Phenolic Compounds Reduce the Fat Content in Caenorhabditis elegans by Affecting Lipogenesis, Lipolysis, and Different Stress Responses
Abstract
1. Introduction
2. Results and Discussion
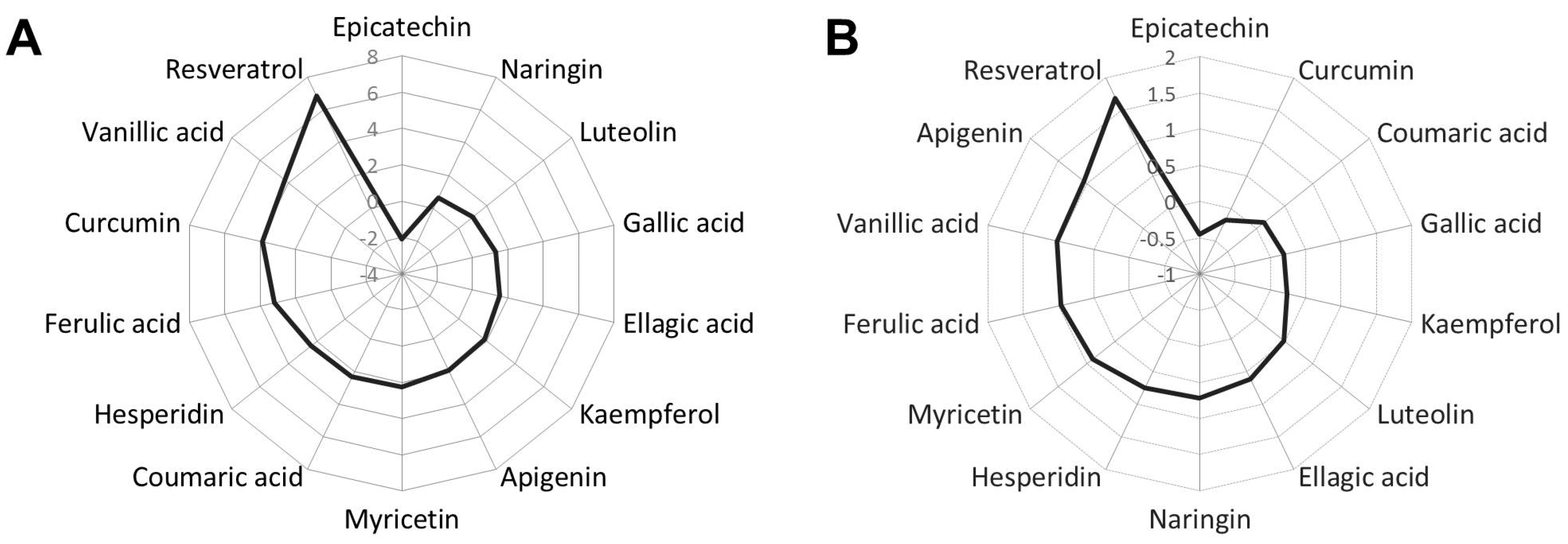
2.1. Phenolic Compounds Reduce Fat Accumulation in Wild-Type C. elegans
2.2. Resveratrol Treatment during the Larval Stages Improves C. elegans Healthspan
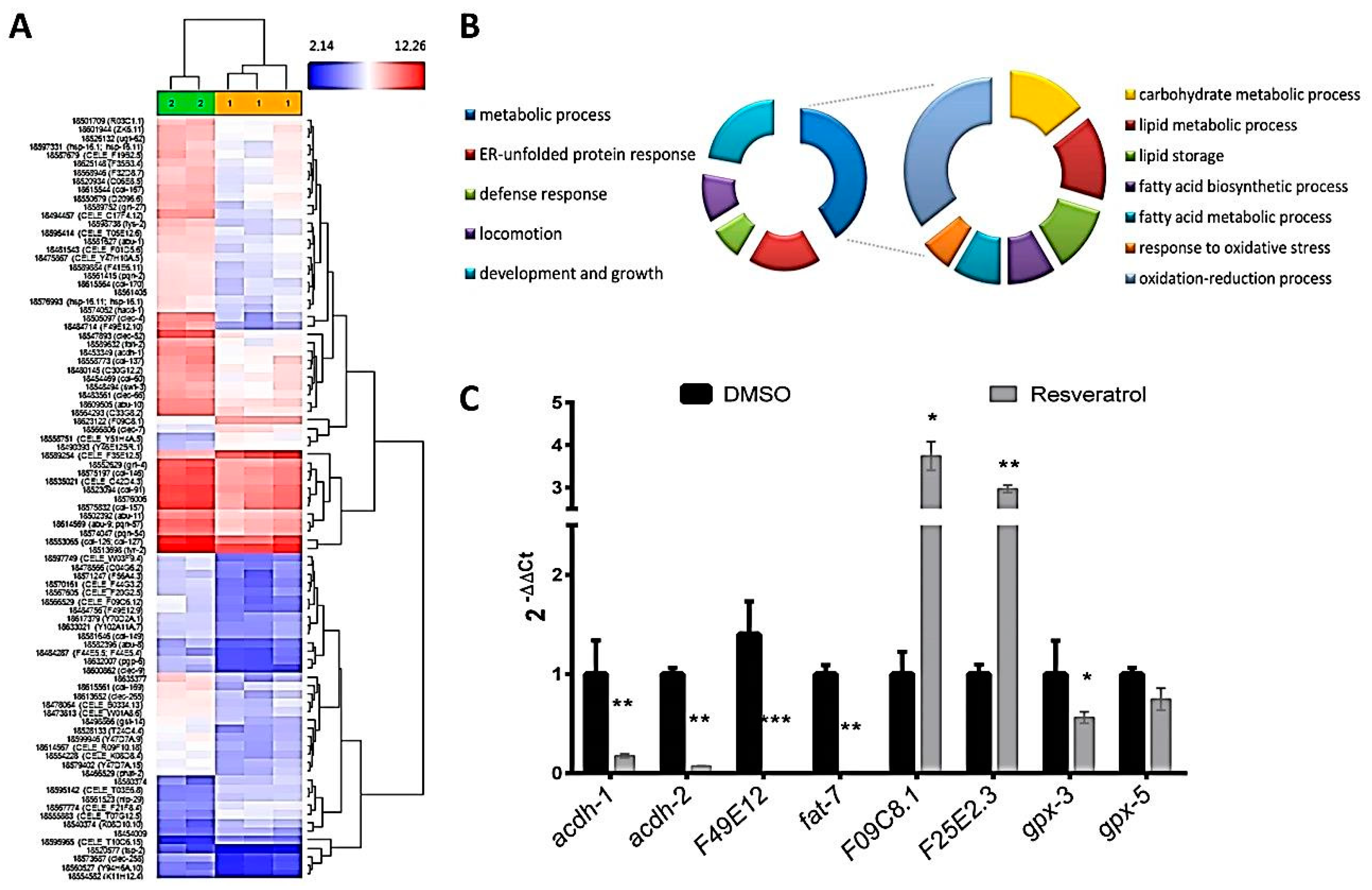
2.3. Resveratrol Affects Lipogenesis-Related Genes, Stress Resistance, Protein Processing in Endoplasmic Reticulum, and Fatty Acid Metabolism-Related Genes by Mimicking Calorie Restriction Status
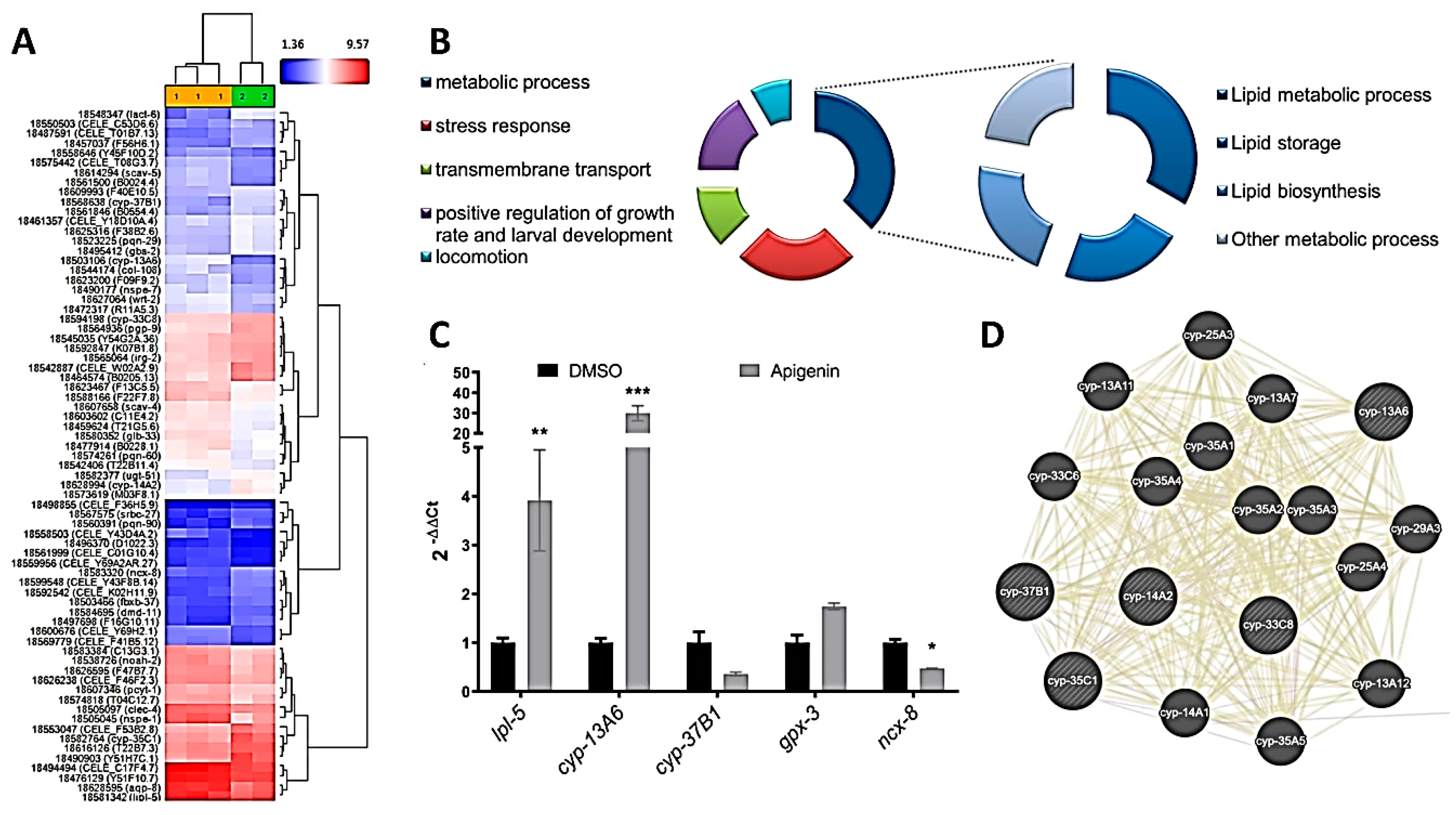
2.4. Apigenin Treatment Induces the Oxidative Stress Response and Lipid Mobilization in C. elegans
2.5. Vanillic Acid Induces the Oxidative Stress Response and Upregulates Heat Shock Proteins in C. elegans
3. Materials and Methods
3.1. Reagents
3.2. C. elegans Culture and Phenolic Compounds Treatment
3.3. Nile Red Staining
3.4. Oil Red O Staining
3.5. DHE Staining
3.6. C. elegans Aging Visualization
3.7. Image Acquisition and Quantification
3.8. Lifespan Analysis
3.9. Egg Lying and Worm Size
3.10. Cohen’s d Effect Size
3.11. RNA Extraction and Quantitative PCR Analysis
3.12. Whole-Genome Transcriptomic Analysis
3.13. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Banerjee, S.; Talukdar, I.; Banerjee, A.; Gupta, A.; Balaji, A.; Aduri, R. Type II diabetes mellitus and obesity: Common links, existing therapeutics and future developments. J. Biosci. 2019, 44, 150. [Google Scholar] [CrossRef] [PubMed]
- Després, J.P. Abdominal Obesity and Cardiovascular Disease: Is Inflammation the Missing Link? Can. J. Cardiol. 2012, 28, 642–652. [Google Scholar] [CrossRef]
- Lanktree, M.B.; Hegele, R.A. Metabolic Syndrome. In Genomic and Precision Medicine: Primary Care, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2017; ISBN 9780128006542. [Google Scholar]
- O’Neill, S.; O’Driscoll, L. Metabolic syndrome: A closer look at the growing epidemic and its associated pathologies. Obes. Rev. 2015. [Google Scholar] [CrossRef]
- Trigueros, L.; Pena, S.; Av, U.; Sayas-Barbera, E.; Ja, P.A.; Sendra, E.; Ugidos, A.V.; Sayas-Barbera, E.; Perez-Alvarez, J.A.; Sendra, E. Food ingredients as anti-obesity agents: A review. Crit. Rev. Food Sci. Nutr. 2013. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.L. Fat synthesis and adiposity regulation in Caenorhabditis elegans. Trends Endocrinol. Metab. 2009, 20, 58–65. [Google Scholar] [CrossRef]
- Hsu, C.L.; Yen, G.C. Effects of flavonoids and phenolic acids on the inhibition of adipogenesis in 3T3-L1 adipocytes. J. Agric. Food Chem. 2007. [Google Scholar] [CrossRef]
- Hsu, C.L.; Huang, S.L.; Yen, G.C. Inhibitory effect of phenolic acids on the proliferation of 3T3-L1 preadipocytes in relation to their antioxidant activity. J. Agric. Food Chem. 2006, 54, 4191–4197. [Google Scholar] [CrossRef]
- Saul, N.; Pietsch, K.; Stürzenbaum, S.R.; Menzel, R.; Steinberg, C.E.W. Diversity of polyphenol action in Caenorhabditis elegans: Between toxicity and longevity. J. Nat. Prod. 2011, 74, 1713–1720. [Google Scholar] [CrossRef]
- Pallauf, K.; Duckstein, N.; Rimbach, G. A literature review of flavonoids and lifespan in model organisms. Proc. Nutr. Soc. 2017, 76, 145–162. [Google Scholar] [CrossRef]
- Shen, P.; Yue, Y.; Zheng, J.; Park, Y. Caenorhabditis elegans: A Convenient in Vivo Model for Assessing the Impact of Food Bioactive Compounds on Obesity, Aging, and Alzheimer’s Disease. Annu. Rev. Food Sci. Technol. 2018. [Google Scholar] [CrossRef]
- Lemieux, G.A.; Ashrafi, K. Insights and challenges in using C. elegans for investigation of fat metabolism. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, K. Obesity and the regulation of Fat Metabolism. In Worm Book; Shields Publications: Eagle River, WI, USA, 2007. [Google Scholar]
- Shen, P.; Yue, Y.; Park, Y. A living model for obesity and aging research: Caenorhabditis elegans. Crit. Rev. Food Sci. Nutr. 2018, 58, 741–754. [Google Scholar] [CrossRef] [PubMed]
- El-Houri, R.B.; Kotowska, D.; Olsen, L.C.B.; Bhattacharya, S.; Christensen, L.P.; Grevsen, K.; Oksbjerg, N.; Færgeman, N.; Kristiansen, K.; Christensen, K.B. Screening for bioactive metabolites in plant extracts modulating glucose uptake and fat accumulation. Evid. Based Complement. Altern. Med. 2014. [Google Scholar] [CrossRef]
- Wählby, C.; Lee Conery, A.; Bray, M.A.; Kamentsky, L.; Larkins-Ford, J.; Sokolnicki, K.L.; Veneskey, M.; Michaels, K.; Carpenter, A.E.; O’Rourke, E.J. High- and low-throughput scoring of fat mass and body fat distribution in C. elegans. Methods 2014. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Christensen, K.B.; Olsen, L.C.B.; Christensen, L.P.; Grevsen, K.; Færgeman, N.J.; Kristiansen, K.; Young, J.F.; Oksbjerg, N. Bioactive components from flowers of Sambucus nigra L. increase glucose uptake in primary porcine myotube cultures and reduce fat accumulation in Caenorhabditis elegans. J. Agric. Food Chem. 2013, 61, 11033–11040. [Google Scholar] [CrossRef]
- Shi, X.; Li, J.; Zou, X.; Greggain, J.; Rødkær, S.V.; Færgeman, N.J.; Liang, B.; Watts, J.L. Regulation of lipid droplet size and phospholipid composition by stearoyl-CoA desaturase. J. Lipid Res. 2013. [Google Scholar] [CrossRef]
- Yen, K.; Le, T.T.; Bansal, A.; Narasimhan, S.D.; Cheng, J.X.; Tissenbaum, H.A. A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods. PLoS ONE 2010, 5, e12810. [Google Scholar] [CrossRef]
- Yang, J.; Della-fera, M.A.; Hartzell, D.L.; Nelson-dooley, C.; Hausman, D.B.; Baile, C.A. Esculetin Induces Apoptosis and Inhibits Adipogenesis in 3T3-L1 Cells. Obesity 2006. [Google Scholar] [CrossRef]
- Pereira-Fernandes, A.; Demaegdt, H.; Vandermeiren, K.; Hectors, T.L.M.; Jorens, P.G.; Blust, R.; Vanparys, C. Evaluation of a Screening System for Obesogenic Compounds: Screening of Endocrine Disrupting Compounds and Evaluation of the PPAR Dependency of the Effect. PLoS ONE 2013, 8, e77481. [Google Scholar] [CrossRef]
- Aranaz, P.; Navarro-Herrera, D.; Zabala, M.; Miguéliz, I.; Romo-Hualde, A.; López-Yoldi, M.; Alfredo Martínez, J.; Vizmanos, J.L.; Milagro, F.I.; González-Navarro, C.J. Phenolic compounds inhibit 3T3-L1 adipogenesis depending on the stage of differentiation and their binding affinity to PPARγ. Molecules 2019, 24, 1045. [Google Scholar] [CrossRef]
- Zheng, J.; Heber, D.; Wang, M.; Gao, C.; Heymsfield, S.B.; Martin, R.J.; Greenway, F.L.; Finley, J.W.; Burton, J.H.; Johnson, W.D.; et al. Pomegranate juice and extract extended lifespan and reduced intestinal fat deposition in Caenorhabditis elegans. Int. J. Vitam. Nutr. Res. 2017. [Google Scholar] [CrossRef]
- Navarro-Herrera, D.; Aranaz, P.; Eder-Azanza, L.; Zabala, M.; Hurtado, C.; Romo-Hualde, A.; Martínez, J.A.; González-Navarro, C.J.; Vizmanos, J.L. Dihomo-gamma-linolenic acid induces fat loss in: C. elegans in an omega-3-independent manner by promoting peroxisomal fatty acid β-oxidation. Food Funct. 2018. [Google Scholar] [CrossRef]
- Aranaz, P.; Navarro-Herrera, D.; Romo-Hualde, A.; Zabala, M.; López-Yoldi, M.; González-Ferrero, C.; Gil, A.G.; Alfredo Martinez, J.; Vizmanos, J.L.; Milagro, F.I.; et al. Broccoli extract improves high fat diet-induced obesity, hepatic steatosis and glucose intolerance in Wistar rats. J. Funct. Foods 2019, 59. [Google Scholar] [CrossRef]
- Wetchakul, P.; Goon, J.A.; Adekoya, A.E.; Olatunji, O.J.; Ruangchuay, S.; Jaisamut, P.; Issuriya, A.; Kunworarath, N.; Limsuwan, S.; Chusri, S. Traditional tonifying polyherbal infusion, Jatu-Phala-Tiga, exerts antioxidant activities and extends lifespan of Caenorhabditis elegans. BMC Complement. Altern. Med. 2019. [Google Scholar] [CrossRef]
- Ayuda-Durán, B.; González-Manzano, S.; Gil-Sánchez, I.; Victoria Moreno-Arribas, M.; Bartolomé, B.; Sanz-Buenhombre, M.; Guadarrama, A.; Santos-Buelga, C.; González-Paramás, A.M. Antioxidant characterization and biological effects of grape pomace extracts supplementation in Caenorhabditis elegans. Foods 2019, 8, 75. [Google Scholar] [CrossRef]
- Sethupathy, S.; Ananthi, S.; Selvaraj, A.; Shanmuganathan, B.; Vigneshwari, L.; Balamurugan, K.; Mahalingam, S.; Pandian, S.K. Vanillic acid from Actinidia deliciosa impedes virulence in Serratia marcescens by affecting S-layer, flagellin and fatty acid biosynthesis proteins. Sci. Rep. 2017. [Google Scholar] [CrossRef]
- Yue, Y.; Shen, P.; Xu, Y.; Park, Y. p-Coumaric acid improves oxidative and osmosis stress responses in Caenorhabditis elegans. J. Sci. Food Agric. 2019. [Google Scholar] [CrossRef]
- Kampkötter, A.; Tanner, S.; Bosbach, H.; Büchter, C.; Sack, M.; Chovolou, Y.; Wätjen, W.; Kahl, R. Flavonoids extent life span, increase stress resistance and modulate intracellular signalling in the model organism Caenorhabditis elegans. Toxicol. Lett. 2009. [Google Scholar] [CrossRef]
- Büchter, C.; Ackermann, D.; Havermann, S.; Honnen, S.; Chovolou, Y.; Fritz, G.; Kampkötter, A.; Wätjen, W. Myricetin-mediated lifespan extension in Caenorhabditis elegans is modulated by DAF-16. Int. J. Mol. Sci. 2013, 14, 11895–11914. [Google Scholar] [CrossRef]
- Grünz, G.; Haas, K.; Soukup, S.; Klingenspor, M.; Kulling, S.E.; Daniel, H.; Spanier, B. Structural features and bioavailability of four flavonoids and their implications for lifespan-extending and antioxidant actions in C. elegans. Mech. Ageing Dev. 2012. [Google Scholar] [CrossRef]
- Koch, K.; Havermann, S.; Büchter, C.; Wätjen, W. Caenorhabditis elegans as model system in pharmacology and toxicology: Effects of flavonoids on redox-sensitive signalling pathways and ageing. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef]
- Martorell, P.; Llopis, S.; González, N.; Montón, F.; Ortiz, P.; Genovés, S.; Ramón, D. Caenorhabditis elegans as a model to study the effectiveness and metabolic targets of dietary supplements used for obesity treatment: The specific case of a conjugated linoleic acid mixture (Tonalin). J. Agric. Food Chem. 2012, 60, 11071–11079. [Google Scholar] [CrossRef]
- Lin, Y.; Yang, N.; Bao, B.; Wang, L.; Chen, J.; Liu, J. Luteolin reduces fat storage in Caenorhabditis elegans by promoting the central serotonin pathway. Food Funct. 2020. [Google Scholar] [CrossRef]
- Peng, H.; Wei, Z.; Luo, H.; Yang, Y.; Wu, Z.; Gan, L.; Yang, X. Inhibition of Fat Accumulation by Hesperidin in Caenorhabditis elegans. J. Agric. Food Chem. 2016. [Google Scholar] [CrossRef]
- Yue, Y.; Shen, P.; Chang, A.L.; Qi, W.; Kim, K.H.; Kim, D.; Park, Y. Trans-Trismethoxy resveratrol decreased fat accumulation dependent on fat-6 and fat-7 in Caenorhabditis elegans. Food Funct. 2019. [Google Scholar] [CrossRef]
- Shen, C.Y.; Wan, L.; Wang, T.X.; Jiang, J.G. Citrus aurantium L. var. amara Engl. inhibited lipid accumulation in 3T3-L1 cells and Caenorhabditis elegans and prevented obesity in high-fat diet-fed mice. Pharm. Res. 2019. [Google Scholar] [CrossRef]
- Shen, P.; Zhang, R.; McClements, D.J.; Park, Y. Nanoemulsion-based delivery systems for testing nutraceutical efficacy using Caenorhabditis elegans: Demonstration of curcumin bioaccumulation and body-fat reduction. Food Res. Int. 2019. [Google Scholar] [CrossRef]
- Li, S.; Bouzar, C.; Cottet-Rousselle, C.; Zagotta, I.; Lamarche, F.; Wabitsch, M.; Tokarska-Schlattner, M.; Fischer-Posovszky, P.; Schlattner, U.; Rousseau, D. Resveratrol inhibits lipogenesis of 3T3-L1 and SGBS cells by inhibition of insulin signaling and mitochondrial mass increase. Biochim. Biophys. Acta 2016, 1857, 643–652. [Google Scholar] [CrossRef]
- Gomez-Zorita, S.; Treguer, K.; Mercader, J.; Carpene, C. Resveratrol directly affects in vitro lipolysis and glucose transport in human fat cells. J. Physiol. Biochem. 2013, 69, 585–593. [Google Scholar] [CrossRef]
- Bird, J.K.; Raederstorff, D.; Weber, P.; Steinert, R.E. Cardiovascular and antiobesity effects of resveratrol mediated through the gut microbiota. Adv. Nutr. 2017. [Google Scholar] [CrossRef]
- Farkhondeh, T.; Folgado, S.L.; Pourbagher-Shahri, A.M.; Ashrafizadeh, M.; Samarghandian, S. The therapeutic effect of resveratrol: Focusing on the Nrf2 signaling pathway. Biomed. Pharm. 2020, 127, 110234. [Google Scholar] [CrossRef]
- Dvorakova, M.; Landa, P. Anti-inflammatory activity of natural stilbenoids: A review. Pharm. Res. 2017, 124, 126–145. [Google Scholar] [CrossRef]
- Tsai, H.Y.; Ho, C.T.; Chen, Y.K. Biological actions and molecular effects of resveratrol, pterostilbene, and 3′-hydroxypterostilbene. J. Food Drug Anal. 2017, 25, 134–147. [Google Scholar] [CrossRef]
- Aguirre, L.; Fernández-Quintela, A.; Arias, N.; Portillo, M.P. Resveratrol: Anti-obesity mechanisms of action. Molecules 2014, 19, 18632–18655. [Google Scholar] [CrossRef]
- Gruber, J.; Soon, Y.T.; Halliwell, B. Evidence for a trade-off between survival and fitness caused by resveratrol treatment of Caenorhabditis elegans. Ann. N. Y. Acad. Sci. 2007, 1100, 530–542. [Google Scholar] [CrossRef]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.L.; et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 2003. [Google Scholar] [CrossRef]
- Wood, J.G.; Regina, B.; Lavu, S.; Hewitz, K.; Helfand, S.L.; Tatar, M.; Sinclair, D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 2004. [Google Scholar] [CrossRef]
- Viswanathan, M.; Kim, S.K.; Berdichevsky, A.; Guarente, L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev. Cell 2005. [Google Scholar] [CrossRef]
- Fischer, N.; Büchter, C.; Koch, K.; Albert, S.; Csuk, R.; Wätjen, W. The resveratrol derivatives trans-3,5-dimethoxy-4-fluoro-4′-hydroxystilbene and trans-2,4′,5-trihydroxystilbene decrease oxidative stress and prolong lifespan in Caenorhabditis elegans. J. Pharm. Pharm. 2017. [Google Scholar] [CrossRef]
- Zhang, J. Resveratrol inhibits insulin responses in a SirT1-independent pathway. Biochem. J. 2006. [Google Scholar] [CrossRef]
- Lee, S.H.; Min, K.J. Lifespan-Extending Effect of Resveratrol and Other Phytochemicals. RSC Drug Discov. Ser. 2017. [Google Scholar] [CrossRef]
- Ye, K.; Ji, C.-B.; Lu, X.-W.; Ni, Y.-H.; Gao, C.-L.; Chen, X.-H.; Zhao, Y.-P.; Gu, G.-X.; Guo, X.-R. Resveratrol Attenuates Radiation Damage in Caenorhabditis elegans by Preventing Oxidative Stress. J. Radiat. Res. 2010. [Google Scholar] [CrossRef]
- Liao, V.H.C.; Yu, C.W.; Chu, Y.J.; Li, W.H.; Hsieh, Y.C.; Wang, T.T. Curcumin-mediated lifespan extension in Caenorhabditis elegans. Mech. Ageing Dev. 2011. [Google Scholar] [CrossRef]
- Liao, V.H.C. Use of Caenorhabditis elegans to Study the Potential Bioactivity of Natural Compounds. J. Agric. Food Chem. 2018, 66, 1737–1742. [Google Scholar] [CrossRef]
- Wei, Y.; Kenyon, C. Roles for ROS and hydrogen sulfide in the longevity response to germline loss in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2016. [Google Scholar] [CrossRef]
- Lee, J.; Kwon, G.; Park, J.; Kim, J.K.; Lim, Y.H. Brief Communication: SIR-2.1-dependent lifespan extension of Caenorhabditis elegans by oxyresveratrol and resveratrol. Exp. Biol. Med. 2016. [Google Scholar] [CrossRef]
- Côté, C.D.; Rasmussen, B.A.; Duca, F.A.; Zadeh-Tahmasebi, M.; Baur, J.A.; Daljeet, M.; Breen, D.M.; Filippi, B.M.; Lam, T.K.T. Resveratrol activates duodenal Sirt1 to reverse insulin resistance in rats through a neuronal network. Nat. Med. 2015. [Google Scholar] [CrossRef]
- Kashyap, S.S.; Johnson, J.R.; McCue, H.V.; Chen, X.; Edmonds, M.J.; Ayala, M.; Graham, M.E.; Jenn, R.C.; Barclay, J.W.; Burgoyne, R.D.; et al. Caenorhabditis elegans dnj-14, the orthologue of the DNAJC5 gene mutated in adult onset neuronal ceroid lipofuscinosis, provides a new platform for neuroprotective drug screening and identifies a SIR-2.1-independent action of resveratrol. Hum. Mol. Genet. 2014. [Google Scholar] [CrossRef]
- Bass, T.M.; Weinkove, D.; Houthoofd, K.; Gems, D.; Partridge, L. Effects of resveratrol on lifespan in Drosophila melanogaster and Caenorhabditis elegans. Mech. Ageing Dev. 2007. [Google Scholar] [CrossRef]
- Brock, T.J.; Browse, J.; Watts, J.L. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2006. [Google Scholar] [CrossRef]
- Watts, J.L.; Ristow, M. Lipid and carbohydrate metabolism in Caenorhabditis elegans. Genetics 2017. [Google Scholar] [CrossRef]
- Van Gilst, M.R.; Hadjivassiliou, H.; Yamamoto, K.R. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. USA 2005. [Google Scholar] [CrossRef] [PubMed]
- Laing, S.T.; Ivens, A.; Butler, V.; Ravikumar, S.P.; Laing, R.; Woods, D.J.; Gilleard, J.S. The transcriptional response of Caenorhabditis elegans to ivermectin exposure identifies novel genes involved in the response to reduced food intake. PLoS ONE 2012, 7, e31367. [Google Scholar] [CrossRef]
- Marchal, J.; Pifferi, F.; Aujard, F. Resveratrol in mammals: Effects on aging biomarkers, age-related diseases, and life span. Ann. N. Y. Acad. Sci. 2013. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. Phospholipases. In Lipid Metabolism in Plants; IntechOpen: London, UK, 2018; ISBN 9781351082525. [Google Scholar]
- Hunt, M.C.; Siponen, M.I.; Alexson, S.E.H. The emerging role of acyl-CoA thioesterases and acyltransferases in regulating peroxisomal lipid metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 2012, 1822, 1397–1410. [Google Scholar] [CrossRef]
- Haskins, K.A.; Russell, J.F.; Gaddis, N.; Dressman, H.K.; Aballay, A. Unfolded Protein Response Genes Regulated by CED-1 Are Required for Caenorhabditis elegans Innate Immunity. Dev. Cell 2008. [Google Scholar] [CrossRef]
- Swindell, W.R. Heat shock proteins in long-lived worms and mice with insulin/insulin-like signaling mutations. Aging 2009. [Google Scholar] [CrossRef]
- Jo, H.; Shim, J.; Lee, J.H.; Lee, J.; Kim, J.B. IRE-1 and HSP-4 Contribute to Energy Homeostasis via Fasting-Induced Lipases in C. elegans. Cell Metab. 2009. [Google Scholar] [CrossRef]
- Qiao, Q.; Bouwman, F.G.; van Baak, M.A.; Roumans, N.J.T.; Vink, R.G.; Coort, S.L.M.; Renes, J.W.; Mariman, E.C.M. Adipocyte abundances of CES1, CRYAB, ENO1 and GANAB are modified in-vitro by glucose restriction and are associated with cellular remodelling during weight regain. Adipocyte 2019. [Google Scholar] [CrossRef]
- Hou, N.S.; Taubert, S. Membrane lipids and the endoplasmic reticulum unfolded protein response: An interesting relationship. Worm 2014. [Google Scholar] [CrossRef]
- Hou, N.S.; Gutschmidt, A.; Choi, D.Y.; Pather, K.; Shi, X.; Watts, J.L.; Hoppe, T.; Taubert, S. Activation of the endoplasmic reticulum unfolded protein response by lipid disequilibrium without disturbed proteostasis in vivo. Proc. Natl. Acad. Sci. USA 2014. [Google Scholar] [CrossRef] [PubMed]
- Buis, A.; Bellemin, S.; Goudeau, J.; Monnier, L.; Loiseau, N.; Guillou, H.; Aguilaniu, H. Coelomocytes Regulate Starvation-Induced Fat Catabolism and Lifespan Extension through the Lipase LIPL-5 in Caenorhabditis elegans. Cell Rep. 2019. [Google Scholar] [CrossRef] [PubMed]
- Macedo, F.; Martins, G.L.; Luévano-Martínez, L.A.; Viana, G.M.; Riske, K.A.; Inague, A.; Yoshinaga, M.Y.; Aguilaniu, H.; Miyamoto, S.; Glezer, I.; et al. Lipase-like 5 enzyme controls mitochondrial activity in response to starvation in Caenorhabditis elegans. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2020. [Google Scholar] [CrossRef]
- Miersch, C.; Döring, F. Sex differences in body composition, fat storage, and gene expression profile in Caenorhabditis elegans in response to dietary restriction. Physiol. Genom. 2013. [Google Scholar] [CrossRef]
- Purroy, R.; Britti, E.; Delaspre, F.; Tamarit, J.; Ros, J. Mitochondrial pore opening and loss of Ca2+ exchanger NCLX levels occur after frataxin depletion. Biochim. Biophys. Acta-Mol. Basis Dis. 2018. [Google Scholar] [CrossRef]
- Bishop-Bailey, D.; Thomson, S.; Askari, A.; Faulkner, A.; Wheeler-Jones, C. Lipid-Metabolizing CYPs in the Regulation and Dysregulation of Metabolism. Annu. Rev. Nutr. 2014. [Google Scholar] [CrossRef]
- Basheer, L.; Kerem, Z. Interactions between CYP3A4 and Dietary Polyphenols. Oxid. Med. Cell. Longev. 2015, 2015. [Google Scholar] [CrossRef]
- Ladage, M.L.; King, S.D.; Burks, D.J.; Quan, D.L.; Garcia, A.M.; Azad, R.K.; Padilla, P.A. Glucose or altered ceramide biosynthesis mediate oxygen deprivation sensitivity through novel pathways revealed by transcriptome analysis in Caenorhabditis elegans. G3 Genes Genomes Genet. 2016. [Google Scholar] [CrossRef]
- Imanikia, S.; Hylands, P.; Stürzenbaum, S.R. The double mutation of cytochrome P450′s and fatty acid desaturases affect lipid regulation and longevity in C. elegans. Biochem. Biophys. Rep. 2015. [Google Scholar] [CrossRef]
- Aarnio, V.; Lehtonen, M.; Storvik, M.; Callaway, J.C.; Lakso, M.; Wong, G. Caenorhabditis elegans mutants predict regulation of fatty acids and endocannabinoids by the CYP-35A gene family. Front. Pharm. 2011. [Google Scholar] [CrossRef]
- Song, S.; Zhang, X.; Wu, H.; Han, Y.; Zhang, J.; Ma, E.; Guo, Y. Molecular basis for antioxidant enzymes in mediating copper detoxification in the nematode Caenorhabditis elegans. PLoS ONE 2014, 9, e107685. [Google Scholar] [CrossRef]
- Cheng, S.C.; Li, W.H.; Shi, Y.C.; Yen, P.L.; Lin, H.Y.; Liao, V.H.C.; Chang, S.T. Antioxidant activity and delayed aging effects of hot water extract from Chamaecyparis obtusa var. formosana leaves. J. Agric. Food Chem. 2014. [Google Scholar] [CrossRef]
- Vinothiya, K.; Ashokkumar, N. Modulatory effect of vanillic acid on antioxidant status in high fat diet-induced changes in diabetic hypertensive rats. Biomed. Pharm. 2017. [Google Scholar] [CrossRef]
- He, L.; He, T.; Farrar, S.; Ji, L.; Liu, T.; Ma, X. Antioxidants Maintain Cellular Redox Homeostasis by Elimination of Reactive Oxygen Species. Cell. Physiol. Biochem. 2017, 44, 532–553. [Google Scholar] [CrossRef]
- Kenet, G.; Freedman, J.; Shenkman, B.; Regina, E.; Brok-Simoni, F.; Holzman, F.; Vavva, F.; Brand, N.; Michelson, A.; Trolliet, M.; et al. Plasma glutathione peroxidase deficiency and platelet insensitivity to nitric oxide in children with familial stroke. Arterioscler. Thromb. Vasc. Biol. 1999. [Google Scholar] [CrossRef]
- Chandler-Brown, D.; Choi, H.; Park, S.; Ocampo, B.R.; Chen, S.; Le, A.; Sutphin, G.L.; Shamieh, L.S.; Smith, E.D.; Kaeberlein, M. Sorbitol treatment extends lifespan and induces the osmotic stress response in Caenorhabditis elegans. Front. Genet. 2015. [Google Scholar] [CrossRef]
- Joshi, M.; Eagan, J.; Desai, N.K.; Newton, S.A.; Towne, M.C.; Marinakis, N.S.; Esteves, K.M.; De Ferranti, S.; Bennett, M.J.; McIntyre, A.; et al. A compound heterozygous mutation in GPD1 causes hepatomegaly, steatohepatitis, and hypertriglyceridemia. Eur. J. Hum. Genet. 2014. [Google Scholar] [CrossRef]
- Basel-Vanagaite, L.; Zevit, N.; Zahav, A.H.; Guo, L.; Parathath, S.; Pasmanik-Chor, M.; McIntyre, A.D.; Wang, J.; Albin-Kaplanski, A.; Hartman, C.; et al. Transient infantile hypertriglyceridemia, fatty liver, and hepatic fibrosis caused by mutated GPD1, encoding glycerol-3-phosphate dehydrogenase 1. Am. J. Hum. Genet. 2012. [Google Scholar] [CrossRef]
- He, H.; Sun, D.; Zeng, Y.; Wang, R.; Zhu, W.; Cao, S.; Bray, G.A.; Chen, W.; Shen, H.; Sacks, F.M.; et al. A systems genetics approach identified GPD1L and its molecular mechanism for obesity in human adipose tissue. Sci. Rep. 2017. [Google Scholar] [CrossRef]
- Fu, S.; Yang, L.; Li, P.; Hofmann, O.; Dicker, L.; Hide, W.; Lin, X.; Watkins, S.M.; Ivanov, A.R.; Hotamisligil, G.S. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature 2011. [Google Scholar] [CrossRef]
- Brenner, S. The genetics of Caenorabditis elegans. Genetics 1974. [Google Scholar] [CrossRef]
- Deline, M.L.; Vrablik, T.L.; Watts, J.L. Dietary supplementation of polyunsaturated fatty acids in Caenorhabditis elegans. J. Vis. Exp. 2013, 81, e50879. [Google Scholar] [CrossRef]
- Pino, E.C.; Webster, C.M.; Carr, C.E.; Soukas, A.A. Biochemical and high throughput microscopic assessment of fat mass in Caenorhabditis elegans. J. Vis. Exp. 2013. [Google Scholar] [CrossRef]
- Tao, J.; Wu, Q.Y.; Ma, Y.C.; Chen, Y.L.; Zou, C.G. Antioxidant response is a protective mechanism against nutrient deprivation in C. elegans. Sci. Rep. 2017. [Google Scholar] [CrossRef]
- Gardner, M.P.; Gems, D.; Viney, M.E. Aging in a very short-lived nematode. Exp. Gerontol. 2004. [Google Scholar] [CrossRef]
- Cohen, J. Statistical power analysis for the behavioral sciences. Stat. Power Anal. Behav. Sci. 1988, 84, 1096–1097. [Google Scholar]
- Nakagawa, S.; Cuthill, I.C. Effect size, confidence interval and statistical significance: A practical guide for biologists. Biol. Rev. 2007, 82, 591–605. [Google Scholar] [CrossRef]
- Wu, H.; Taki, F.A.; Zhang, Y.; Dobbins, D.L.; Pan, X. Evaluation and identification of reliable reference genes for toxicological study in Caenorhabditis elegans. Mol. Biol. Rep. 2014. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010. [Google Scholar] [CrossRef]



| Treatment | Mean Fat Content | Integrated Density | Volume | Sample Size (n) |
|---|---|---|---|---|
| NGM | 100.0 ± 0.7 | 100.0 ± 1.3 | 100.0 ± 1.3 | 490 |
| Orlistat | 76.3 ± 0.9 *** | 54.5 ± 1.4 *** | 59.4 ± 1.9 *** | 246 |
| Hesperidin | ||||
| 10 µM | 95.0 ± 1.0 *** | 90.8 ± 1.6 *** | 94.8 ± 1.5 ** | 201 |
| 100 µm | 94.7 ± 0.9 *** | 87.4 ± 1.4 *** | 90.2 ± 1.4 *** | 204 |
| 500 µM | 92.6 ± 1.5 *** | 87.3 ± 1.8 *** | 90.9 ± 1.2 *** | 105 |
| Naringin | ||||
| 10 µM | 101.3 ± 1.6 | 105.6 ± 2.4 * | 108.2 ± 1.8 ** | 118 |
| 100 µM | 95.1 ± 1.1 *** | 96.9 ± 2.5 | 102.3 ± 3.9 | 146 |
| 500 µM | 99.4 ± 2.4 | 99.4 ± 3.2 | 99.2 ± 3.2 | 98 |
| Curcumin | ||||
| 10 µM | 91.6 ± 1.3 *** | 79.5 ± 2.4 *** | 81.9 ± 2.5 *** | 115 |
| 100 µM | 101.5 ± 1.6 | 86.3 ± 3.0 *** | 82.5 ± 2.9 *** | 124 |
| 500 µM | 85.8 ± 0.8 *** | 53.7 ± 0.9 *** | 51.9 ± 1.1 *** | 110 |
| Resveratrol | ||||
| 10 µM | 101.7 ± 1.1 | 105.9 ± 1.7 ** | 103.9 ± 1.4 | 125 |
| 100 µM | 88.1 ± 0.5 *** | 77.0 ± 0.9 *** | 82.0 ± 1.1 *** | 211 |
| 500 µM | 68.2 ± 1.0 *** | 56.4 ± 1.9 *** | 63.6 ± 1.0 *** | 190 |
| Myricetin | ||||
| 10 µM | 96.5 ± 0.8 *** | 91.7 ± 1.4 *** | 92.9 ± 1.5 *** | 167 |
| 100 µM | 94.2 ± 0.7 *** | 85.3 ± 1.3 *** | 86.7 ± 1.1 *** | 207 |
| 500 µM | 95.4 ± 0.8 *** | 89.7 ± 1.3 *** | 91.8 ± 1.2 *** | 100 |
| Kaempferol | ||||
| 10 µM | 96.3 ± 0.7 *** | 91.0 ± 1.3 *** | 92.1 ± 1.3 *** | 157 |
| 100 µM | 98.4 ± 0.8 | 91.3 ± 1.5 *** | 90.4 ± 1.3 *** | 166 |
| 500 µM | 94.1 ± 0.8 *** | 86.1 ± 1.3 *** | 88.7 ± 1.3 *** | 125 |
| Epicatechin | ||||
| 10 µM | 105.3 ± 1.6 *** | 111.8 ± 2.7 *** | 109.6 ± 2.4 *** | 110 |
| 100 µM | 103.5 ± 0.9 ** | 103.2 ± 1.7 | 99.3 ± 1.5 | 150 |
| 500 µM | 107.1 ± 1.3 *** | 110.4 ± 2.3 *** | 101.5 ± 1.8 | 104 |
| Apigenin | ||||
| 10 µM | 98.30 ± 0.78 | 97.82 ± 1.34 | 98.60 ± 1.15 | 183 |
| 100 µM | 93.39 ± 0.84 *** | 89.52 ± 1.55 *** | 92.83 ± 1.56 *** | 218 |
| 500 µM | 95.96 ± 0.87 *** | 88.74 ± 1.52 *** | 89.33 ± 1.30 *** | 130 |
| Luteolin | ||||
| 10 µM | 101.0 ± 0.8 | 103.9 ± 1.6 * | 104.1 ± 1.4 * | 172 |
| 100 µM | 97.0 ± 0.8 *** | 96.6 ± 1.4 * | 99.0 ± 1.4 | 210 |
| 500 µM | 96.0 ± 0.8 *** | 92.6 ± 1.4 *** | 95.6 ± 1.3 * | 126 |
| Treatment | Mean Fat Content | Integrated Density | Volume | Sample Size (n) |
|---|---|---|---|---|
| NGM | 100.0 ± 0.9 | 100.0 ± 1.6 | 100.0 ± 1.4 | 353 |
| Orlistat | 75.3 ± 1.1 *** | 62.3 ± 1.8 *** | 72.0 ± 2.2 *** | 169 |
| p-Coumaric acid | ||||
| 10 µM | 97.8 ± 0.9 | 103.0 ± 2.0 | 108.7 ± 2.3 ** | 124 |
| 100 µM | 97.0 ± 0.9 | 97.0 ± 1.7 | 99.7 ± 1.8 | 180 |
| 500 µM | 91.9 ± 0.9 *** | 94.6 ± 1.8 ** | 103.4 ± 2.2 | 92 |
| Ellagic acid | ||||
| 10 µM | 97.7 ± 1.2 * | 102.0 ± 2.0 | 108.8 ± 2.5 ** | 133 |
| 100 µM | 93.6 ± 1.0 ** | 94.6 ± 1.9 | 100.5 ± 2.2 | 174 |
| 500 µM | 96.8 ± 1.1 | 96.3 ± 1.9 | 98.8 ± 2.1 | 103 |
| Ferulic acid | ||||
| 10 µM | 94.3 ± 0.9 *** | 95.0 ± 2.0 | 101.8 ± 2.2 | 182 |
| 100 µM | 92.1 ± 1.0 *** | 88.7 ± 2.1 *** | 95.3 ± 2.3 | 190 |
| 500 µM | 91.3 ± 0.9 *** | 91.4 ± 1.7 ** | 100.3 ± 1.7 | 119 |
| Gallic acid | ||||
| 10 µM | 94.2 ± 0.9 *** | 90.8 ± 1.2 *** | 93.5 ± 0.9 *** | 142 |
| 100 µM | 96.0 ± 1.4 ** | 94.9 ± 2.0 ** | 96.0 ± 1.6 * | 136 |
| 500 µM | 96.5 ± 1.1 ** | 92.9 ± 1.6 *** | 94.2 ± 1.2 *** | 120 |
| Vanillic acid | ||||
| 10 µM | 94.8 ± 1.1 *** | 92.9 ± 1.9 *** | 97.4 ± 1.6 | 126 |
| 100 µM | 89.2 ± 1.1 *** | 86.3 ± 1.7 *** | 94.5 ± 1.4 * | 187 |
| 500 µM | 86.6 ± 0.8 *** | 80.6 ± 1.4 *** | 89.3 ± 1.3 *** | 134 |
| Gene | Description | Human Ortholog | logFC | ANOVA p Value |
|---|---|---|---|---|
| Fatty acid and sterol biosynthesis | ||||
| F49E12.10 | Sterol biosynthesis | FAXDC2 (fatty acid hydroxylase domain containing 2) | −13.27 | 0.0007 |
| F49E12.9/drd-1 | Dietary restriction downregulated-1 | FAXDC2 (fatty acid hydroxylase domain containing 2) | −4.16 | 0.0054 |
| Lipid biosynthesis and fatty acid elongation in mitochondria | ||||
| acdh-1 | Oxidoreductase activity | ACADSB (acyl-CoA dehydrogenase short/branched chain) | −3.90 | 0.0010 |
| W03F9.4 | Carnitine O-palmitoyltransferase activity | −3.64 | 0.0035 | |
| C06E8.5 | Lipid binding activity | BPIFA2 (BPI fold containing family A member 2) | −3.21 | 0.0249 |
| hacd-1 | 3-hydroxyacyl-CoA dehydrogenase activity and NAD+ binding activity | HADH (Hydroxy-Acyl-CoA Dehydrogenase) | −2.90 | 0.0276 |
| cpt-4 | Carnitine palmitoyl transferase | CPT1A, CPT1B, and CPT1C | −2.27 | 0.0098 |
| W02F12.2 | N-acylsphingosine amidohydrolase activity. Sphingosine and ceramide biosinthesis | ACER1 (alkaline ceramidase 1) | −2.22 | 0.0465 |
| fat-7 | Fatty acid and lipid biosynthesis. stearoyl-CoA 9-desaturase activity | SCD (stearoyl-CoA desaturase) and SCD5 | −2.15 | 0.0210 |
| C06G1.1 | Lipid binding activity | BPIFA3 (BPI fold containing family A member 3) | −2.14 | 0.0147 |
| acdh-2 | Oxidoreductase activity | ACADSB (acyl-CoA dehydrogenase short/branched chain) | −2.08 | 0.0304 |
| Fatty acid oxidation and hydrolysis | ||||
| F09C8.1 | phospholipase activity | PLB1 (phospholipase B1) | 3.16 | 0.0050 |
| Y51H4A.5 | hydrolase activity | ND | 3.01 | 0.0064 |
| F25E2.3 | acyl-CoA hydrolase activity | ACOT8 (acyl-CoA thioesterase 8) | 2.16 | 0.0111 |
| Fatty acid and gluthathione metabolism | ||||
| F56A4.3 | glutathione transferase activity | GSTA5 (glutathione S-transferase alpha 5); GSTM1 (glutathione S-transferase mu 1); and GSTM3 (glutathione S-transferase mu 3) | −2.66 | 0.0397 |
| gst-14 | glutathione transferase activity | GSTA4 (glutathione S-transferase alpha 4), GSTA5 (glutathione S-transferase alpha 5), and HPGDS (hematopoietic prostaglandin D synthase) | −2.53 | 0.0154 |
| tyr-2 | Oxidoreductase activity | DCT (dopachrome tautomerase) | −2.44 | 0.0280 |
| mlt-7 | peroxidase activity | EPX (eosinophil peroxidase) | −2.33 | 0.0447 |
| C11E4.1/gpx-5 | glutathione peroxidase activity | GPX5 (glutathione peroxidase 5) and GPX6 (glutathione peroxidase 6) | −2.10 | 0.0186 |
| Gene | Description | Human Ortholog | logFC | ANOVA p Value |
|---|---|---|---|---|
| abu-10 | Activated in Blocked Unfolded protein response | ND * | −3.06 | 0.0017 |
| F44E5.5 | Heat shock protein 70 | HSPA14 (heat shock protein family A (Hsp70) member 14) HSPA4 (heat shock protein family A (Hsp70) member 4) HSPH1 (heat shock protein family H (Hsp110) member 1) | −3.00 | 0.0367 |
| abu-1 | Activated in Blocked Unfolded protein response | ND | −2.99 | 0.0231 |
| abu-9 | Activated in Blocked Unfolded protein response | ND | −2.94 | 0.0121 |
| hsp-16.1 | Heat Shock Protein | HSPB2 (heat shock protein family B (small) member 2) HSPB7 (heat shock protein family B (small) member 7) CRYAA (crystallin alpha A) | −2.92 | 0.0404 |
| abu-8 | Activated in Blocked Unfolded protein response | ND | −2.91 | 0.0301 |
| F32D8.7 | AMBP (alpha-1-microglobulin/bikunin precursor) LRP11 (LDL receptor related protein 11) SPINT1 (serine peptidase inhibitor, Kunitz type 1) | −2.87 | 0.0479 | |
| D2096.6 | ND | −2.67 | 0.0229 | |
| F41E6.11 | ND | −2.58 | 0.0120 | |
| hsp-16.11 | Heat Shock Protein | HSPB2 (heat shock protein family B (small) member 2) HSPB7 (heat shock protein family B (small) member 7) CRYAA (crystallin alpha A) | −2.45 | 0.0296 |
| abu-11 | Activated in Blocked Unfolded protein response | ND | −2.45 | 0.0135 |
| CELE_T06E4.8 | ND | -2.27 | 0.0126 | |
| abu-6 | Activated in Blocked Unfolded protein response | ND | −2.22 | 0.0192 |
| abu-5 | Activated in Blocked Unfolded protein response | KRTAP10-1 (keratin associated protein 10-1) KRTAP10-4 (keratin associated protein 10-4) KRTAP10-7 (keratin associated protein 10-7) | −2.18 | 0.0204 |
| pqn-74 | ND | −2.03 | 0.0282 | |
| aagr-4 | carbohydrate binding activity and hydrolase activity, hydrolyzing O-glycosyl compounds | GANAB (glucosidase II alpha subunit) | −2.01 | 0.0493 |
| Gene | Description | Human Ortholog | logFC | ANOVA p Value |
|---|---|---|---|---|
| Lipid metabolism and transmembrane transport | ||||
| lipl-5 | Lipoprotein lipase-5. Lipid storage, degradation, lipid homeostasis and localization. TAG catabolism | LIPA (lipase A, lysosomal acid type); LIPF (lipase F, gastric type) LIPM (lipase family member M) | 1.73 | 0.0431 |
| pgp-9 | ATP-binding cassette (ABC) transporter superfamily. Transmembrane transport | ABCB1 (ATP binding cassette subfamily B member 1) ABCB11 (ATP binding cassette subfamily B member 11) | −1.77 | 0.0037 |
| ncx-8 | Na/Ca exchangers. Lipid storage Lipid storage, localization, and transmembrane transport. | SLC8B1 (solute carrier family 8 member B1) | −1.67 | 0.0097 |
| pcyt-1 | Phosphocholine cytidylyltransferase. choline-phosphate. Phosphatidylcholine biosynthetic process, lipid metabolic process, phospholipid biosynthetic process | PCYT1B (phosphate cytidylyltransferase 1, choline, beta). | 1.5 | 0.0070 |
| gba-2 | Beta-Glucocere Brosidase with hydrolase activity. Lipid metabolic process, sphingolipid metabolic process and carbohydrate metabolic process | GBA (glucosylceramidase beta) | −1.53 | 0.0487 |
| Stress response and detoxification | ||||
| cyp-35C1 | Cytochrome P450 family with oxidation-reduction function, xenobiotic metabolic process | CYP2E1 (cytochrome P450 family 2 subfamily E member 1) CYP2C18 (cytochrome P450 family 2 subfamily C member 18) CYP2D7 (cytochrome P450 family 2 subfamily D member 7 (gene/pseudogene)) | −1.94 | 0.0194 |
| cyp-37B1 | Cytochrome P450 family with oxidation-reduction function | CYP4A11 (cytochrome P450 family 4 subfamily A member 11) CYP4B1 (cytochrome P450 family 4 subfamily B member 1) CYP4F3 (cytochrome P450 family 4 subfamily F member 3) | −1.65 | 0.0124 |
| irg-2 | Infection response gene. Innate immune response | ND * | −1.62 | 0.0229 |
| cyp-14A2 | Cytochrome P450 family with oxidation-reduction function. Monooxygenase activity | CYP2U1 (cytochrome P450 family 2 subfamily U member 1) | −1.61 | 0.0252 |
| cyp-33C8 | Cytochrome P450 family with oxidation-reduction function. Fatty acid metabolism | CYP2J2 (cytochrome P450 family 2 subfamily J member 2) | −1.56 | 0.0003 |
| gpx-3 (C11E4.2) | Glutathione peroxidase activity Glutathione metabolism Arachidonic acid metabolism Response to oxidative stress Stress response | GPX3 (glutathione peroxidase 3) | 1.73 | 0.0010 |
| cyp-13A6 | Cytochrome P450 family. Stress response. monooxygenase activity involved in oxidation-reduction process. Xenobiotic response | CYP3A4 (cytochrome P450 family 3 subfamily A member 4) | 2.81 | 0.0005 |
| Gene | Function | Human Ortholog | logFC | ANOVA p-Value |
|---|---|---|---|---|
| UPRER response genes | ||||
| pqn-57 | Prion-like-(Q/N-rich)-domain-bearing protein. Involved in endoplasmic reticulum unfolded protein response. | ND * | −1.78 | 0.0317 |
| hsp-16.48 | Unfolded protein binding activity. IRE1-mediated unfolded protein response | HSPB1 (heat shock protein family B (small) member 1), HSPB2 (heat shock protein family B (small) member 2) and CRYAA (crystallin alpha A) | 1.51 | 0.0110 |
| hsp-16.2 | Unfolded protein binding activity | HSPB1 (heat shock protein family B (small) member 1), CRYAA (crystallin alpha A), CRYAB (crystallin alpha B) | 1.98 | 0.0111 |
| F44E5.5 | Unfolded protein binding activity, misfolded protein binding activity, ATP binding activity | HSPA14 (heat shock protein family A (Hsp70) member 14); HSPA4 (heat shock protein family A (Hsp70) member 4); and HSPH1 (heat shock protein family H (Hsp110) member 1 | 2.04 | 0.0341 |
| Other stress-response genes | ||||
| C11E4.2/gpx-3 | Glutathione peroxidase activity | GPX3 (glutathione peroxidase 3) | 1.84 | 0.0027 |
| cyp-37B1 | Cytochrome P450 family member with oxidoreductase activity involved in defense response to Gram-positive bacterium. | CYP4A11 (cytochrome P450 family 4 subfamily A member 11); CYP4B1 (cytochrome P450 family 4 subfamily B member 1); and CYP4F3 (cytochrome P450 family 4 subfamily F member 3). | −1.63 | 0.0172 |
| fmo-3 | Monooxygenase activity | FMO5 (flavin containing dimethylaniline monoxygenase 5) | 1.71 | 0.0099 |
| irg-5/CELE_F35E12.5 | Defense response to Gram-positive bacterium. | ND | 1.53 | 0.0393 |
| Other genes | ||||
| gpdh-1 | glycerol-3-phosphate dehydrogenase (quinone) activity and glycerol-3-phosphate dehydrogenase [NAD+] activity | GPD1 (glycerol-3-phosphate dehydrogenase 1) and GPD1L (glycerol-3-phosphate dehydrogenase 1 like) | 1.7 | 0.0130 |
| arl-6 | Exhibits GTPase activity. Is involved in intracellular transport | ARL6 (ADP ribosylation factor like GTPase 6) | −1.75 | 0.0267 |
| dop-3 | Exhibits dopamine neurotransmitter receptor activity, coupled via Gi/Go | DRD2 (dopamine receptor D2) | −1.53 | 0.0205 |
| aat-7 | l-amino acid transmembrane transporter activity. | SLC7A11 (solute carrier family 7 member 11); SLC7A7 (solute carrier family 7 member 7); and SLC7A9 (solute carrier family 7 member 9) | 1.52 | 0.0027 |
| ugt-63 | UDP-glycosyltransferase activity | ND | 1.52 | 0.0076 |
| glb-33 | Heme binding activity and oxygen binding activity | ND | 1.54 | 0.0222 |
| F35B12.4/piit-1 | Serine-type endopeptidase inhibitor activity | TFPI2 (tissue factor pathway inhibitor 2) | 1.63 | 0.0420 |
| str-130 | G protein-coupled olfactory receptor activity | ND | 1.7 | 0.0226 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aranaz, P.; Navarro-Herrera, D.; Zabala, M.; Romo-Hualde, A.; López-Yoldi, M.; Vizmanos, J.L.; Milagro, F.I.; González-Navarro, C.J. Phenolic Compounds Reduce the Fat Content in Caenorhabditis elegans by Affecting Lipogenesis, Lipolysis, and Different Stress Responses. Pharmaceuticals 2020, 13, 355. https://doi.org/10.3390/ph13110355
Aranaz P, Navarro-Herrera D, Zabala M, Romo-Hualde A, López-Yoldi M, Vizmanos JL, Milagro FI, González-Navarro CJ. Phenolic Compounds Reduce the Fat Content in Caenorhabditis elegans by Affecting Lipogenesis, Lipolysis, and Different Stress Responses. Pharmaceuticals. 2020; 13(11):355. https://doi.org/10.3390/ph13110355
Chicago/Turabian StyleAranaz, Paula, David Navarro-Herrera, María Zabala, Ana Romo-Hualde, Miguel López-Yoldi, José Luis Vizmanos, Fermín I. Milagro, and Carlos J. González-Navarro. 2020. "Phenolic Compounds Reduce the Fat Content in Caenorhabditis elegans by Affecting Lipogenesis, Lipolysis, and Different Stress Responses" Pharmaceuticals 13, no. 11: 355. https://doi.org/10.3390/ph13110355
APA StyleAranaz, P., Navarro-Herrera, D., Zabala, M., Romo-Hualde, A., López-Yoldi, M., Vizmanos, J. L., Milagro, F. I., & González-Navarro, C. J. (2020). Phenolic Compounds Reduce the Fat Content in Caenorhabditis elegans by Affecting Lipogenesis, Lipolysis, and Different Stress Responses. Pharmaceuticals, 13(11), 355. https://doi.org/10.3390/ph13110355






