Role of the Metal Center in the Modulation of the Aggregation Process of Amyloid Model Systems by Square Planar Complexes Bearing 2-(2′-pyridyl)benzimidazole Ligands
Abstract
1. Introduction
2. Materials and Methods
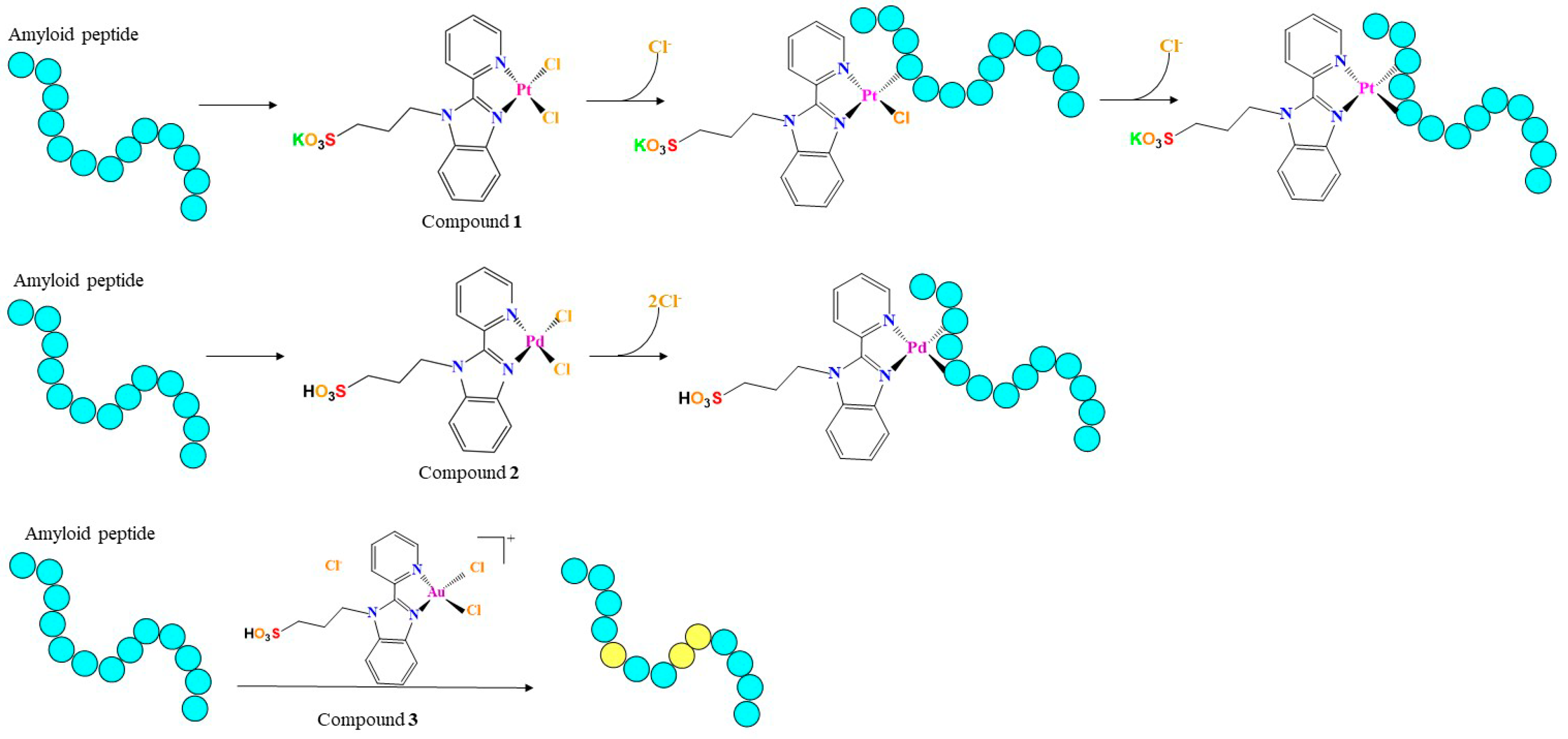
2.1. Peptide and Metal Compound Synthesis
2.2. Interaction of the Metal Compounds with Amyloid Peptides
2.2.1. UV-Vis Absorption Spectroscopy
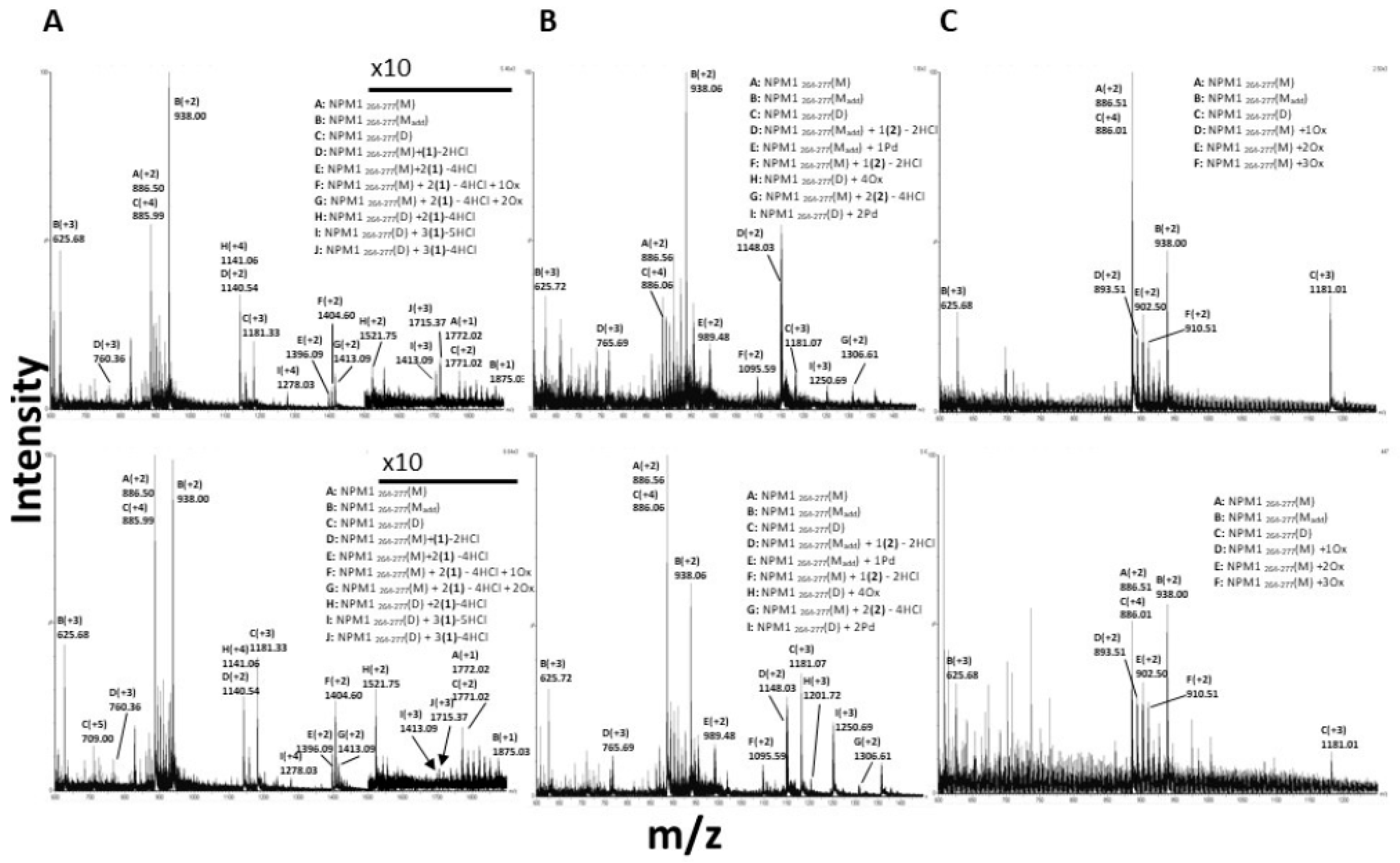
2.2.2. ESI-MS Analyses of the Adducts among Metal Complexes and Amyloid Peptides
2.3. Fluorescence Assays
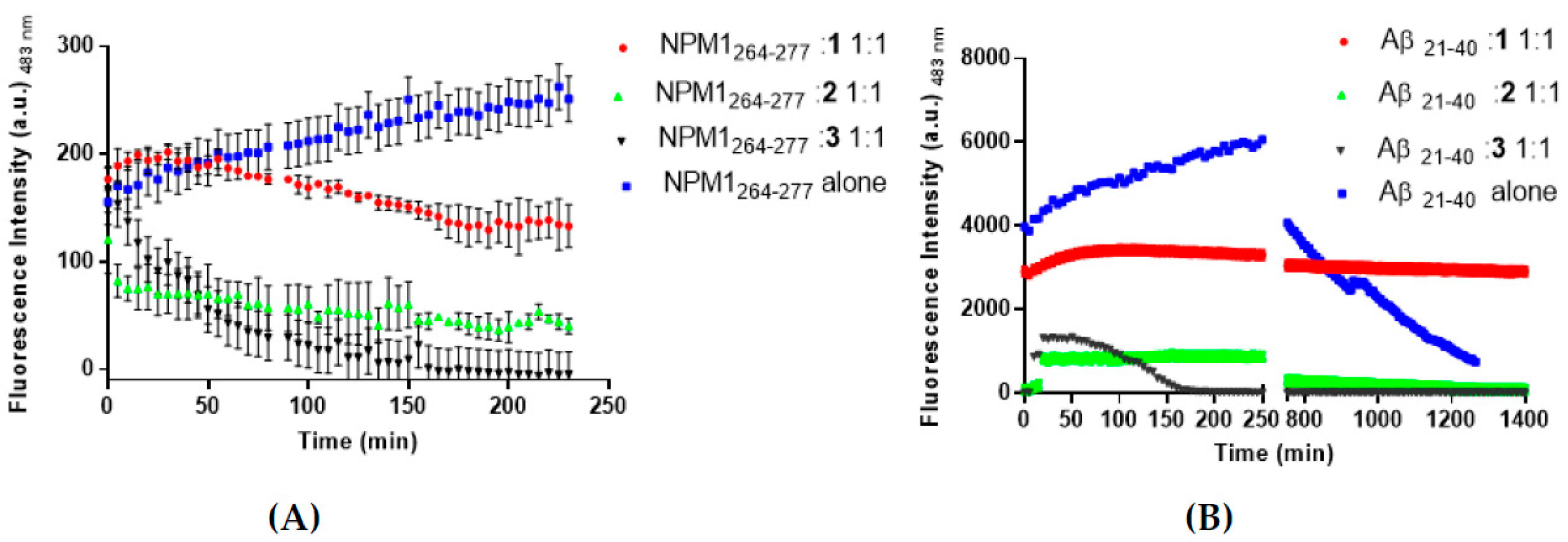
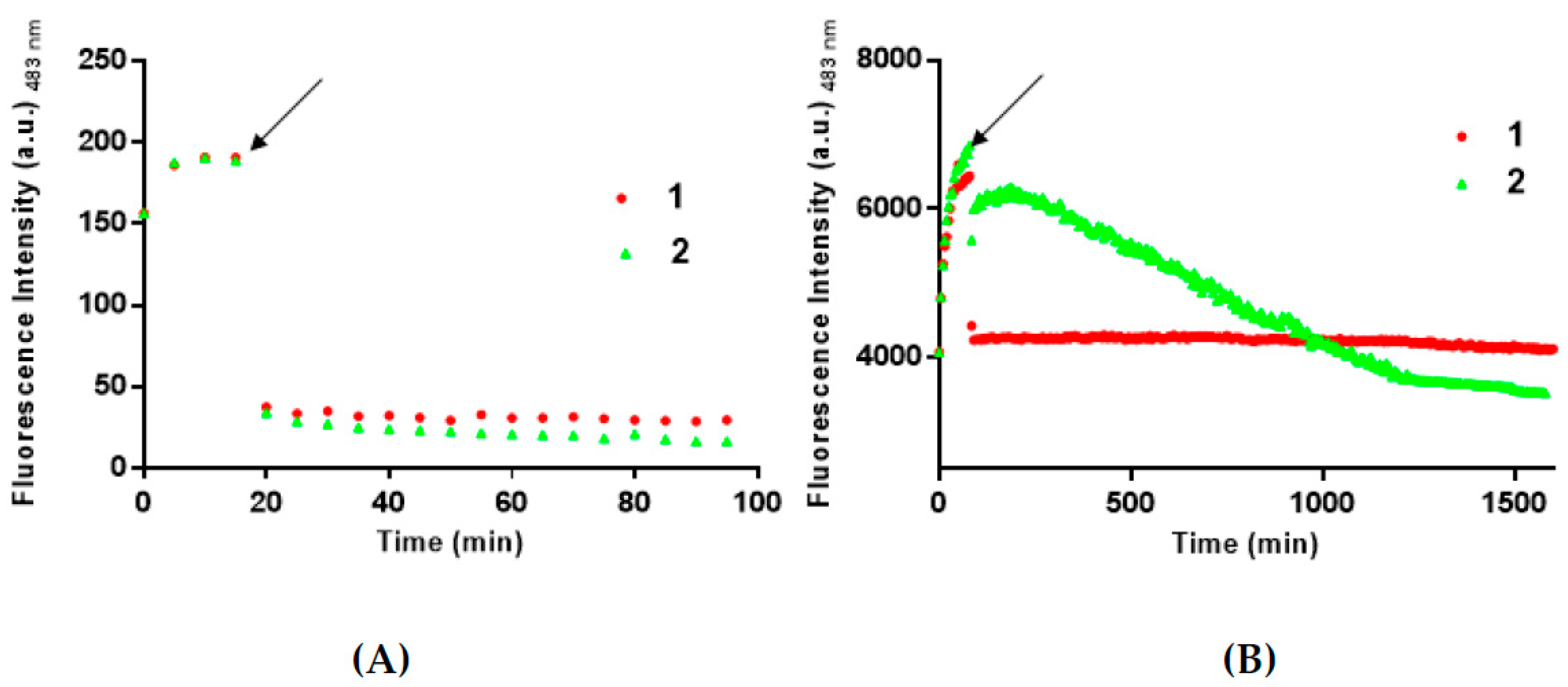
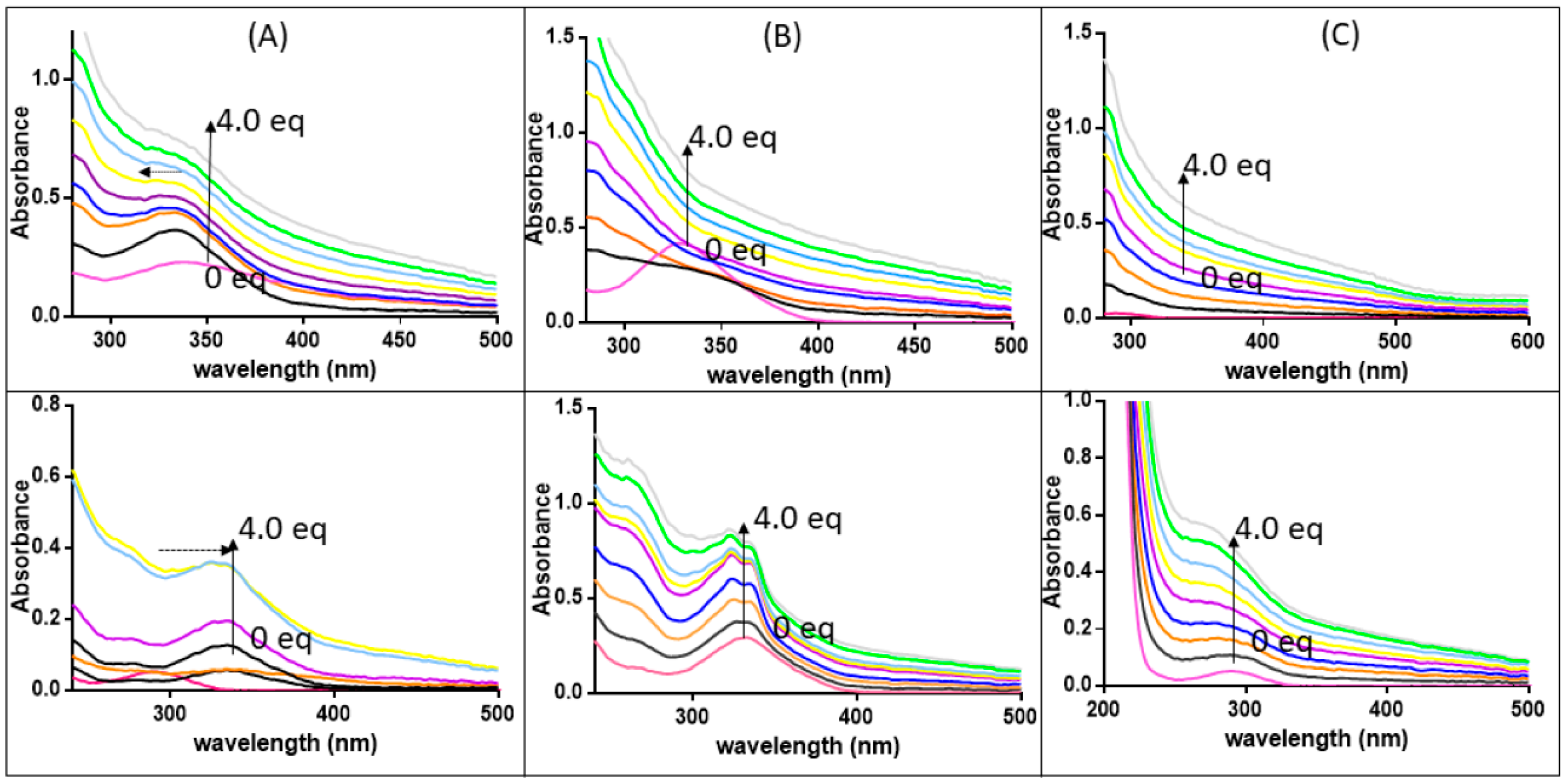
3. Results and Discussion
3.1. The Aggregation Propensity of Amyloidogenic Peptides is Affected by the Presence of 1, 2, and 3
3.2. 1 and 2 Are Able to Disaggregate Amyloid Assemblies
3.3. The Ligand Fields of 1 and 2 Are Tuned by the Presence of Amyloidogenic Peptides
3.4. 1 and 2 Form Adducts with NPM1264–277
3.5. 3 Does Not Form Adducts with NPM1264–277, But It Significantly Affects the Number of Oxidized Forms of NPM1264-277
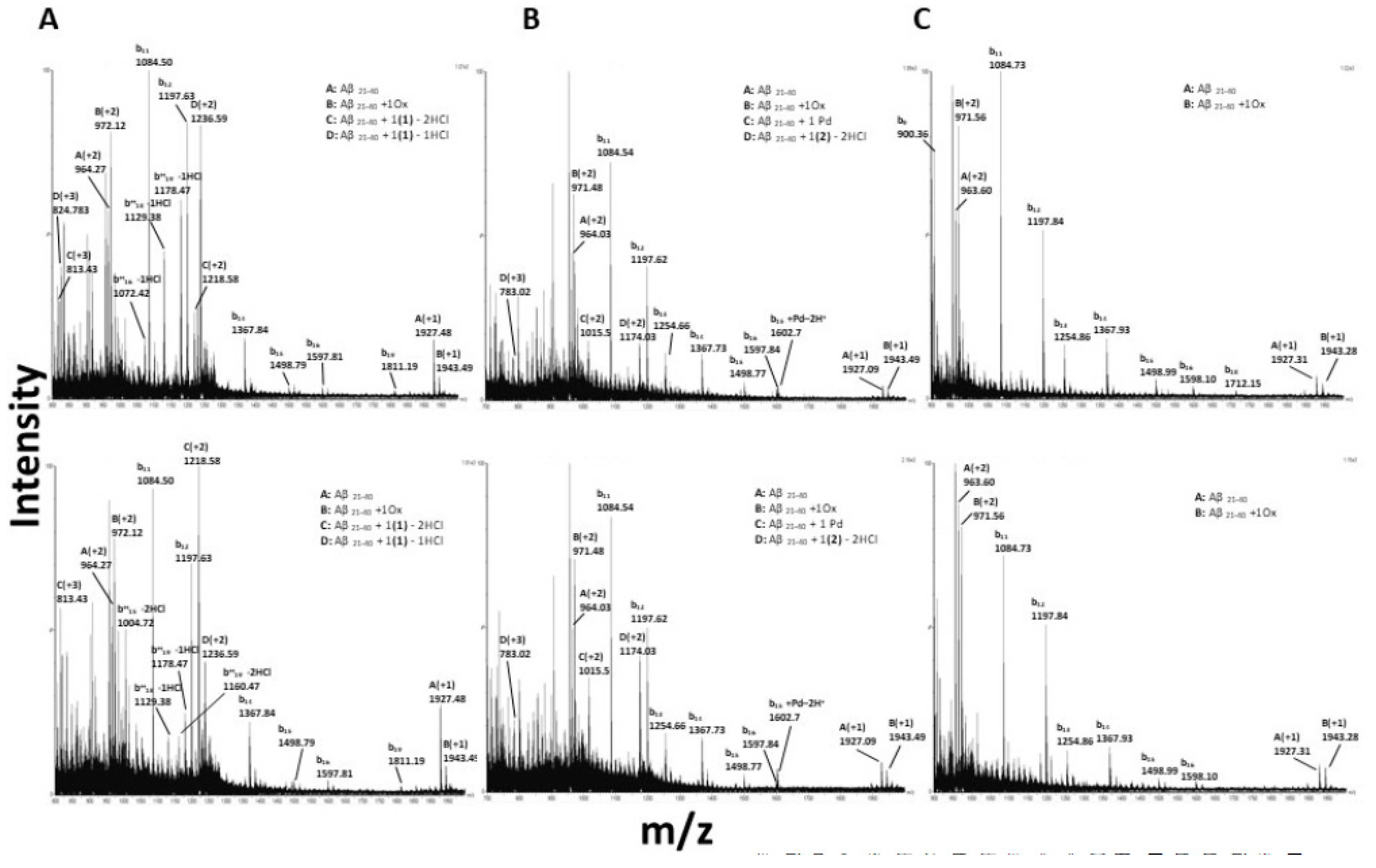
3.6. Interactions of the Metal Complexes with Aβ21–40
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hayne, D.J.; Lim, S.; Donnelly, P.S. Metal complexes designed to bind to amyloid-beta for the diagnosis and treatment of Alzheimer’s disease. Chem. Soc. Rev. 2014, 43, 6701–6715. [Google Scholar] [CrossRef]
- Mjos, K.D.; Orvig, C. Metallodrugs in medicinal inorganic chemistry. Chem. Rev. 2014, 114, 4540–4563. [Google Scholar] [CrossRef] [PubMed]
- Son, G.; Lee, B.I.; Chung, Y.J.; Park, C.B. Light-triggered dissociation of self-assembled beta-amyloid aggregates into small, nontoxic fragments by ruthenium (II) complex. Acta Biomater. 2018, 67, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.M.; Kim, G.; Kang, J.; Lim, M.H. Strategies Employing Transition Metal Complexes to Modulate Amyloid-beta Aggregation. Inorg. Chem. 2019, 58, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Florio, D.; Malfitano, A.M.; Di Somma, S.; Mugge, C.; Weigand, W.; Ferraro, G.; Iacobucci, I.; Monti, M.; Morelli, G.; Merlino, A.; et al. Platinum(II) O,S Complexes Inhibit the Aggregation of Amyloid Model Systems. Int. J. Mol. Sci. 2019, 20, 829. [Google Scholar] [CrossRef] [PubMed]
- Barnham, K.J.; Kenche, V.B.; Ciccotosto, G.D.; Smith, D.P.; Tew, D.J.; Liu, X.; Perez, K.; Cranston, G.A.; Johanssen, T.J.; Volitakis, I.; et al. Platinum-based inhibitors of amyloid-beta as therapeutic agents for Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2008, 105, 6813–6818. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Huang, F.; Pu, X.; Jia, L.; Jiang, T.; Li, L.; Liu, Y. Identification of [PtCl2(phen)] binding modes in amyloid-beta peptide and the mechanism of aggregation inhibition. Chemistry 2011, 17, 11657–11666. [Google Scholar] [CrossRef]
- Ma, G.; Wang, E.; Wei, H.; Wei, K.; Zhu, P.; Liu, Y. PtCl2(phen) disrupts the metal ions binding to amyloid-beta peptide. Metall. Integr. Biomet. Sci. 2013, 5, 879–887. [Google Scholar] [CrossRef]
- Cheignon, C.; Tomas, M.; Bonnefont-Rousselot, D.; Faller, P.; Hureau, C.; Collin, F. Oxidative stress and the amyloid beta peptide in Alzheimer’s disease. Redox Biol. 2018, 14, 450–464. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, W.; Sun, J.; Guo, S. Triplet photosensitizers: From molecular design to applications. Chem. Soc. Rev. 2013, 42, 5323–5351. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J.; Wang, L.; Zhang, B.; Du, W. Interaction of the human prion protein PrP106-126 with metal complexes: Potential therapeutic agents against prion disease. Chemistry 2010, 16, 13339–13342. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhu, D.; Zhao, C.; Jia, X.; Wang, X.; Du, W. Effects of gold complexes on the assembly behavior of human islet amyloid polypeptide. J. Inorg. Biochem. 2015, 152, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, C.; Zhu, D.; Gong, G.; Du, W. Inhibition of amyloid peptide fibril formation by gold-sulfur complexes. J. Inorg. Biochem. 2017, 171, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, X.; He, L.; Zhu, D.; Wang, B.; Du, W. Influence of gold-bipyridyl derivants on aggregation and disaggregation of the prion neuropeptide PrP106-126. Metall. Integr. Biomet. Sci. 2014, 6, 2117–2125. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, L.; Zhang, B.; Wang, X.; Huang, C.; Li, Y.; Du, W. Palladium complexes affect the aggregation of human prion protein PrP106-126. Inorg. Chem. 2011, 50, 4340–4348. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, M.L.; Kukushkin, V.Y.; Pombeiro, A.J. Reactivity of Pt- and Pd-bound nitriles towards nitrile oxides and nitrones: Substitution vs. cycloaddition. Dalton Trans. 2008, 10, 1312–1322. [Google Scholar] [CrossRef] [PubMed]
- Ghani, N.T.A.; Mansour, A.M. Novel palladium(II) and platinum(II) complexes with 1H-benzimidazol-2-ylmethyl-N-(4-bromo-phenyl)-amine: Structural studies and anticancer activity. Eur. J. Med. Chem. 2012, 47, 399–411. [Google Scholar] [CrossRef]
- Mansour, A.M.; Abdel-Ghani, N.T. Synthesis, spectroscopic, DFT, cytotoxicity and antimicrobial activity of Pd(II) and Pt(II) complexes of N,N-chelated benzimidazole derivatives. Inorg. Chim. Acta 2015, 438, 76–84. [Google Scholar] [CrossRef]
- Kumar, R.; Nevado, C. Cyclometalated Gold(III) Complexes: Synthesis, Reactivity, and Physicochemical Properties. Angew. Chem. 2017, 56, 1994–2015. [Google Scholar] [CrossRef]
- Ronga, L.; Langella, E.; Palladino, P.; Marasco, D.; Tizzano, B.; Saviano, M.; Pedone, C.; Improta, R.; Ruvo, M. Does tetracycline bind helix 2 of prion? An integrated spectroscopical and computational study of the interaction between the antibiotic and alpha helix 2 human prion protein fragments. Proteins 2007, 66, 707–715. [Google Scholar] [CrossRef]
- Tizzano, B.; Palladino, P.; De Capua, A.; Marasco, D.; Rossi, F.; Benedetti, E.; Pedone, C.; Ragone, R.; Ruvo, M. The human prion protein alpha2 helix: A thermodynamic study of its conformational preferences. Proteins 2005, 59, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.M.; Shehab, O.R. Lysozyme and DNA binding affinity of Pd(II) and Pt(II) complexes bearing charged N,N-pyridylbenzimidazole bidentate ligands. Dalton Trans. 2018, 47, 3459–3468. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.D.; Jia, W.L.; Wang, S. Blue luminescent 2-(2′-pyridyl) benzimidazole derivative ligands and their orange luminescent mononuclear and polynuclear organoplatinum(II) complexes. Inorg. Chem. 2005, 44, 1332–1343. [Google Scholar] [CrossRef] [PubMed]
- Shavaleev, N.M.; Bell, Z.R.; Easun, T.L.; Rutkaite, R.; Swanson, L.; Ward, M.D. Complexes of substituted derivatives of 2-(2-pyridyl)benzimidazole with Re(I), Ru(II) and Pt(II): Structures, redox and luminescence properties. Dalton Trans. 2004, 21, 3678–3688. [Google Scholar] [CrossRef] [PubMed]
- Marasco, D.; Messori, L.; Marzo, T.; Merlino, A. Oxaliplatin vs. cisplatin: Competition experiments on their binding to lysozyme. Dalton Trans. 2015, 44, 10392–10398. [Google Scholar] [CrossRef] [PubMed]
- Krauss, I.R.; Messori, L.; Cinellu, M.A.; Marasco, D.; Sirignano, R.; Merlino, A. Interactions of gold-based drugs with proteins: The structure and stability of the adduct formed in the reaction between lysozyme and the cytotoxic gold(III) compound Auoxo3. Dalton Trans. 2014, 43, 17483–17488. [Google Scholar] [CrossRef]
- Ferraro, G.; Mansour, A.M.; Merlino, A. Exploring the interactions between model proteins and Pd(II) or Pt(II) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography. Dalton Trans. 2018, 47, 10130–10138. [Google Scholar] [CrossRef]
- Di Natale, C.; Scognamiglio, P.L.; Cascella, R.; Cecchi, C.; Russo, A.; Leone, M.; Penco, A.; Relini, A.; Federici, L.; Di Matteo, A.; et al. Nucleophosmin contains amyloidogenic regions that are able to form toxic aggregates under physiological conditions. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2015, 29, 3689–3701. [Google Scholar] [CrossRef]
- Russo, A.; Diaferia, C.; La Manna, S.; Giannini, C.; Sibillano, T.; Accardo, A.; Morelli, G.; Novellino, E.; Marasco, D. Insights into amyloid-like aggregation of H2 region of the C-terminal domain of nucleophosmin. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 176–185. [Google Scholar] [CrossRef]
- Scognamiglio, P.L.; Di Natale, C.; Leone, M.; Cascella, R.; Cecchi, C.; Lirussi, L.; Antoniali, G.; Riccardi, D.; Morelli, G.; Tell, G.; et al. Destabilisation, aggregation, toxicity and cytosolic mislocalisation of nucleophosmin regions associated with acute myeloid leukemia. Oncotarget 2016, 7, 59129–59143. [Google Scholar] [CrossRef]
- De Santis, A.; La Manna, S.; Krauss, I.R.; Malfitano, A.M.; Novellino, E.; Federici, L.; De Cola, A.; Di Matteo, A.; D’Errico, G.; Marasco, D. Nucleophosmin-1 regions associated with acute myeloid leukemia interact differently with lipid membranes. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 967–978. [Google Scholar] [CrossRef] [PubMed]
- La Manna, S.; Roviello, V.; Scognamiglio, P.L.; Diaferia, C.; Giannini, C.; Sibillano, T.; Morelli, G.; Novellino, E.; Marasco, D. Amyloid fibers deriving from the aromatic core of C-terminal domain of nucleophosmin 1. Int. J. Biol. Macromol. 2018, 122, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Di Natale, C.; La Manna, S.; Malfitano, A.M.; Di Somma, S.; Florio, D.; Scognamiglio, P.L.; Novellino, E.; Netti, P.A.; Marasco, D. Structural insights into amyloid structures of the C-terminal region of nucleophosmin 1 in type A mutation of acute myeloid leukemia. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Iversen, L.L.; Mortishire-Smith, R.J.; Pollack, S.J.; Shearman, M.S. The toxicity in vitro of beta-amyloid protein. Biochem. J. 1995, 311, 1–16. [Google Scholar] [CrossRef]
- La Manna, S.; Scognamiglio, P.L.; Roviello, V.; Borbone, F.; Florio, D.; Di Natale, C.; Bigi, A.; Cecchi, C.; Cascella, R.; Giannini, C.; et al. The acute myeloid leukemia-associated Nucleophosmin 1 gene mutations dictate amyloidogenicity of the C-terminal domain. FEBS J. 2019, 286, 2311–2328. [Google Scholar] [CrossRef]
- Joseph, R.; Han, E. Amyloid beta-protein fragment 25-35 causes activation of cytoplasmic calcium in neurons. Biochem. Biophys. Res. Commun. 1992, 184, 1441–1447. [Google Scholar] [CrossRef]
- Giuffrida, M.L.; Grasso, G.; Ruvo, M.; Pedone, C.; Saporito, A.; Marasco, D.; Pignataro, B.; Cascio, C.; Copani, A.; Rizzarelli, E. Abeta(25-35) and its C-and/or N-blocked derivatives: Copper driven structural features and neurotoxicity. J. Neurosci. Res. 2007, 85, 623–633. [Google Scholar] [CrossRef]
- Saporito, A.; Marasco, D.; Chambery, A.; Botti, P.; Monti, S.M.; Pedone, C.; Ruvo, M. The chemical synthesis of the GstI protein by NCL on a X-Met site. Biopolymers 2006, 83, 508–518. [Google Scholar] [CrossRef]
- Mansour, A.M.; Shehab, O.R. Pyridylbenzimidazole-Based Gold(III) Complexes: Lysozyme Metalation, DNA Binding Studies, and Biological Activity. Eur. J. Inorg. Chem. 2019, 2019, 2830–2838. [Google Scholar] [CrossRef]
- Messori, L.; Merlino, A. Cisplatin binding to proteins: A structural perspective. Coord. Chem. Rev. 2016, 315, 67–89. [Google Scholar] [CrossRef]
- Ferraro, G.; De Benedictis, I.; Malfitano, A.; Morelli, G.; Novellino, E.; Marasco, D. Interactions of cisplatin analogues with lysozyme: A comparative analysis. Biomet. Int. J. Role Met. Ions Biol. Biochem. Med. 2017, 30, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Ferraro, G.; Merlino, A. Cisplatin-Protein Interactions: Unexpected Drug Binding to N-Terminal Amine and Lysine Side Chains. Inorg. Chem. 2016, 55, 7814–7816. [Google Scholar] [CrossRef] [PubMed]
- Messori, L.; Marzo, T.; Merlino, A. The X-ray structure of the complex formed in the reaction between oxaliplatin and lysozyme. Chem. Commun. (Camb.) 2014, 50, 8360–8362. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, G.; Giorgio, A.; Mansour, A.M.; Merlino, A. Protein-mediated disproportionation of Au(i): Insights from the structures of adducts of Au(III) compounds bearing N,N-pyridylbenzimidazole derivatives with lysozyme. Dalton Trans. 2019, 48, 14027–14035. [Google Scholar] [CrossRef] [PubMed]
- Durovic, M.D.; Bugarcic, Z.D.; Heinemann, F.W.; van Eldik, R. Substitution versus redox reactions of gold(III) complexes with L-cysteine, L-methionine and glutathione. Dalton Trans. 2014, 43, 3911–3921. [Google Scholar] [CrossRef]
- Bitan, G.; Tarus, B.; Vollers, S.S.; Lashuel, H.A.; Condron, M.M.; Straub, J.E.; Teplow, D.B. A molecular switch in amyloid assembly: Met35 and amyloid beta-protein oligomerization. J. Am. Chem. Soc. 2003, 125, 15359–15365. [Google Scholar] [CrossRef]

| Peptide | Sequence | pI | Net Charge at pH7 |
|---|---|---|---|
| NPM1264–277 | VEAKFINYVKNCFR | 10.15 | 1.9 |
| Aβ21–40 | AEDVGSNKGAIIGLMVGGVV | 3.93 | −1 |
| Complex | Signal (m/z) | Charged Species | Exp MW (Da) | Theor MW (Da) | Species | Time (h) |
|---|---|---|---|---|---|---|
| 1 | 1772.02 886.50 | A (+1) A (+2) | 1770.99 ± 0.02 | 1770.91 | Monomer (NPM1264–277(M)) | 0,3,17 |
| 1875.03 938.00 625.68 | B (+1) B (+2) B (+3) | 1874.02 ± 0.02 | 1873.91 | Monomer Adduct (NPM1264–277(Madd)) | 0,3,17 | |
| 1771.02 1181.33 885.99 709.00 | C (+2) C (+3) C (+4) C (+5) | 3540.22 ± 0.43 | 3539.82 | Dimer (NPM1264–277(D)) | 0,3,17 | |
| 1140.539 760.360 | D (+2) D (+3) | 2278.81 ± 0.52 | 2281.45 | NPM1264–277(M) + (1)−2HCl | 0,3,17 | |
| 1396.095 931.070 | E (+2) E (+3) | 2789.960 ± 0.31 | 2791.9 | NPM1264–277(M) + 2(1)−4HCl | 0,3,17 | |
| 3,17 | ||||||
| 1404.60 936.73 | F (+2) F (+3) | 2806.96 ± 0.30 | 2807.9 | NPM1264–277(M) + 2(1)−4HCl + 1Ox † | 0,3,17 | |
| 3,17 | ||||||
| 1413.099 942.72 | G (+2) G (+3) | 2824.98 ± 0.60 | 2823.9 | NPM1264–277(M) + 2(1)−4HCl + 2Ox | 0,3,17 | |
| 1521.75 1141.06 | H (+2) H (+4) | 4561.57 ± 1.00 | 4560.9 | NPM1264–277(D) +2(1)−4HCl | 0,3,17 | |
| 1704.337 1278.03 | I (+3) I (+4) | 5108.63 ± 0.50 | 5107.8 | NPM1264-277(D) + 3(1)−5HCl | 0,3 | |
| 0,3,17 | ||||||
| 1715.37 | J (+3) | 5143.11 * | 5144.25 | NPM1264–277(D) + 3(1)−4HCl | 0,3 | |
| 2 | 1772.146 886.560 | A (+1) A (+2) | 1771.12 ± 0.02 | 1770.91 | Monomer (NPM1264–277(M)) | 0,3,17 |
| 1875.100 938.060 625.720 | B (+1) B (+2) B (+3) | 1874.11 ± 0.02 | 1873.91 | Monomer Adduct (NPM1264–277(Madd)) | 0,3,17 | |
| 1771.14 1181.07 886.06 709.25 | C (+2) C (+3) C (+4) C (+5) | 3540.47 ± 0.44 | 3539.82 | Dimer (NPM1264–277(D)) | 0,3,17 | |
| 1148.026 765.69 | D (+2) D (+3) | 2294.11 ± 0.09 | 2295.96 | NPM1264–277(Madd) + 1(2)−2HCl | 0,3,17 | |
| 989.476 | E (+2) | 1976.952 * | 1976.32 | NPM1264–277(Madd) + 1Pd | 0,3,17 | |
| 1095.587 | F (+2) | 2189.17 * | 2192.79 | NPM1264–277(M) + 1(2)−2HCl | 0,3,17 | |
| 1306.606 | G (+2) | 2611.21 * | 2614.58 | NPM1264–277(M) + 2(2)-4HCl | 0,3,17 | |
| 1201.721 | H (+3) | 3602.19 ± 0.04 | 3604.00 | NPM1264–277(D) + 4Ox | 3,17 | |
| 1250.694 | I (+3) | 3749.082 * | 3748.84 | NPM1264–277(D) + 2Pd | 0,3,17 | |
| 3 | 1772.03 886.51 | A (+1) A (+2) | 1771.02 ± 0.01 | 1770.91 | Monomer (NPM1264–277(M)) | 0,3,17 |
| 938.00 625.68 | B (+2) B (+3) | 1874.98 ± 0.05 | 1873.91 | Monomer Adduct (NPM1264–277(Madd)) | 0,3,17 | |
| 1771.52 1181.01 886.01 | C (+2) C (+3) C (+4) | 3540.35 ± 0.48 | 3539.82 | Dimer (NPM1264–277(D)) | 0,3,17 | |
| 893.51 | D (+2) | 1785.02 * | 1786.91 | NPM1264–277(M) +1Ox | 0,3,17 | |
| 902.50 | E (+2) | 1803.00 * | 1802.91 | NPM1264–277(M) +2Ox | 0,3,17 | |
| 910.51 | F (+2) | 1819.02 * | 1818.91 | NPM1264–277(M) +3Ox | 0,3,17 |
| Complex | Signal (m/z) | Charged Species | Exp MW (Da) | Theor MW (Da) | Species | Time (h) |
|---|---|---|---|---|---|---|
| 1 | 1927.48 964.27 | A (+1) A (+2) | 1926.50 ± 0.02 | 1926.00 | Aβ21–40 | 0,3,17 |
| 1943.49 972.12 | B (+1) B (+2) | 1942.35 ± 0.13 | 1942.00 | Aβ21–40+ 1Ox † | 0,3,17 | |
| 1218.583 813.429 | C (+2) C (+3) | 2435.59 ± 0.56 | 2436.45 | Aβ21–40 + 1(1)-2HCl | 0,3,17 | |
| 1236.599 824.641 | D (+2) D (+3) | 2471.54 ± 0.36 | 2472.9 | Aβ21–40 + 1(1)-1HCl | 0,3,17 | |
| 0,3 | ||||||
| 2 | 1927.09 964.03 | A (+1) A (+2) | 1926.07 ± 0.02 | 1926.00 | Aβ21–40 | 0,3,17 |
| 1943.07 971.48 | B (+1) B (+2) | 1941.50 ± 0.56 | 1942.00 | Aβ21–40 + 1Ox | 0,3,17 | |
| 2030.029 1015.500 | C (+1) C (+2) | 2029.00 ± 0.02 | 2028.42 | Aβ21–40 + 1 Pd | 0,3,17 | |
| 1174.035 783.022 | D (+2) D (+3) | 2346.03 ± 0.02 | 2347.79 | Aβ21–40 + 1(2)-2HCl | 0,3,17 | |
| 3 | 1927.31 963.60 | A (+1) A (+2) | 1925.74 ± 0.56 | 1926.00 | Aβ21–40 | 0,3,17 |
| 1943.28 971.56 | B (+1) B (+2) | 1941.68 ± 0.58 | 1942.00 | Aβ21–40 +1Ox | 0,3,17 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Florio, D.; Iacobucci, I.; Ferraro, G.; Mansour, A.M.; Morelli, G.; Monti, M.; Merlino, A.; Marasco, D. Role of the Metal Center in the Modulation of the Aggregation Process of Amyloid Model Systems by Square Planar Complexes Bearing 2-(2′-pyridyl)benzimidazole Ligands. Pharmaceuticals 2019, 12, 154. https://doi.org/10.3390/ph12040154
Florio D, Iacobucci I, Ferraro G, Mansour AM, Morelli G, Monti M, Merlino A, Marasco D. Role of the Metal Center in the Modulation of the Aggregation Process of Amyloid Model Systems by Square Planar Complexes Bearing 2-(2′-pyridyl)benzimidazole Ligands. Pharmaceuticals. 2019; 12(4):154. https://doi.org/10.3390/ph12040154
Chicago/Turabian StyleFlorio, Daniele, Ilaria Iacobucci, Giarita Ferraro, Ahmed M. Mansour, Giancarlo Morelli, Maria Monti, Antonello Merlino, and Daniela Marasco. 2019. "Role of the Metal Center in the Modulation of the Aggregation Process of Amyloid Model Systems by Square Planar Complexes Bearing 2-(2′-pyridyl)benzimidazole Ligands" Pharmaceuticals 12, no. 4: 154. https://doi.org/10.3390/ph12040154
APA StyleFlorio, D., Iacobucci, I., Ferraro, G., Mansour, A. M., Morelli, G., Monti, M., Merlino, A., & Marasco, D. (2019). Role of the Metal Center in the Modulation of the Aggregation Process of Amyloid Model Systems by Square Planar Complexes Bearing 2-(2′-pyridyl)benzimidazole Ligands. Pharmaceuticals, 12(4), 154. https://doi.org/10.3390/ph12040154











