.; Pantziarka, P.; Meheus, L.; Sukhatme, V.P.; Bouche, G. Repurposing Drugs in Oncology (ReDO)-clarithromycin as an anti-cancer agent. Ecancermedicalscience 2015, 9, 513. [Google Scholar] [CrossRef]
- de Jong, J.; Hellemans, P.; De Wilde, S.; Patricia, D.; Masterson, T.; Manikhas, G.; Myasnikov, A.; Osmanov, D.; Cordoba, R.; Panizo, C.; et al. A drug-drug interaction study of ibrutinib with moderate/strong CYP3A inhibitors in patients with B-cell malignancies. Leuk. Lymphoma 2018, 59, 2888–2895. [Google Scholar] [CrossRef]
- Markowska, A.; Kaysiewicz, J.; Markowska, J.; Huczynski, A. Doxycycline, salinomycin, monensin and ivermectin repositioned as cancer drugs. Bioorg. Med. Chem. Lett. 2019, 29, 1549–1554. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Felix, E.; Magarinos, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrian-Uhalte, E.; et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. [Google Scholar] [CrossRef]
- Chem Axon J. Chem for Office. Available online: https://chemaxon.com (accessed on 12 March 2020).
- Chem Axon Chemaxon Standardizer. Available online: http://www.chemaxon.com (accessed on 24 March 2020).
- Varnek, A.; Fourches, D.; Horvath, D.; Klimchuk, O.; Gaudin, C.; Vayer, P.; Hoonakker, F.; Tetko, I.V.; Marcou, G. ISIDA-Platform for virtual screening based on fragment and pharmacophoric descriptors. Curr. Comput. Aided Drug Des. 2008, 4, 191. [Google Scholar] [CrossRef]
- Ruggiu, F.; Marcou, G.; Varnek, A.; Horvath, D. ISIDA Property-Labelled Fragment Descriptors. Mol. Inform. 2010, 29, 855–868. [Google Scholar] [CrossRef]
- Varnek, A.; Fourches, D.; Hoonakker, F.; Solovev, V.P. Substructural fragments: An universal language to encode reactions, molecular and supramolecular structures. J. Comput. Aided Mol. Des. 2005, 19, 693–703. [Google Scholar] [CrossRef]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Potter, T.; Matter, H. Random or rational design? Evaluation of diverse compound subsets from chemical structure databases. J. Med. Chem 1998, 41, 478–488. [Google Scholar] [CrossRef]
- Serra, M.; Hattinger, C.M. The pharmacogenomics of osteosarcoma. Pharmacogenom. J. 2017, 17, 11–20. [Google Scholar] [CrossRef]
- Hattinger, C.M.; Vella, S.; Tavanti, E.; Fanelli, M.; Picci, P.; Serra, M. Pharmacogenomics of second-line drugs used for treatment of unresponsive or relapsed osteosarcoma patients. Pharmacogenomics 2016, 17, 2097–2114. [Google Scholar] [CrossRef]
- Schwartz, G.K.; Tap, W.D.; Qin, L.X.; Livingston, M.B.; Undevia, S.D.; Chmielowski, B.; Agulnik, M.; Schuetze, S.M.; Reed, D.R.; Okuno, S.H.; et al. Cixutumumab and temsirolimus for patients with bone and soft-tissue sarcoma: A multicentre, open-label, phase 2 trial. Lancet Oncol. 2013, 14, 371–382. [Google Scholar] [CrossRef]
- Trucco, M.M.; Meyer, C.F.; Thornton, K.A.; Shah, P.; Chen, A.R.; Wilky, B.A.; Carrera-Haro, M.A.; Boyer, L.C.; Ferreira, M.F.; Shafique, U.; et al. A phase II study of temsirolimus and liposomal doxorubicin for patients with recurrent and refractory bone and soft tissue sarcomas. Clin. Sarcoma Res. 2018, 8, 21. [Google Scholar] [CrossRef]
- Demetri, G.D.; Chawla, S.P.; Ray-Coquard, I.; Le Cesne, A.; Staddon, A.P.; Milhem, M.M.; Penel, N.; Riedel, R.F.; Bui-Nguyen, B.; Cranmer, L.D.; et al. Results of an international randomized phase III trial of the mammalian target of rapamycin inhibitor ridaforolimus versus placebo to control metastatic sarcomas in patients after benefit from prior chemotherapy. J. Clin. Oncol. 2013, 31, 2485–2492. [Google Scholar] [CrossRef]
- Chawla, S.P.; Staddon, A.P.; Baker, L.H.; Schuetze, S.M.; Tolcher, A.W.; D’Amato, G.Z.; Blay, J.Y.; Mita, M.M.; Sankhala, K.K.; Berk, L.; et al. Phase II study of the mammalian target of rapamycin inhibitor ridaforolimus in patients with advanced bone and soft tissue sarcomas. J. Clin. Oncol. 2012, 30, 78–84. [Google Scholar] [CrossRef]
- Qayed, M.; Cash, T.; Tighiouart, M.; MacDonald, T.J.; Goldsmith, K.C.; Tanos, R.; Kean, L.; Watkins, B.; Suessmuth, Y.; Wetmore, C.; et al. A phase I study of sirolimus in combination with metronomic therapy (CHOAnome) in children with recurrent or refractory solid and brain tumors. Pediatr. Blood Cancer 2020, 67, e28134. [Google Scholar] [CrossRef]
- van der Graaf, W.T.; Blay, J.Y.; Chawla, S.P.; Kim, D.W.; Bui-Nguyen, B.; Casali, P.G.; Schoffski, P.; Aglietta, M.; Staddon, A.P.; Beppu, Y.; et al. Pazopanib for metastatic soft-tissue sarcoma (PALETTE): A randomised, double-blind, placebo-controlled phase 3 trial. Lancet 2012, 379, 1879–1886. [Google Scholar] [CrossRef]
- Longhi, A.; Paioli, A.; Palmerini, E.; Cesari, M.; Abate, M.E.; Setola, E.; Spinnato, P.; Donati, D.; Hompland, I.; Boye, K. Pazopanib in relapsed osteosarcoma patients: Report on 15 cases. Acta Oncol. 2019, 58, 124–128. [Google Scholar] [CrossRef]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef]
- Truchon, J.F.; Bayly, C.I. Evaluating virtual screening methods: Good and bad metrics for the “early recognition” problem. J. Chem. Inf. Model. 2007, 47, 488–508. [Google Scholar] [CrossRef]
- Kirchmair, J.; Markt, P.; Distinto, S.; Wolber, G.; Langer, T. Evaluation of the performance of 3D virtual screening protocols: RMSD comparisons, enrichment assessments, and decoy selection—What can we learn from earlier mistakes? J. Comput. Aided. Mol. Des. 2008, 22, 213–228. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabrera-Andrade, A.; López-Cortés, A.; Jaramillo-Koupermann, G.; González-Díaz, H.; Pazos, A.; Munteanu, C.R.; Pérez-Castillo, Y.; Tejera, E. A Multi-Objective Approach for Anti-Osteosarcoma Cancer Agents Discovery through Drug Repurposing. Pharmaceuticals 2020, 13, 409. https://doi.org/10.3390/ph13110409
Cabrera-Andrade A, López-Cortés A, Jaramillo-Koupermann G, González-Díaz H, Pazos A, Munteanu CR, Pérez-Castillo Y, Tejera E. A Multi-Objective Approach for Anti-Osteosarcoma Cancer Agents Discovery through Drug Repurposing. Pharmaceuticals. 2020; 13(11):409. https://doi.org/10.3390/ph13110409
Chicago/Turabian StyleCabrera-Andrade, Alejandro, Andrés López-Cortés, Gabriela Jaramillo-Koupermann, Humberto González-Díaz, Alejandro Pazos, Cristian R. Munteanu, Yunierkis Pérez-Castillo, and Eduardo Tejera. 2020. "A Multi-Objective Approach for Anti-Osteosarcoma Cancer Agents Discovery through Drug Repurposing" Pharmaceuticals 13, no. 11: 409. https://doi.org/10.3390/ph13110409
APA StyleCabrera-Andrade, A., López-Cortés, A., Jaramillo-Koupermann, G., González-Díaz, H., Pazos, A., Munteanu, C. R., Pérez-Castillo, Y., & Tejera, E. (2020). A Multi-Objective Approach for Anti-Osteosarcoma Cancer Agents Discovery through Drug Repurposing. Pharmaceuticals, 13(11), 409. https://doi.org/10.3390/ph13110409








