C3orf70 Is Involved in Neural and Neurobehavioral Development
Abstract
1. Introduction
2. Results
2.1. Comparative Transcriptome Analysis Reveals Common Target Genes of Neurog1/2 and Ascl1 in Stem Cells
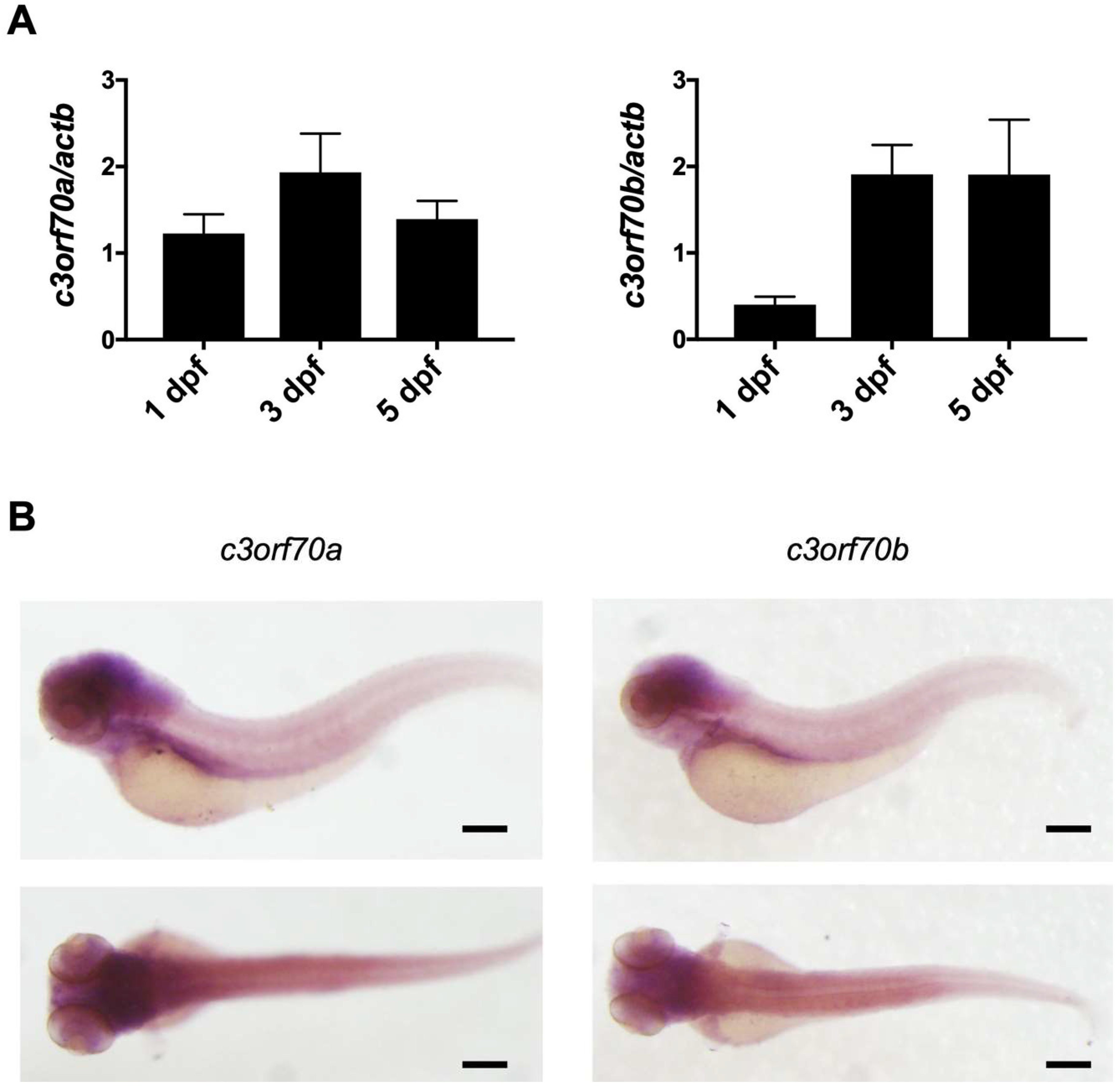
2.2. Zebrafish Orthologs of C3orf70 Are Expressed in the Larval Midbrain and Hindbrain
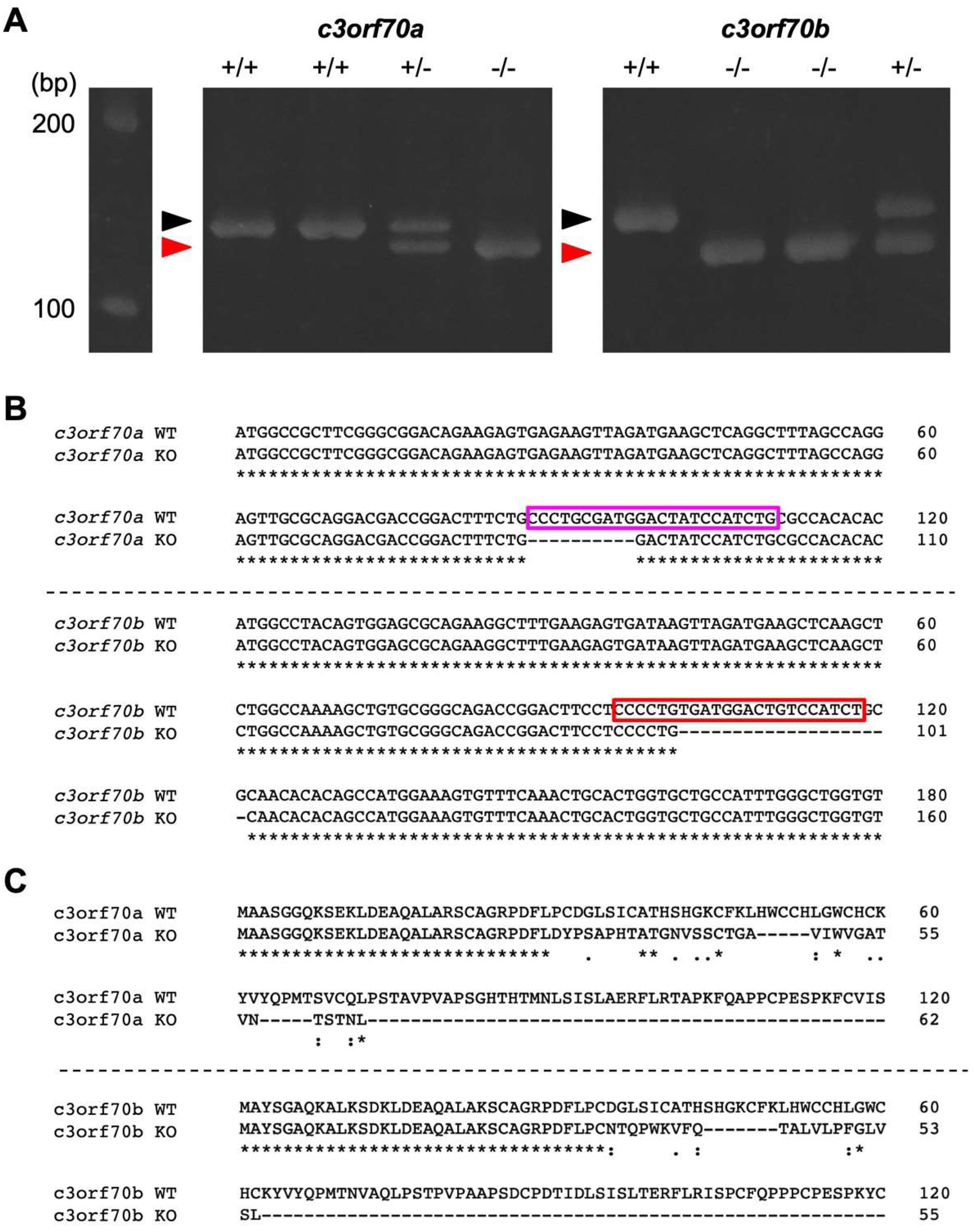
2.3. Generation of c3orf70-KO Zebrafish
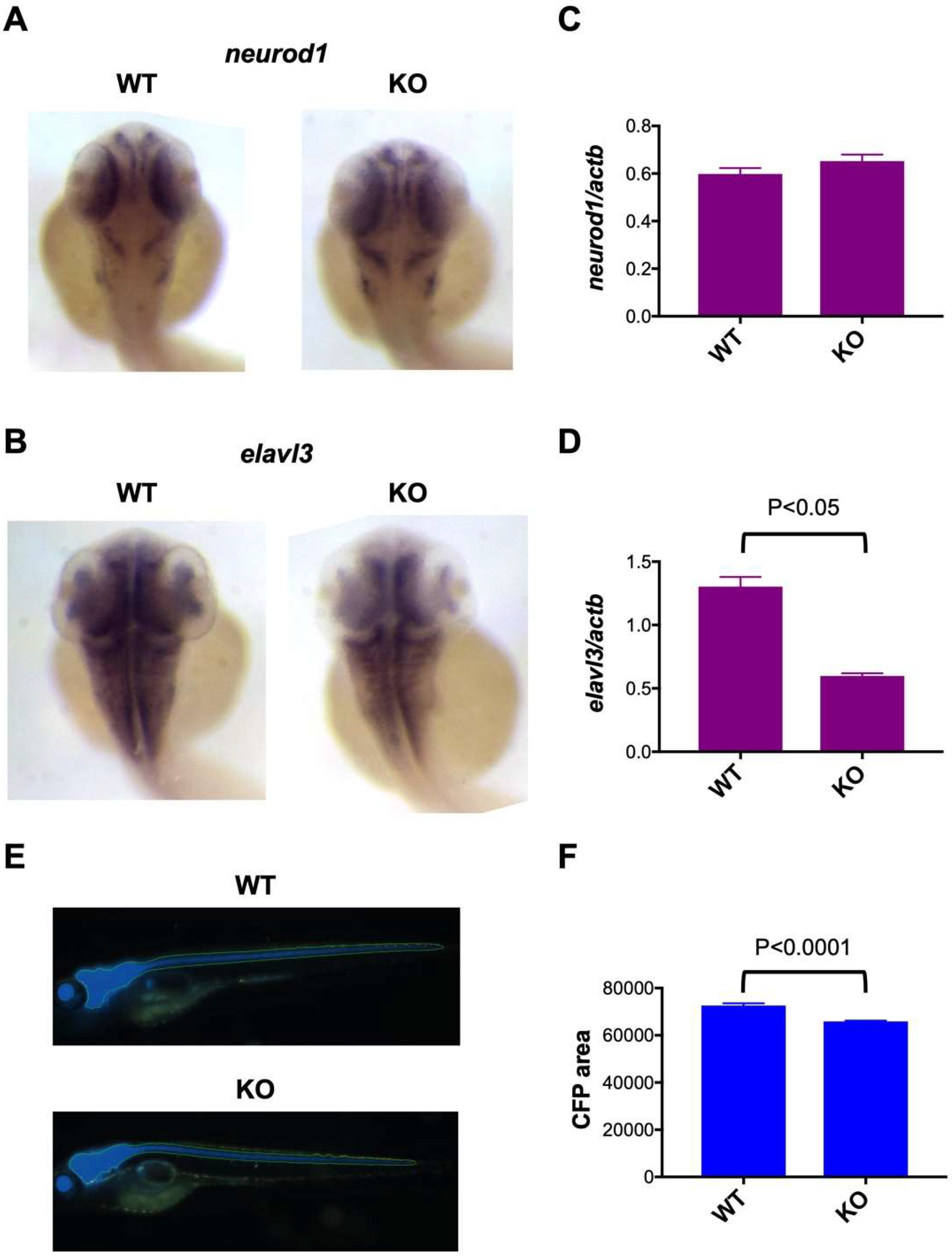
2.4. Impaired Neuronal Marker Expression in c3orf70-KO Zebrafish
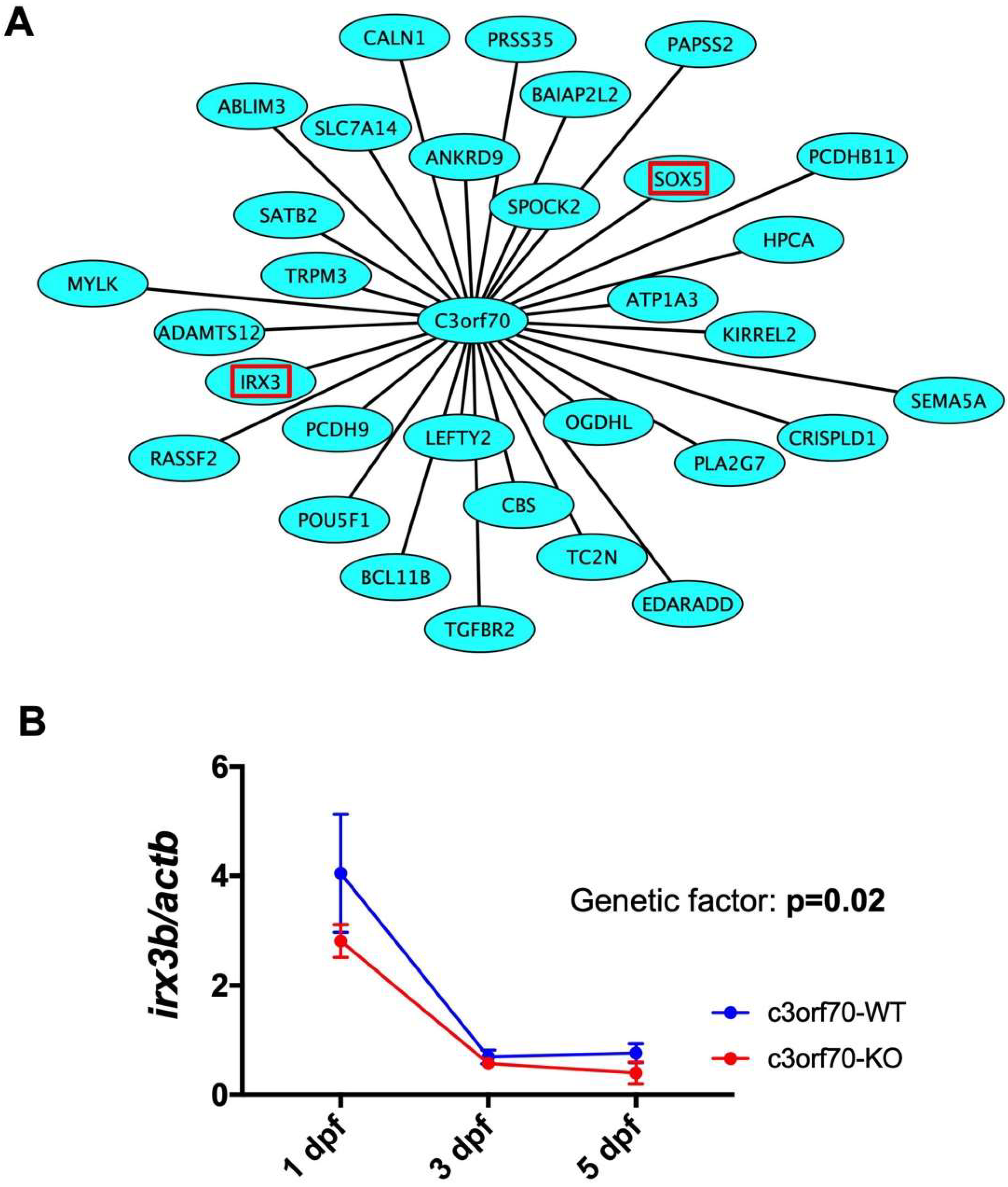
2.5. WGCNA Identifies IRX3 as a Gene Coexpressed with C3orf70 During Neurogenesis
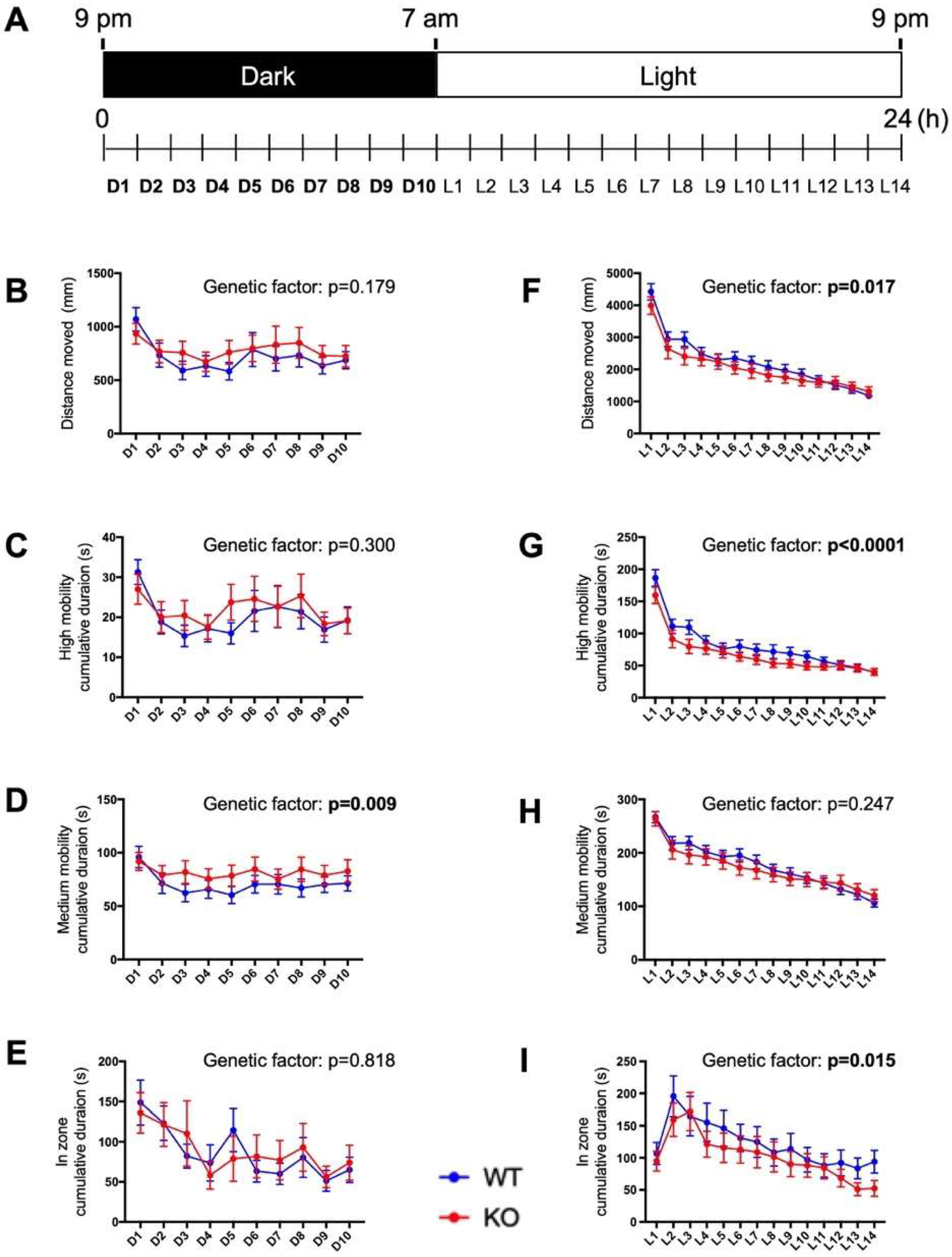
2.6. Circadian Behavioral Responses Are Impaired in c3orf70-KO Zebrafish
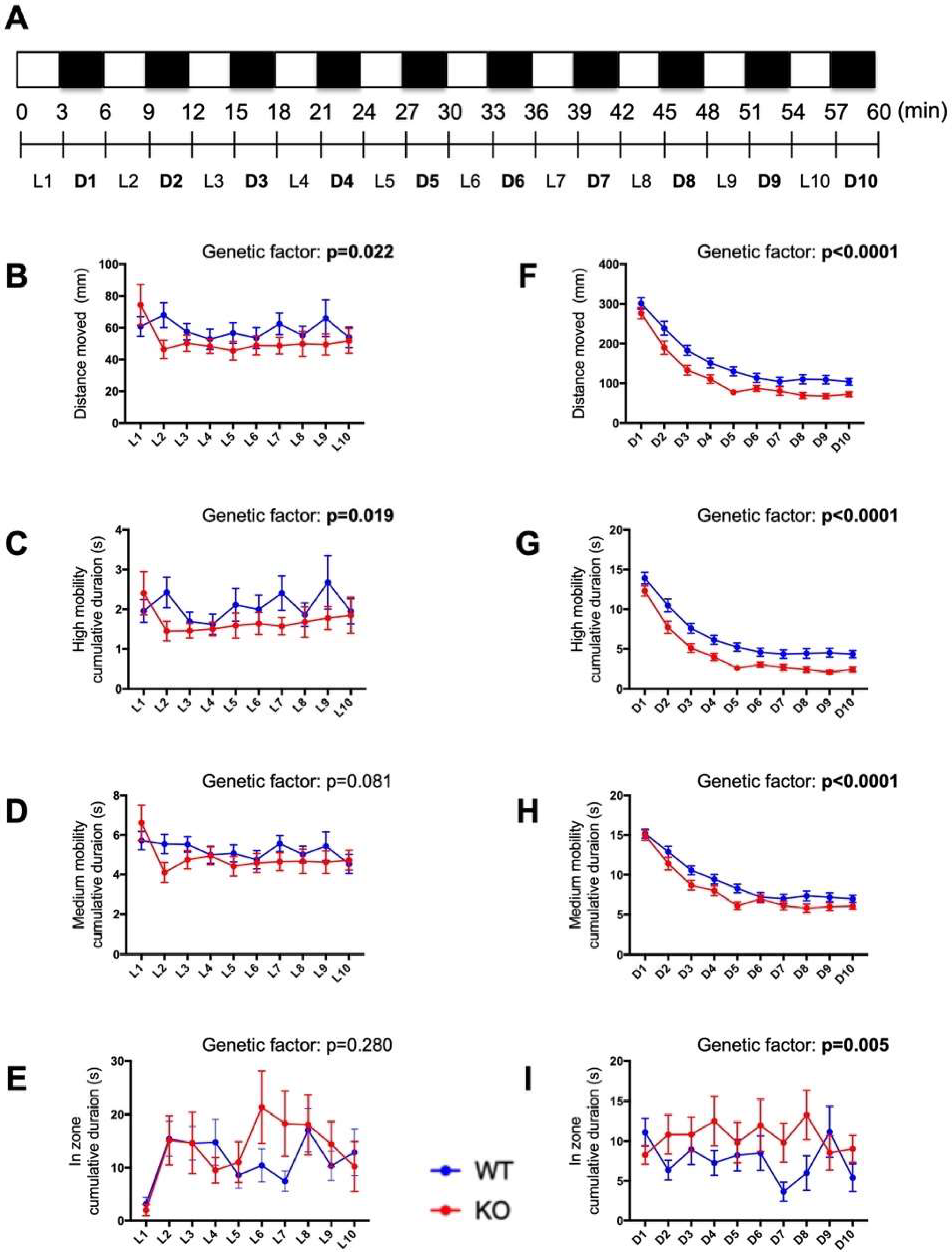
2.7. Behavioral Responses to Alternating Light–Dark Cycles Is Impaired in c3orf70-KO Zebrafish
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Comparative Transcriptome Analysis
4.3. Bioinformatic Analysis of Common DEGs
4.4. Zebrafish Husbandry
4.5. Generation of c3orf70-KO Zebrafish
4.6. Whole-Mount In Situ Hybridization of Neuronal Markers
4.7. qPCR Analysis
4.8. Weighted Gene Coexpression Network Analysis
4.9. In Vivo Imaging of Tg (eno2: Cerulean) Zebrafish
4.10. Behavioral Analysis
4.11. Statistical analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marchetto, M.C.; Belinson, H.; Tian, Y.; Freitas, B.C.; Fu, C.; Vadodaria, K.; Beltrao-Braga, P.; Trujillo, C.A.; Mendes, A.P.D.; Padmanabhan, K.; et al. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol. Psychiatry 2017, 22, 820–835. [Google Scholar] [CrossRef] [PubMed]
- Sacco, R.; Cacci, E.; Novarino, G. Neural stem cells in neuropsychiatric disorders. Curr. Opin. Neurobiol. 2018, 48, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Adhya, D.; Swarup, V.; Nagy, R.; Shum, C.; Nowosiad, P.; Jozwik, K.; Lee, I.; Skuse, D.; Flinter, F.A.; McAlonan, G.; et al. Atypical neurogenesis and excitatory-inhibitory progenitor generation in induced pluripotent stem cell (iPSC) from autistic individuals. bioRxiv 2019. [Google Scholar] [CrossRef]
- Nakano-Kobayashi, A.; Awaya, T.; Kii, I.; Sumida, Y.; Okuno, Y.; Yoshida, S.; Sumida, T.; Inoue, H.; Hosoya, T.; Hagiwara, M. Prenatal neurogenesis induction therapy normalizes brain structure and function in Down syndrome mice. Proc. Natl. Acad. Sci. USA 2017, 114, 10268–10273. [Google Scholar] [CrossRef] [PubMed]
- Bardoni, B.; Capovilla, M.; Lalli, E. Modeling Fragile X syndrome in neurogenesis: An unexpected phenotype and a novel tool for future therapies. Neurogenesis 2017, 4, e1270384. [Google Scholar] [CrossRef][Green Version]
- Chaudhury, D.; Liu, H.; Han, M.H. Neuronal correlates of depression. Cell. Mol. Life Sci. 2015, 72, 4825–4848. [Google Scholar] [CrossRef]
- Baptista, P.; Andrade, J.P. Adult Hippocampal Neurogenesis: Regulation and Possible Functional and Clinical Correlates. Front. Neuroanat. 2018, 12, 44. [Google Scholar] [CrossRef]
- Inta, D.; Lang, U.E.; Borgwardt, S.; Meyer-Lindenberg, A.; Gass, P. Microglia Activation and Schizophrenia: Lessons From the Effects of Minocycline on Postnatal Neurogenesis, Neuronal Survival and Synaptic Pruning. Schizophr. Bull. 2017, 43, 493–496. [Google Scholar] [CrossRef]
- Hartenstein, V.; Stollewerk, A. The evolution of early neurogenesis. Dev. Cell 2015, 32, 390–407. [Google Scholar] [CrossRef]
- Wilkinson, G.; Dennis, D.; Schuurmans, C. Proneural genes in neocortical development. Neuroscience 2013, 253, 256–273. [Google Scholar] [CrossRef]
- Guillemot, F.; Hassan, B.A. Beyond proneural: Emerging functions and regulations of proneural proteins. Curr. Opin. Neurobiol. 2017, 42, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Yamamizu, K.; Piao, Y.; Sharov, A.A.; Zsiros, V.; Yu, H.; Nakazawa, K.; Schlessinger, D.; Ko, M.S. Identification of transcription factors for lineage-specific ESC differentiation. Stem Cell Rep. 2013, 1, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Busskamp, V.; Lewis, N.E.; Guye, P.; Ng, A.H.; Shipman, S.L.; Byrne, S.M.; Sanjana, N.E.; Murn, J.; Li, Y.; Li, S.; et al. Rapid neurogenesis through transcriptional activation in human stem cells. Mol. Syst. Biol. 2014, 10, 760. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Troup, D.B.; Wilhite, S.E.; Ledoux, P.; Rudnev, D.; Evangelista, C.; Kim, I.F.; Soboleva, A.; Tomashevsky, M.; Marshall, K.A.; et al. NCBI GEO: Archive for high-throughput functional genomic data. Nucleic Acids Res. 2009, 37, D885–D890. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Hardwick, L.J.; Philpott, A. Multi-site phosphorylation regulates NeuroD4 activity during primary neurogenesis: A conserved mechanism amongst proneural proteins. Neural Dev. 2015, 10, 15. [Google Scholar] [CrossRef]
- Huang, H.S.; Redmond, T.M.; Kubish, G.M.; Gupta, S.; Thompson, R.C.; Turner, D.L.; Uhler, M.D. Transcriptional regulatory events initiated by Ascl1 and Neurog2 during neuronal differentiation of P19 embryonic carcinoma cells. J. Mol. Neurosci. 2015, 55, 684–705. [Google Scholar] [CrossRef]
- Khan, A.; Fornes, O.; Stigliani, A.; Gheorghe, M.; Castro-Mondragon, J.A.; van der Lee, R.; Bessy, A.; Cheneby, J.; Kulkarni, S.R.; Tan, G.; et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018, 46, D260–D266. [Google Scholar] [CrossRef]
- Cho, J.H.; Tsai, M.J. The role of BETA2/NeuroD1 in the development of the nervous system. Mol. Neurobiol. 2004, 30, 35–47. [Google Scholar] [CrossRef]
- Ince-Dunn, G.; Okano, H.J.; Jensen, K.B.; Park, W.Y.; Zhong, R.; Ule, J.; Mele, A.; Fak, J.J.; Yang, C.; Zhang, C.; et al. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron 2012, 75, 1067–1080. [Google Scholar] [CrossRef]
- Isgro, M.A.; Bottoni, P.; Scatena, R. Neuron-Specific Enolase as a Biomarker: Biochemical and Clinical Aspects. Adv. Exp. Med. Biol. 2015, 867, 125–143. [Google Scholar] [PubMed]
- Oldham, M.C.; Konopka, G.; Iwamoto, K.; Langfelder, P.; Kato, T.; Horvath, S.; Geschwind, D.H. Functional organization of the transcriptome in human brain. Nat. Neurosci. 2008, 11, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, G.Z. Understanding Molecular Mechanisms of the Brain Through Transcriptomics. Front. Physiol. 2019, 10, 214. [Google Scholar] [CrossRef] [PubMed]
- Bosse, A.; Zulch, A.; Becker, M.B.; Torres, M.; Gomez-Skarmeta, J.L.; Modolell, J.; Gruss, P. Identification of the vertebrate Iroquois homeobox gene family with overlapping expression during early development of the nervous system. Mech. Dev. 1997, 69, 169–181. [Google Scholar] [CrossRef]
- Robertshaw, E.; Matsumoto, K.; Lumsden, A.; Kiecker, C. Irx3 and Pax6 establish differential competence for Shh-mediated induction of GABAergic and glutamatergic neurons of the thalamus. Proc. Natl. Acad. Sci. USA 2013, 110, E3919–E3926. [Google Scholar] [CrossRef]
- Cooper, J.M.; Halter, K.A.; Prosser, R.A. Circadian rhythm and sleep-wake systems share the dynamic extracellular synaptic milieu. Neurobiol. Sleep Circadian Rhythm. 2018, 5, 15–36. [Google Scholar] [CrossRef]
- MacPhail, R.C.; Brooks, J.; Hunter, D.L.; Padnos, B.; Irons, T.D.; Padilla, S. Locomotion in larval zebrafish: Influence of time of day, lighting and ethanol. Neurotoxicology 2009, 30, 52–58. [Google Scholar] [CrossRef]
- Nishimura, Y.; Murakami, S.; Ashikawa, Y.; Sasagawa, S.; Umemoto, N.; Shimada, Y.; Tanaka, T. Zebrafish as a systems toxicology model for developmental neurotoxicity testing. Congenit. Anom. 2015, 55, 1–16. [Google Scholar] [CrossRef]
- Venkatraman, A.; Edlow, B.L.; Immordino-Yang, M.H. The Brainstem in Emotion: A Review. Front. Neuroanat. 2017, 11, 15. [Google Scholar] [CrossRef]
- Lecaudey, V.; Anselme, I.; Dildrop, R.; Ruther, U.; Schneider-Maunoury, S. Expression of the zebrafish Iroquois genes during early nervous system formation and patterning. J. Comp. Neurol. 2005, 492, 289–302. [Google Scholar] [CrossRef]
- Rzehak, P.; Covic, M.; Saffery, R.; Reischl, E.; Wahl, S.; Grote, V.; Weber, M.; Xhonneux, A.; Langhendries, J.P.; Ferre, N.; et al. DNA-Methylation and Body Composition in Preschool Children: Epigenome-Wide-Analysis in the European Childhood Obesity Project (CHOP)-Study. Sci. Rep. 2017, 7, 14349. [Google Scholar] [CrossRef] [PubMed]
- De Araujo, T.M.; Razolli, D.S.; Correa-da-Silva, F.; De Lima-Junior, J.C.; Gaspar, R.S.; Sidarta-Oliveira, D.; Victorio, S.C.; Donato, J., Jr.; Kim, Y.B.; Velloso, L.A. The partial inhibition of hypothalamic IRX3 exacerbates obesity. EBioMedicine 2019, 39, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Lai, T.; Jabaudon, D.; Molyneaux, B.J.; Azim, E.; Arlotta, P.; Menezes, J.R.; Macklis, J.D. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron 2008, 57, 232–247. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, J.A.; Ballif, B.C.; Torchia, B.S.; Sahoo, T.; Ravnan, J.B.; Schultz, R.; Lamb, A.; Bejjani, B.A.; Shaffer, L.G. Copy number variations associated with autism spectrum disorders contribute to a spectrum of neurodevelopmental disorders. Genet. Med. 2010, 12, 694–702. [Google Scholar] [CrossRef]
- Lamb, A.N.; Rosenfeld, J.A.; Neill, N.J.; Talkowski, M.E.; Blumenthal, I.; Girirajan, S.; Keelean-Fuller, D.; Fan, Z.; Pouncey, J.; Stevens, C.; et al. Haploinsufficiency of SOX5 at 12p12.1 is associated with developmental delays with prominent language delay, behavior problems, and mild dysmorphic features. Hum. Mutat. 2012, 33, 728–740. [Google Scholar] [CrossRef]
- Lee, J.J.; Wedow, R.; Okbay, A.; Kong, E.; Maghzian, O.; Zacher, M.; Nguyen-Viet, T.A.; Bowers, P.; Sidorenko, J.; Karlsson Linner, R.; et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018, 50, 1112–1121. [Google Scholar] [CrossRef]
- Power, R.A.; Tansey, K.E.; Buttenschon, H.N.; Cohen-Woods, S.; Bigdeli, T.; Hall, L.S.; Kutalik, Z.; Lee, S.H.; Ripke, S.; Steinberg, S.; et al. Genome-wide Association for Major Depression Through Age at Onset Stratification: Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Biol. Psychiatry 2017, 81, 325–335. [Google Scholar] [CrossRef]
- Lane, J.M.; Jones, S.E.; Dashti, H.S.; Wood, A.R.; Aragam, K.G.; Van Hees, V.T.; Strand, L.B.; Winsvold, B.S.; Wang, H.; Bowden, J.; et al. Biological and clinical insights from genetics of insomnia symptoms. Nat. Genet. 2019, 51, 387–393. [Google Scholar] [CrossRef]
- Guthrie, S. Patterning and axon guidance of cranial motor neurons. Nat. Rev. Neurosci. 2007, 8, 859–871. [Google Scholar] [CrossRef]
- Kasahara, K.; Aoki, H.; Kiyono, T.; Wang, S.; Kagiwada, H.; Yuge, M.; Tanaka, T.; Nishimura, Y.; Mizoguchi, A.; Goshima, N.; et al. EGF receptor kinase suppresses ciliogenesis through activation of USP8 deubiquitinase. Nat. Commun. 2018, 9, 758. [Google Scholar] [CrossRef]
- Naert, T.; Vleminckx, K. CRISPR/Cas9 disease models in zebrafish and Xenopus: The genetic renaissance of fish and frogs. Drug Discov. Today Technol. 2018, 28, 41–52. [Google Scholar] [CrossRef]
- Ashikawa, Y.; Nishimura, Y.; Okabe, S.; Sasagawa, S.; Murakami, S.; Yuge, M.; Kawaguchi, K.; Kawase, R.; Tanaka, T. Activation of Sterol Regulatory Element Binding Factors by Fenofibrate and Gemfibrozil Stimulates Myelination in Zebrafish. Front. Pharmacol. 2016, 7, 206. [Google Scholar] [CrossRef] [PubMed]
- Cornet, C.; Di Donato, V.; Terriente, J. Combining Zebrafish and CRISPR/Cas9: Toward a More Efficient Drug Discovery Pipeline. Front. Pharmacol. 2018, 9, 703. [Google Scholar] [CrossRef]
- Mueller, T. What is the Thalamus in Zebrafish? Front. Neurosci. 2012, 6, 64. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Strähle, U.; Scholpp, S. Neurogenesis in zebrafish—From embryo to adult. Neural Dev. 2013, 8, 3. [Google Scholar] [CrossRef]
- Gunnarsson, L.; Jauhiainen, A.; Kristiansson, E.; Nerman, O.; Larsson, D.G. Evolutionary conservation of human drug targets in organisms used for environmental risk assessments. Environ. Sci. Technol. 2008, 42, 5807–5813. [Google Scholar] [CrossRef] [PubMed]
- Rihel, J.; Schier, A.F. Behavioral screening for neuroactive drugs in zebrafish. Dev. Neurobiol. 2012, 72, 373–385. [Google Scholar] [CrossRef]
- Perkins, E.J.; Ankley, G.T.; Crofton, K.M.; Garcia-Reyero, N.; LaLone, C.A.; Johnson, M.S.; Tietge, J.E.; Villeneuve, D.L. Current perspectives on the use of alternative species in human health and ecological hazard assessments. Environ. Health Perspect. 2013, 121, 1002–1010. [Google Scholar] [CrossRef]
- MacRae, C.A.; Peterson, R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015, 14, 721–731. [Google Scholar] [CrossRef]
- Nishimura, Y.; Inoue, A.; Sasagawa, S.; Koiwa, J.; Kawaguchi, K.; Kawase, R.; Maruyama, T.; Kim, S.; Tanaka, T. Using zebrafish in systems toxicology for developmental toxicity testing. Congenit. Anom. 2016, 56, 18–27. [Google Scholar] [CrossRef]
- Lam, P.Y.; Peterson, R.T. Developing zebrafish disease models for in vivo small molecule screens. Curr. Opin. Chem. Biol. 2019, 50, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets–Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Breitling, R.; McEntee, C.W.; Wittner, B.S.; Nemhauser, J.L.; Chory, J. RankProd: A bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 2006, 22, 2825–2827. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Sasagawa, S.; Nishimura, Y.; Hayakawa, Y.; Murakami, S.; Ashikawa, Y.; Yuge, M.; Okabe, S.; Kawaguchi, K.; Kawase, R.; Tanaka, T. E2F4 promotes neuronal regeneration and functional recovery after spinal cord injury in zebrafish. Front. Pharmacol. 2016, 7, 119. [Google Scholar]
- Sasagawa, S.; Nishimura, Y.; Sawada, H.; Zhang, E.; Murakami, S.; Ashikawa, Y.; Yuge, M.; Okabe, S.; Kawaguchi, K.; Kawase, R.; et al. Comparative transcriptome analysis identifies CCDC80 as a novel gene associated with pulmonary arterial hypertension. Front. Pharmacol. 2016, 7, 142. [Google Scholar]
- Sasagawa, S.; Nishimura, Y.; Okabe, S.; Murakami, S.; Ashikawa, Y.; Yuge, M.; Kawaguchi, K.; Kawase, R.; Okamoto, R.; Ito, M.; et al. Downregulation of GSTK1 Is a Common Mechanism Underlying Hypertrophic Cardiomyopathy. Front. Pharmacol. 2016, 7, 162. [Google Scholar] [CrossRef]
- Kotani, H.; Taimatsu, K.; Ohga, R.; Ota, S.; Kawahara, A. Efficient Multiple Genome Modifications Induced by the crRNAs, tracrRNA and Cas9 Protein Complex in Zebrafish. PLoS ONE 2015, 10, e0128319. [Google Scholar] [CrossRef]
- Nishimura, Y.; Okabe, S.; Sasagawa, S.; Murakami, S.; Ashikawa, Y.; Yuge, M.; Kawaguchi, K.; Kawase, R.; Tanaka, T. Pharmacological profiling of zebrafish behavior using chemical and genetic classification of sleep-wake modifiers. Front. Pharmacol. 2015, 6, 257. [Google Scholar] [CrossRef]
- Ashikawa, Y.; Nishimura, Y.; Okabe, S.; Sato, Y.; Yuge, M.; Tada, T.; Miyao, H.; Murakami, S.; Kawaguchi, K.; Sasagawa, S.; et al. Potential protective function of the sterol regulatory element binding factor 1-fatty acid desaturase 1/2 axis in early-stage age-related macular degeneration. Heliyon 2017, 3, e00266. [Google Scholar] [CrossRef]
- Matsui, T.; Thitamadee, S.; Murata, T.; Kakinuma, H.; Nabetani, T.; Hirabayashi, Y.; Hirate, Y.; Okamoto, H.; Bessho, Y. Canopy1, a positive feedback regulator of FGF signaling, controls progenitor cell clustering during Kupffer’s vesicle organogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 9881–9886. [Google Scholar] [CrossRef] [PubMed]
- Thisse, B.; Thisse, C. In situ hybridization on whole-mount zebrafish embryos and young larvae. Methods Mol. Biol. 2014, 1211, 53–67. [Google Scholar] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinf. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
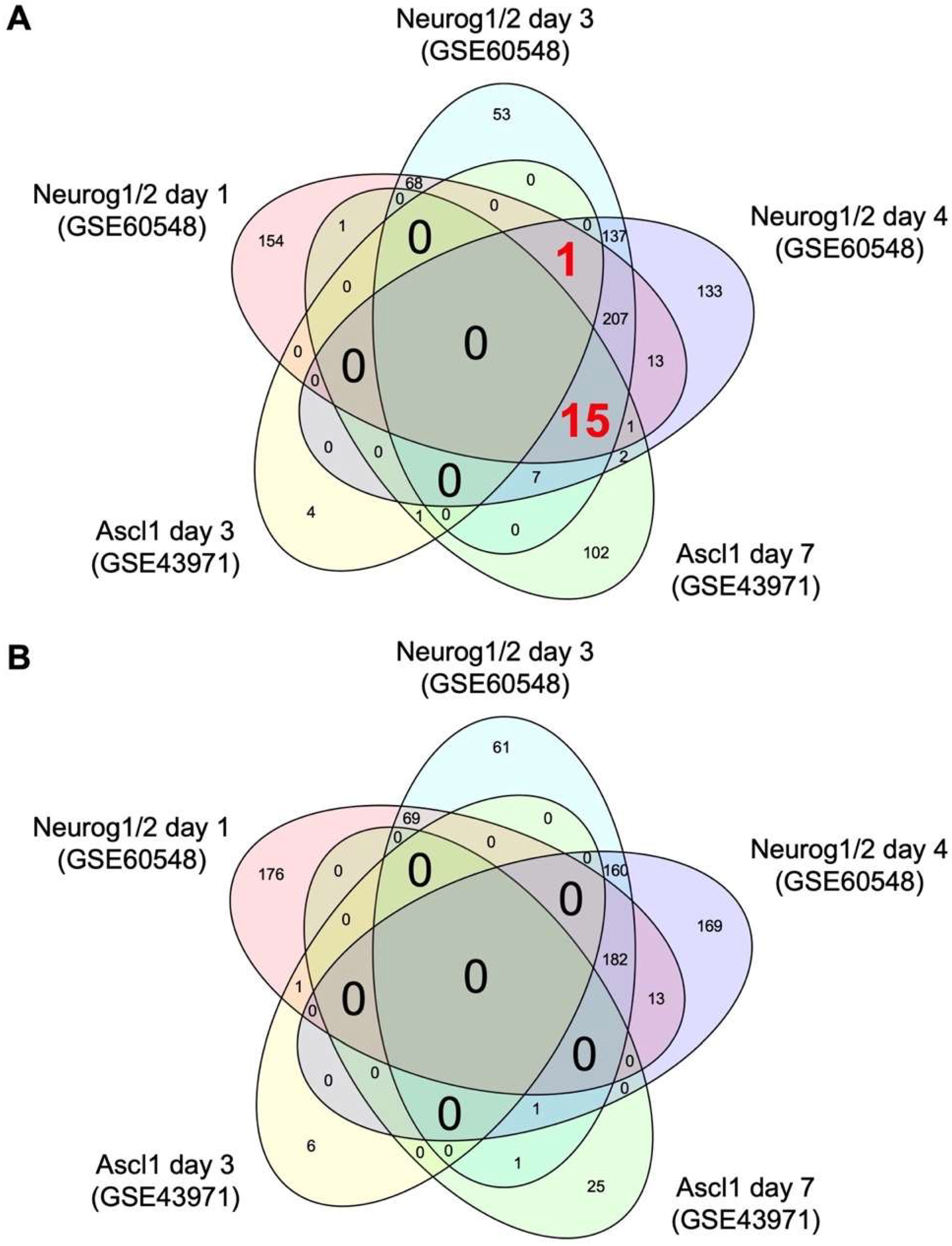
| Symbol | Neurog1/2 Day 1 | Neurog1/2 Day 3 | Neurog1/2 Day 4 | Ascl1 Day 3 | Ascl1 Day 7 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| FC | FDR | FC | FDR | FC | FDR | FC | FDR | FC | FDR | |
| C3orf70 | 8.13 | 2.761 × 10−2 | 24.41 | 1.11 × 10−2 | 23.27 | 1.91 × 10−2 | 1.54 | 3.67 × 10−2 | ||
| CHGB | 4.80 | 7.97 × 10−2 | 12.25 | 4.08 × 10−2 | 14.02 | 4.98 × 10−2 | 1.89 | 2.21 × 10−2 | ||
| CHRNA3 | 26.20 | 2.17 × 10−3 | 23.90 | 1.13 × 10−2 | 39.49 | 7.76 × 10−3 | 1.72 | 3.30 × 10−2 | ||
| DCX | 11.89 | 1.22 × 10−2 | 38.49 | 4.20 × 10−3 | 102.50 | 9.74 × 10−4 | 2.91 | 0.0000 | ||
| EBF2 | 90.33 | 3.33 × 10−5 | 24.69 | 1.08 × 10−2 | 37.51 | 8.50 × 10−3 | 1.84 | 2.51 × 10−2 | ||
| ELAVL3 | 100.60 | 9.81 × 10−6 | 360.60 | 3.26 × 10−6 | 509.80 | 8.26 × 10−6 | 1.50 | 7.38 × 10−2 | ||
| ELAVL4 | 10.17 | 1.78 × 10−2 | 100.80 | 5.48 × 10−4 | 172.60 | 2.61 × 10−4 | 1.62 | 4.85 × 10−2 | ||
| GFRA1 | 40.96 | 5.87 × 10−4 | 54.30 | 2.23 × 10−3 | 120.00 | 6.45 × 10−4 | 1.65 | 4.44 × 10−2 | ||
| INSM1 | 222.40 | 1.23 × 10−7 | 290.70 | 1.35 × 10−5 | 461.80 | 1.47 × 10−5 | 1.50 | 7.42 × 10−2 | ||
| ISL1 | 24.59 | 2.51 × 10−3 | 97.31 | 5.67 × 10−4 | 227.50 | 1.15 × 10−4 | 1.59 | 5.34 × 10−2 | ||
| MDGA1 | 110.60 | 2.84 × 10−6 | 291.10 | 3.54 × 10−6 | 224.50 | 4.52 × 10−5 | 1.54 | 6.46 × 10−2 | ||
| MYT1 | 5.63 | 5.69 × 10−2 | 45.99 | 3.05 × 10−3 | 74.14 | 2.16 × 10−3 | 2.09 | 1.05 × 10−2 | ||
| ONECUT2 | 11.57 | 1.27 × 10−2 | 85.82 | 7.27 × 10−4 | 112.50 | 7.48 × 10−4 | 1.48 | 8.22 × 10−2 | ||
| PCDH9 | 8.05 | 2.91 × 10−2 | 79.33 | 9.67 × 10−4 | 94.84 | 1.24 × 10−3 | 1.49 | 7.73 × 10−2 | ||
| POU3F2 | 38.74 | 6.63 × 10−4 | 256.10 | 2.35 × 10−5 | 189.20 | 1.85 × 10−4 | 1.70 | 3.55 × 10−2 | ||
| ROBO2 | 12.58 | 1.05 × 10−2 | 27.02 | 9.23 × 10−3 | 73.36 | 2.16 × 10−3 | 1.49 | 7.67 × 10−2 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashikawa, Y.; Shiromizu, T.; Miura, K.; Adachi, Y.; Matsui, T.; Bessho, Y.; Tanaka, T.; Nishimura, Y. C3orf70 Is Involved in Neural and Neurobehavioral Development. Pharmaceuticals 2019, 12, 156. https://doi.org/10.3390/ph12040156
Ashikawa Y, Shiromizu T, Miura K, Adachi Y, Matsui T, Bessho Y, Tanaka T, Nishimura Y. C3orf70 Is Involved in Neural and Neurobehavioral Development. Pharmaceuticals. 2019; 12(4):156. https://doi.org/10.3390/ph12040156
Chicago/Turabian StyleAshikawa, Yoshifumi, Takashi Shiromizu, Koki Miura, Yuka Adachi, Takaaki Matsui, Yasumasa Bessho, Toshio Tanaka, and Yuhei Nishimura. 2019. "C3orf70 Is Involved in Neural and Neurobehavioral Development" Pharmaceuticals 12, no. 4: 156. https://doi.org/10.3390/ph12040156
APA StyleAshikawa, Y., Shiromizu, T., Miura, K., Adachi, Y., Matsui, T., Bessho, Y., Tanaka, T., & Nishimura, Y. (2019). C3orf70 Is Involved in Neural and Neurobehavioral Development. Pharmaceuticals, 12(4), 156. https://doi.org/10.3390/ph12040156







