Radiolabeling of Nucleic Acid Aptamers for Highly Sensitive Disease-Specific Molecular Imaging
Abstract
1. Introduction
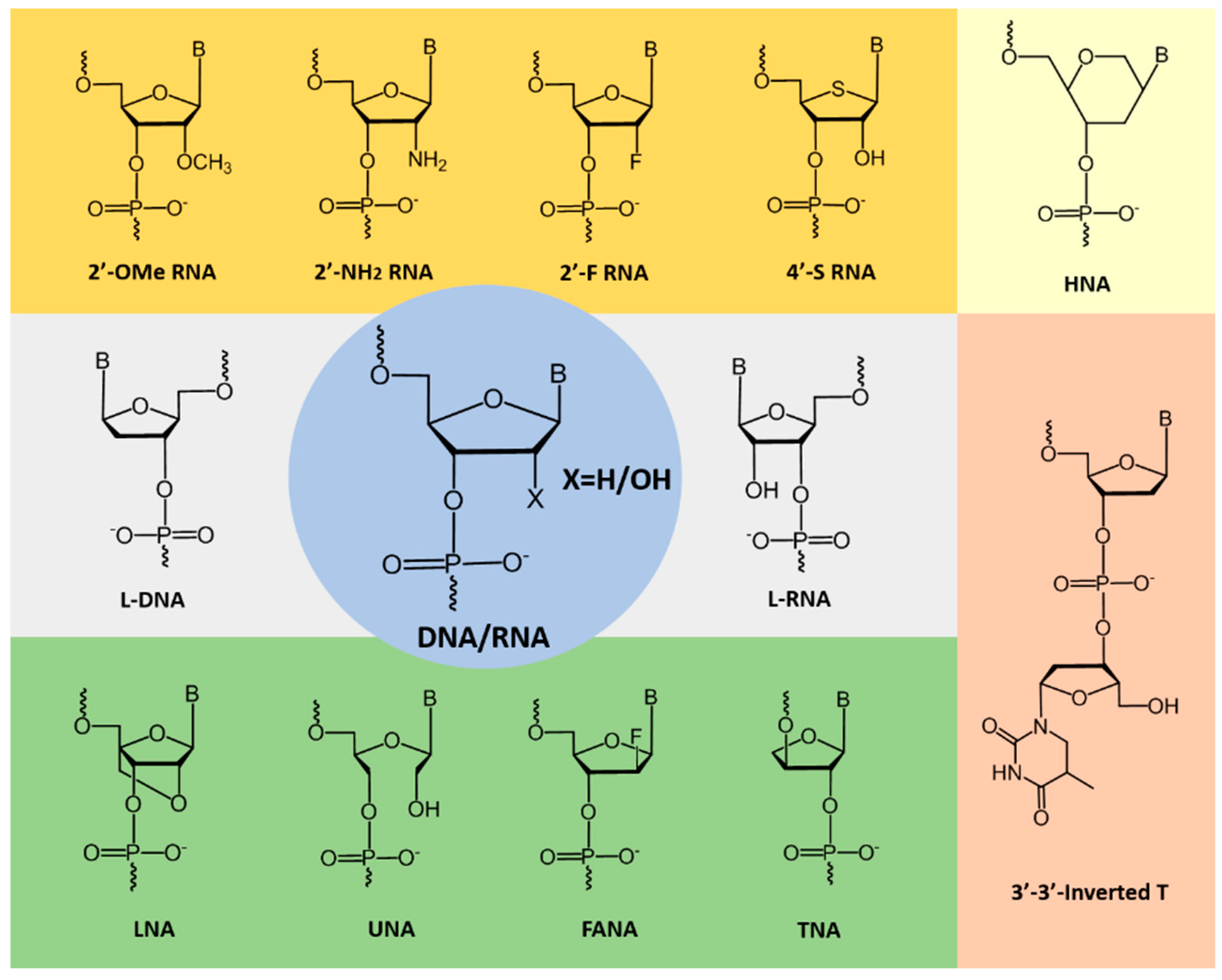
2. Improving the Stability of Aptamers
3. SPECT and PET Imaging Techniques
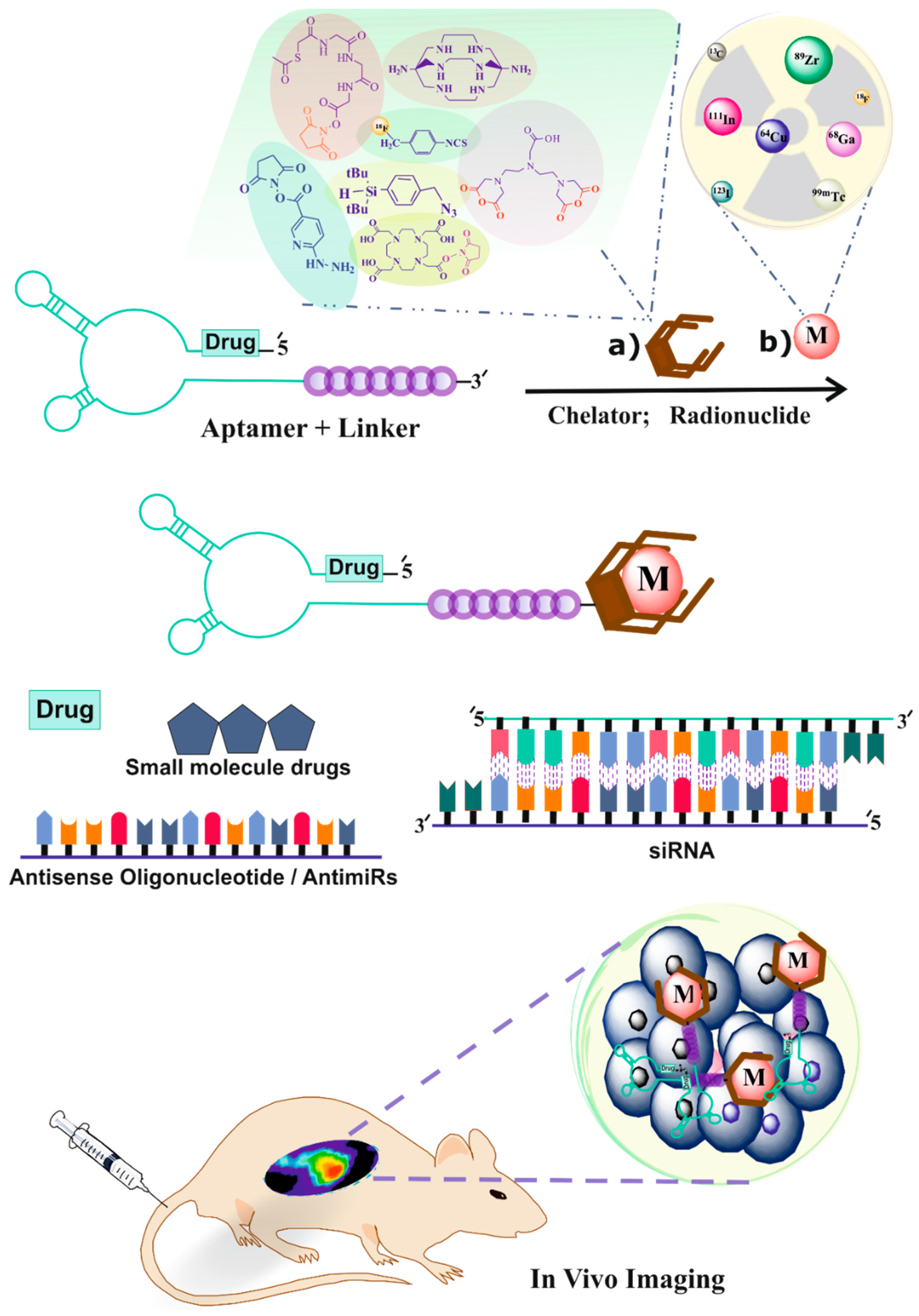
4. Radiolabeling of Ligands for SPECT Imaging
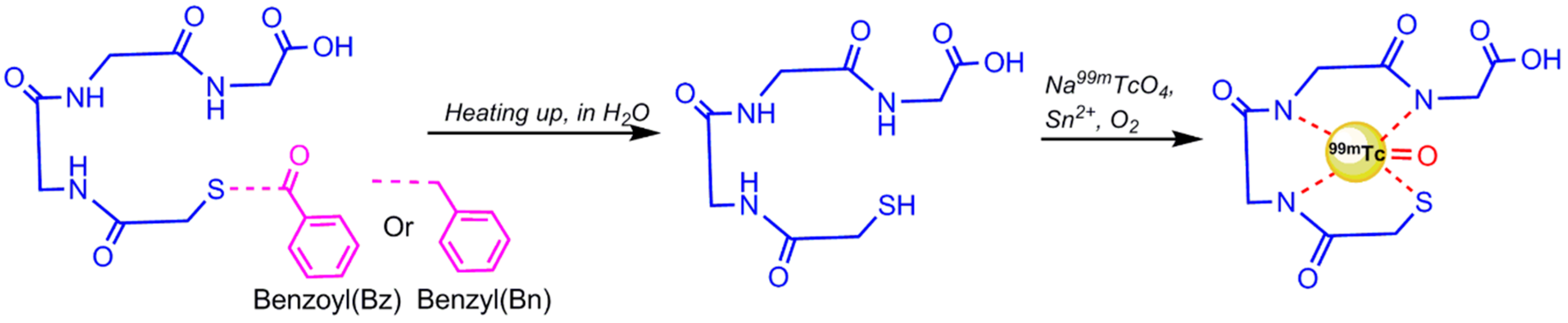
4.1. 99mTc-Radiolabeling
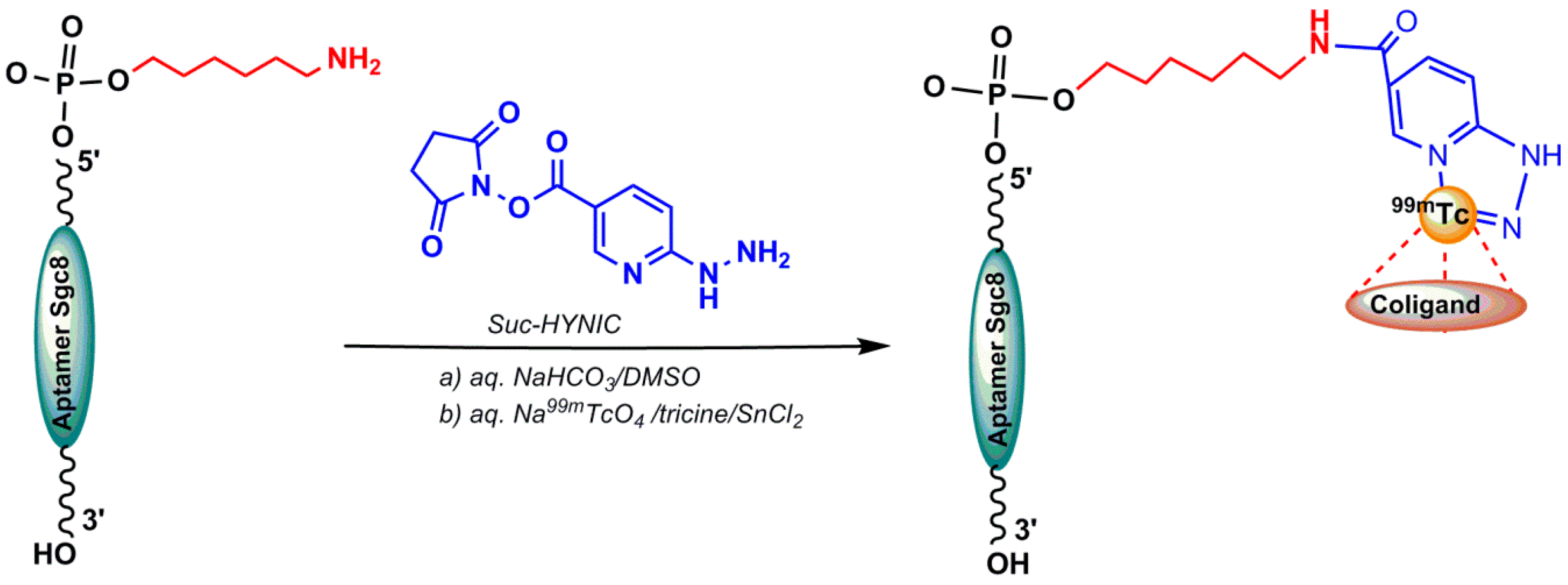
4.1.1. Direct 99mTc/188Re Radiolabeling of Aptamers
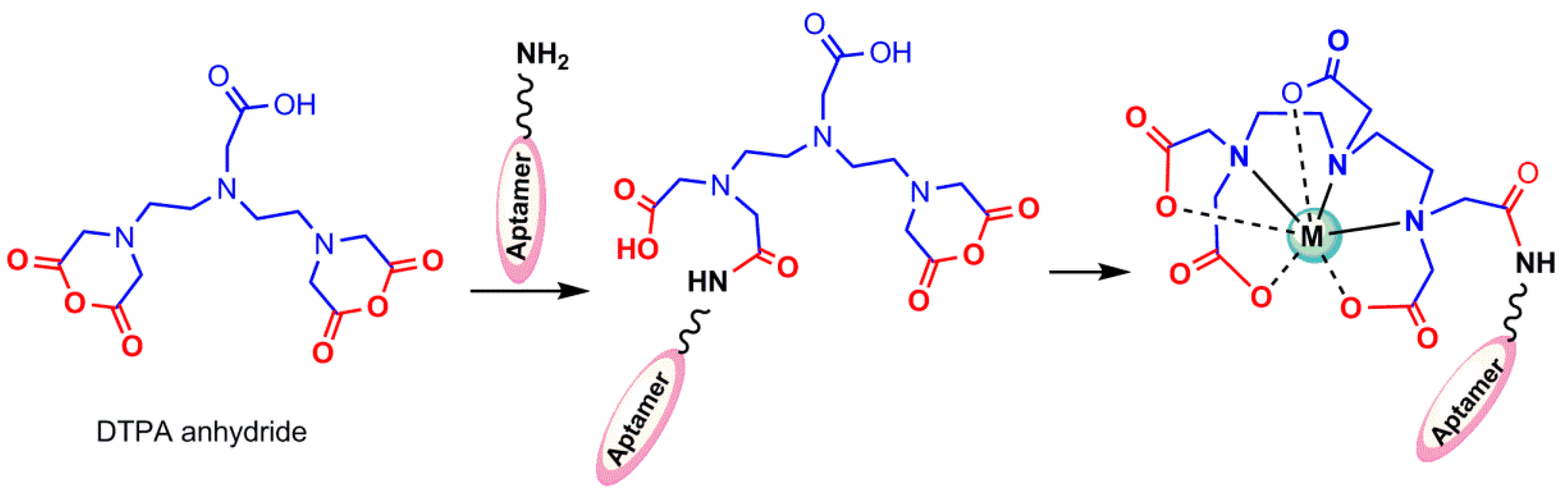
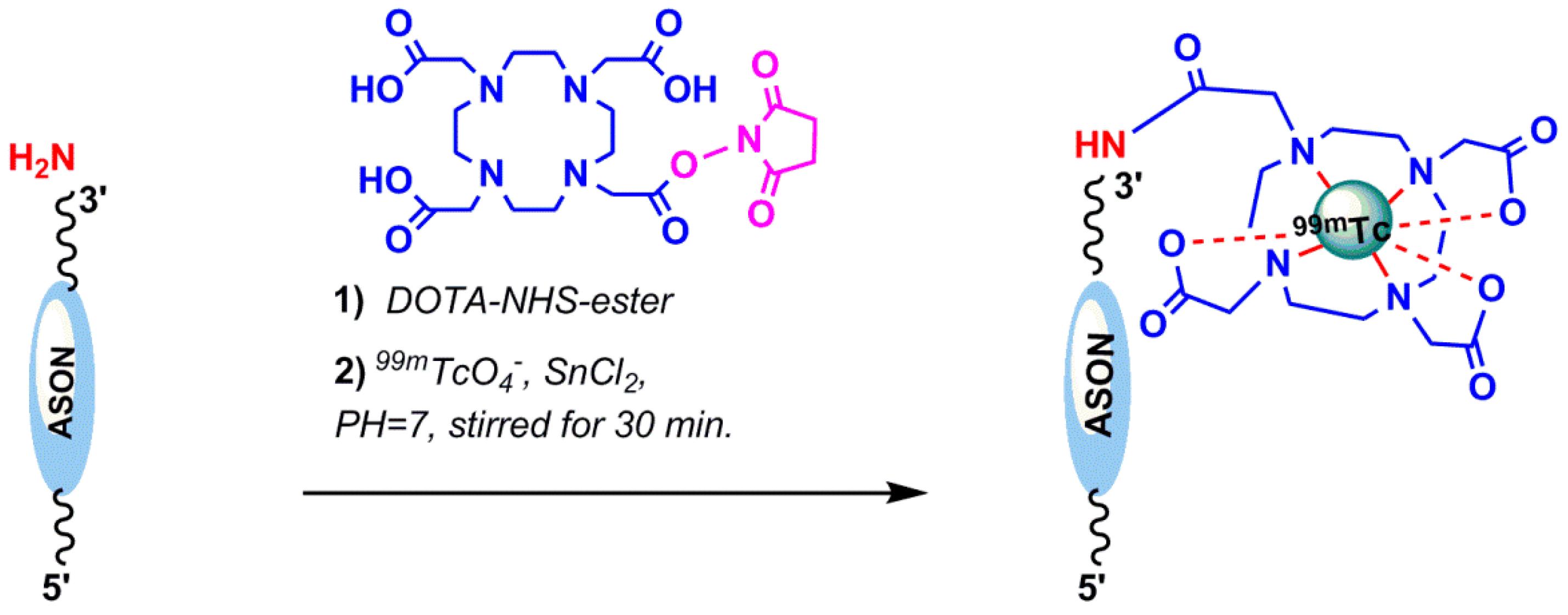
4.1.2. Indirect 99mTc-Labeling of Aptamers
5. Radiolabeling of Ligands for PET Imaging
5.1. 18F Radiolabeling
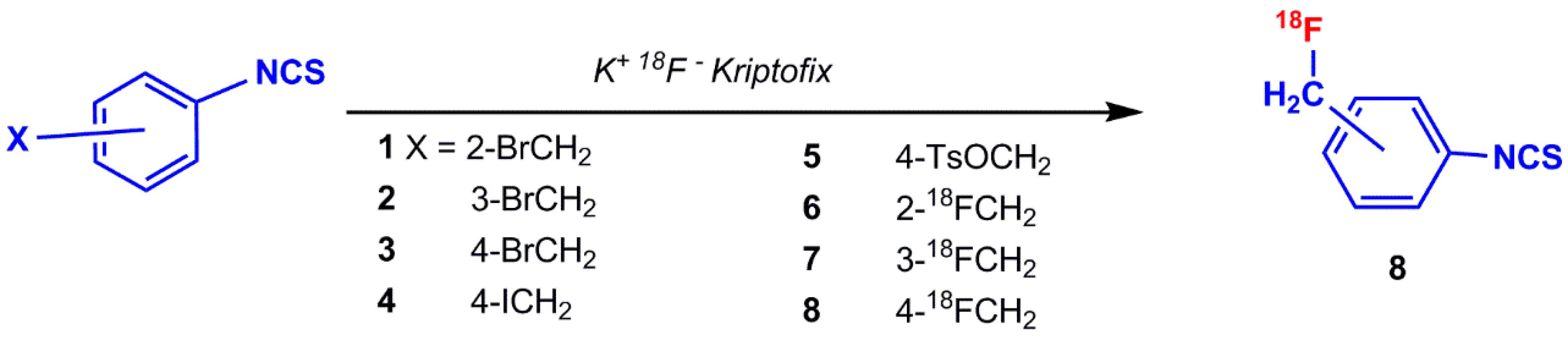
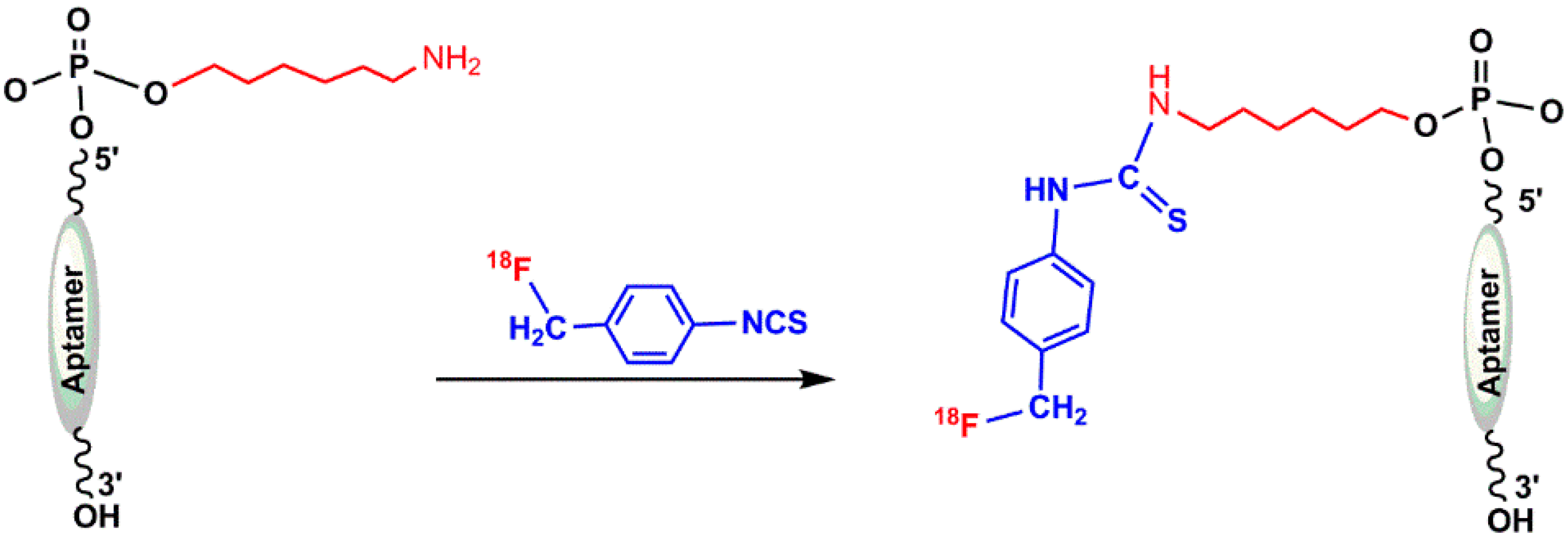
5.1.1. ([18F]fluoromethyl)phenyl Isothiocyanate
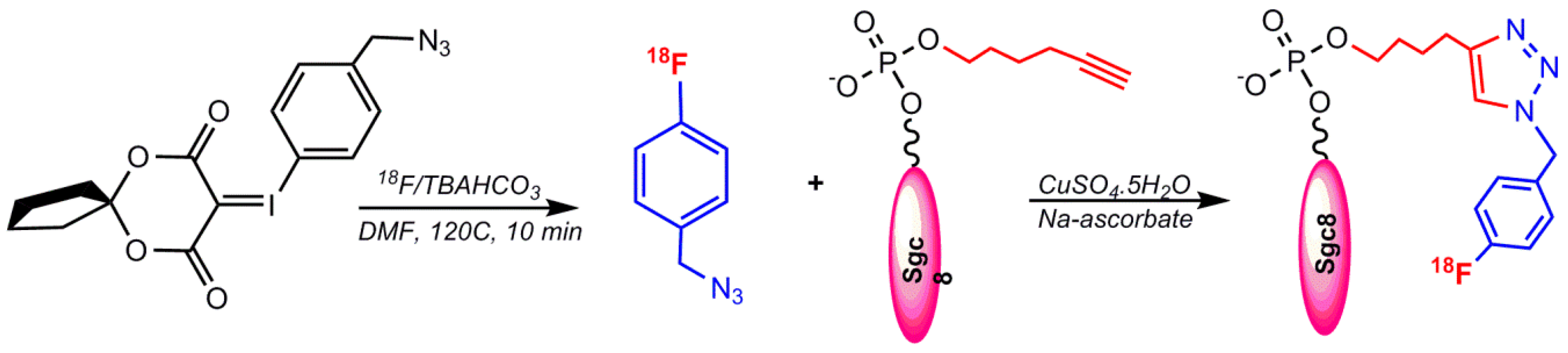
5.1.2. Labeling via Click Chemistry Using 1-(Azidomethyl)–4-(Fluoro–18F) Benzene
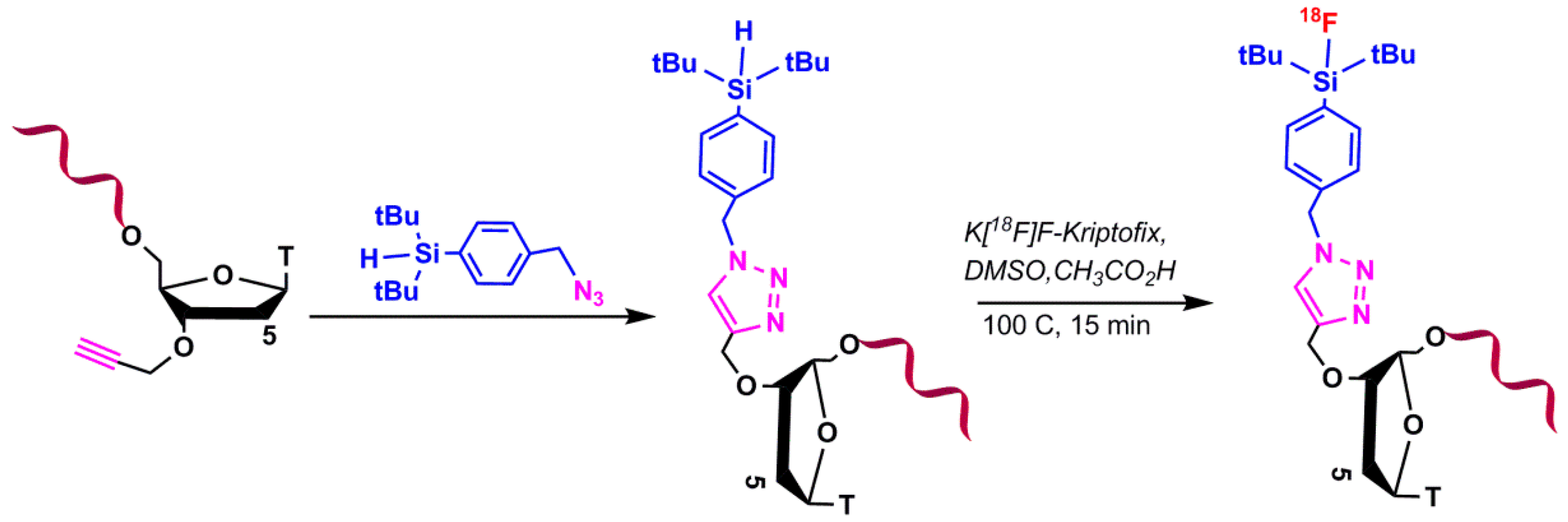
5.1.3. Silicon-Based Chemistry for 18F-Radiolabeling
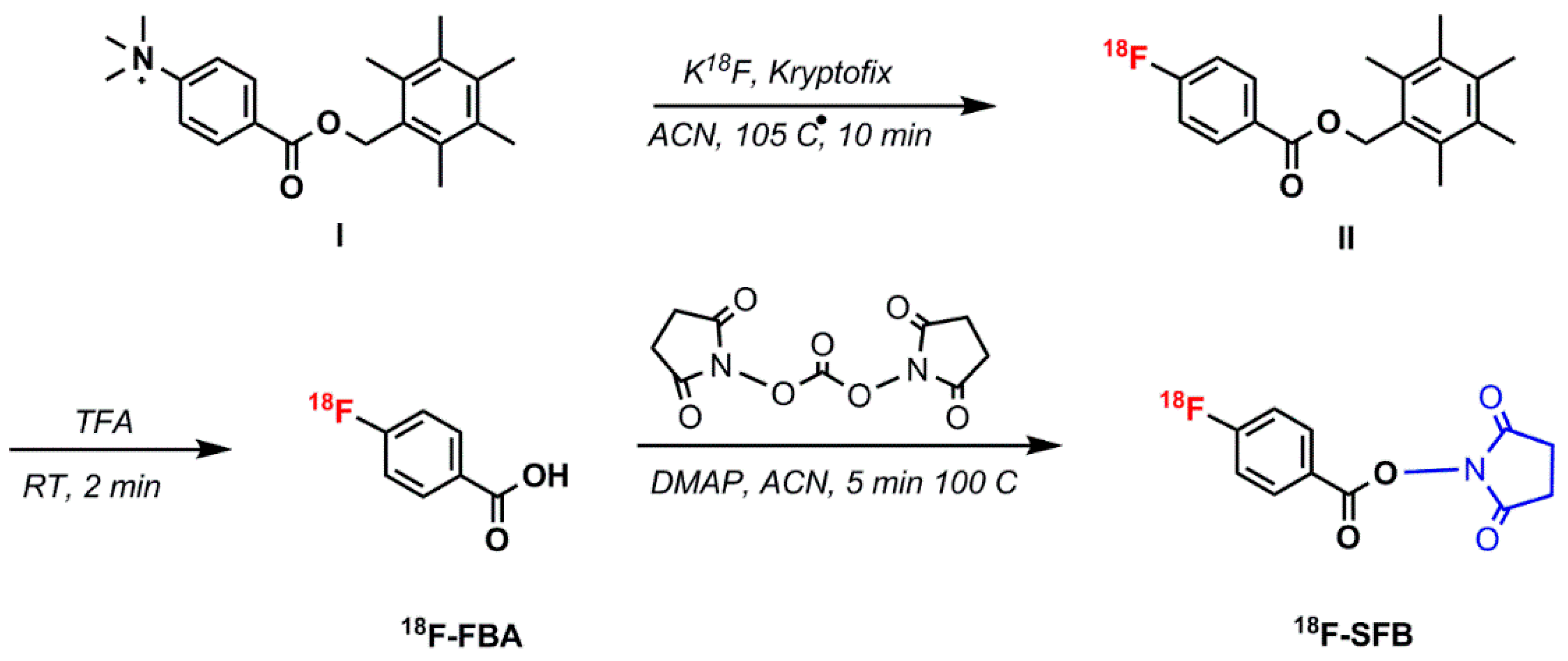
5.1.4. N–Succinimidyl 4-18F–Fluorobenzoate (18F–SFB)
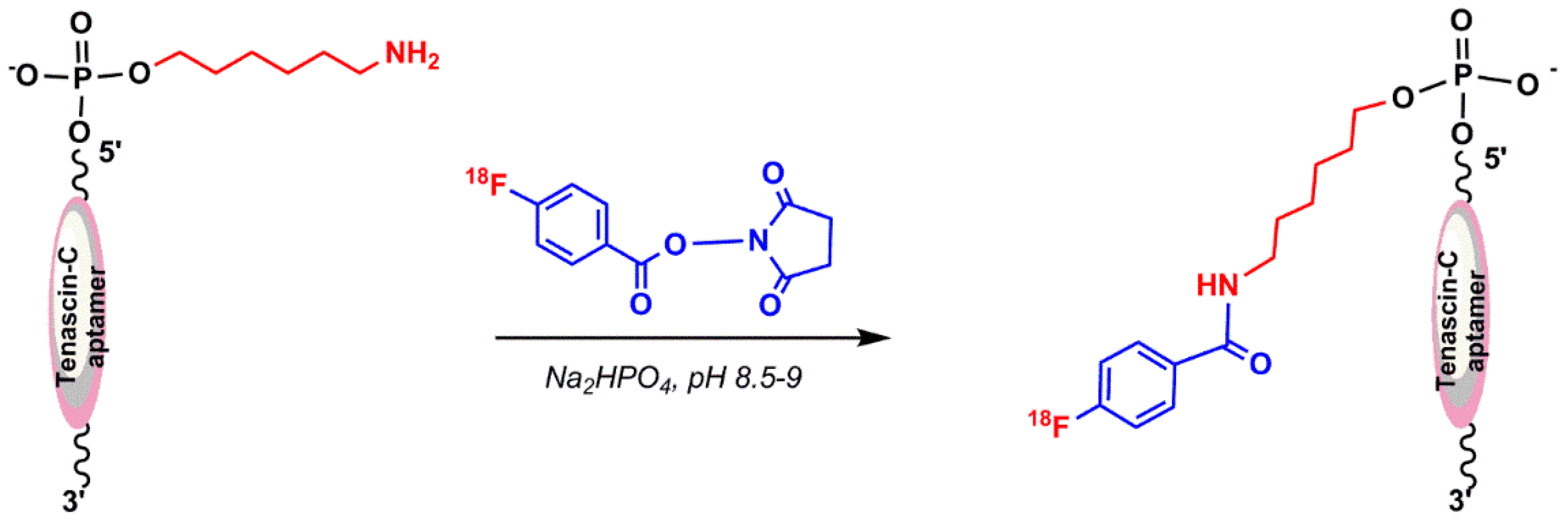
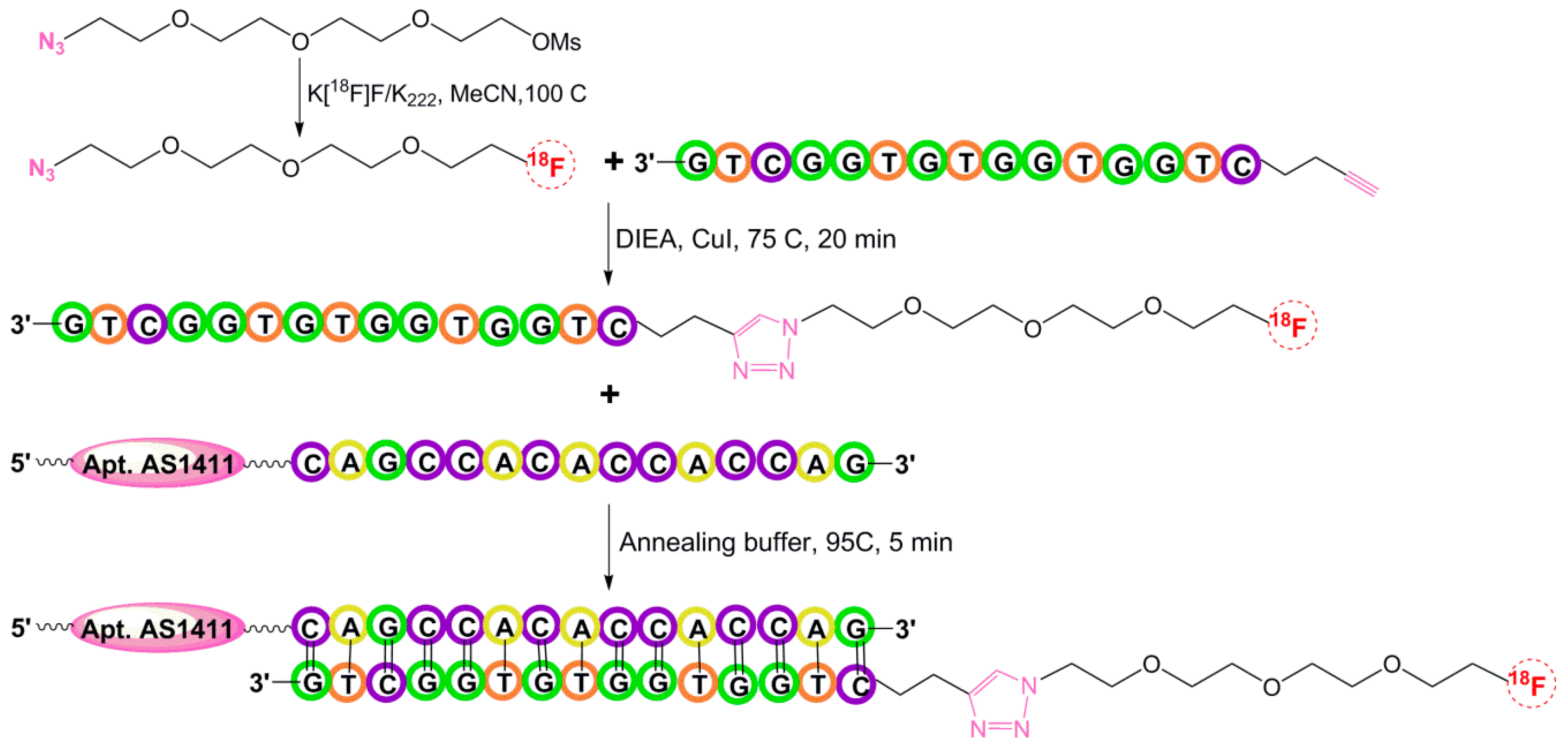
5.1.5. Hybridization-Based 18F-Radiolabeling of Aptamers:
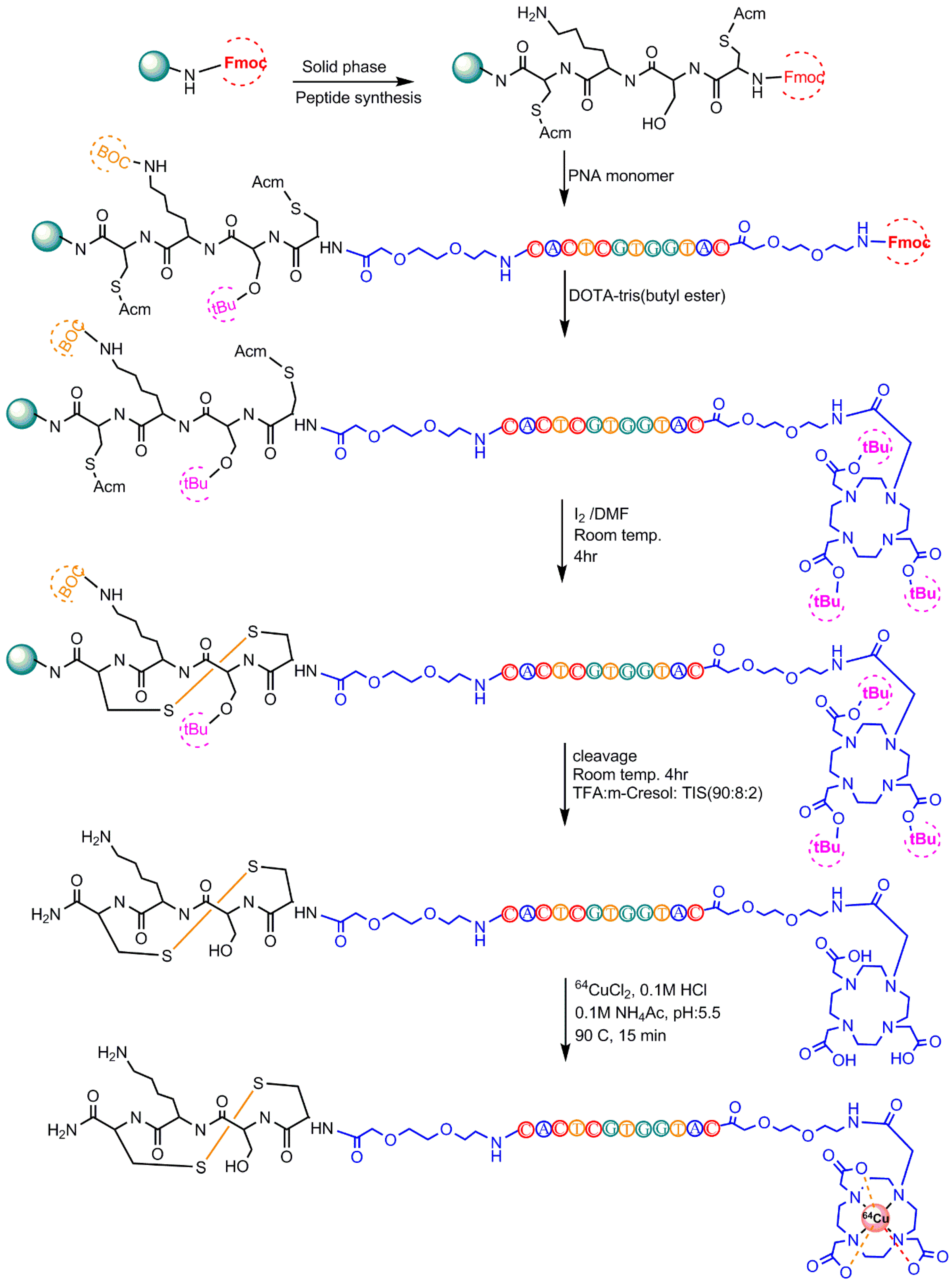
5.2. 64Cu-Radiolabeling
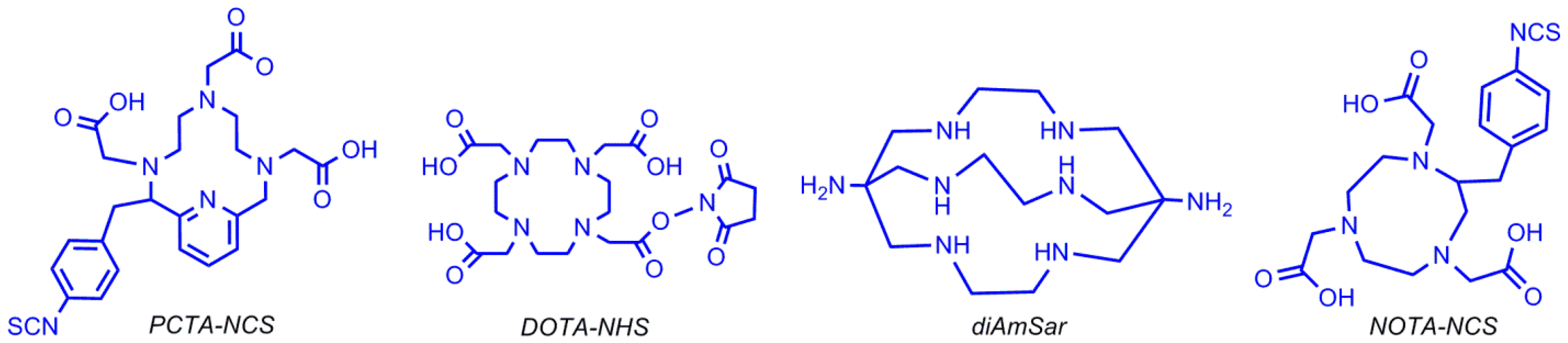
5.3. Zirconium-89 Labeling
5.3.1. Desferrioxamine B Chelating Agent
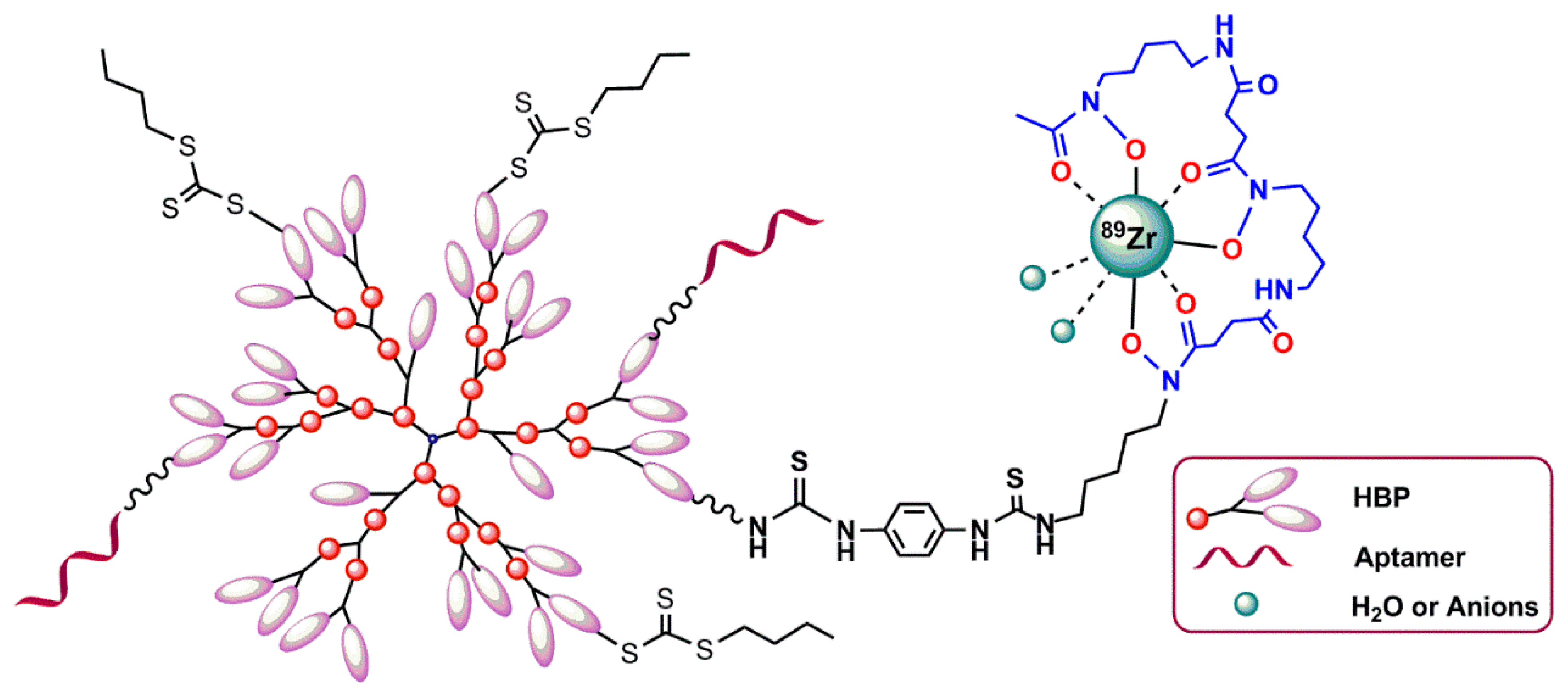
5.3.2. Hyperbranched Polymers (HBPs)
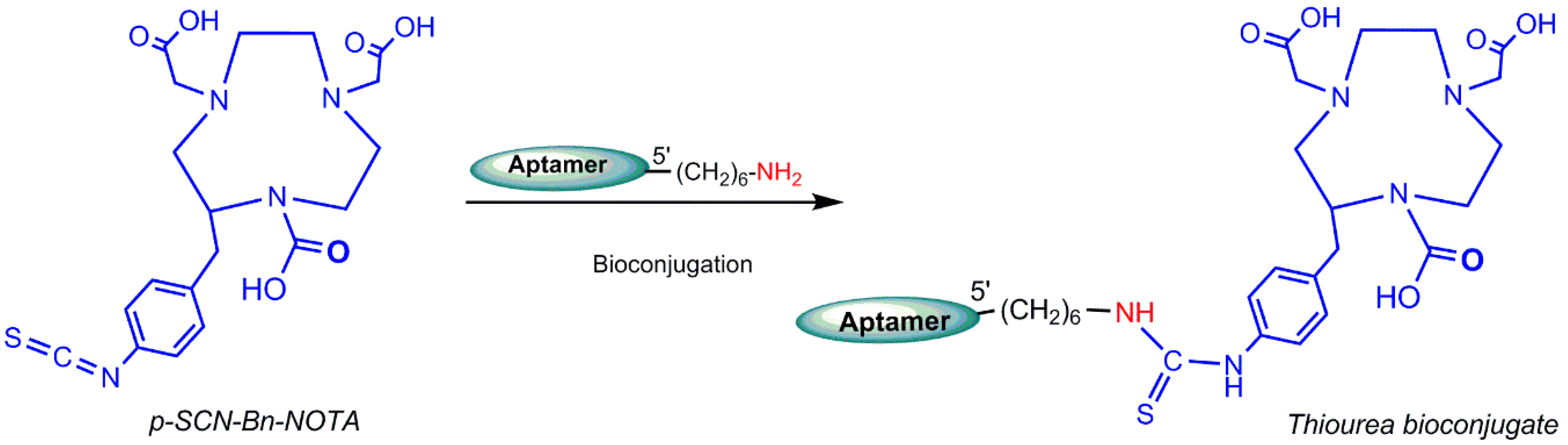
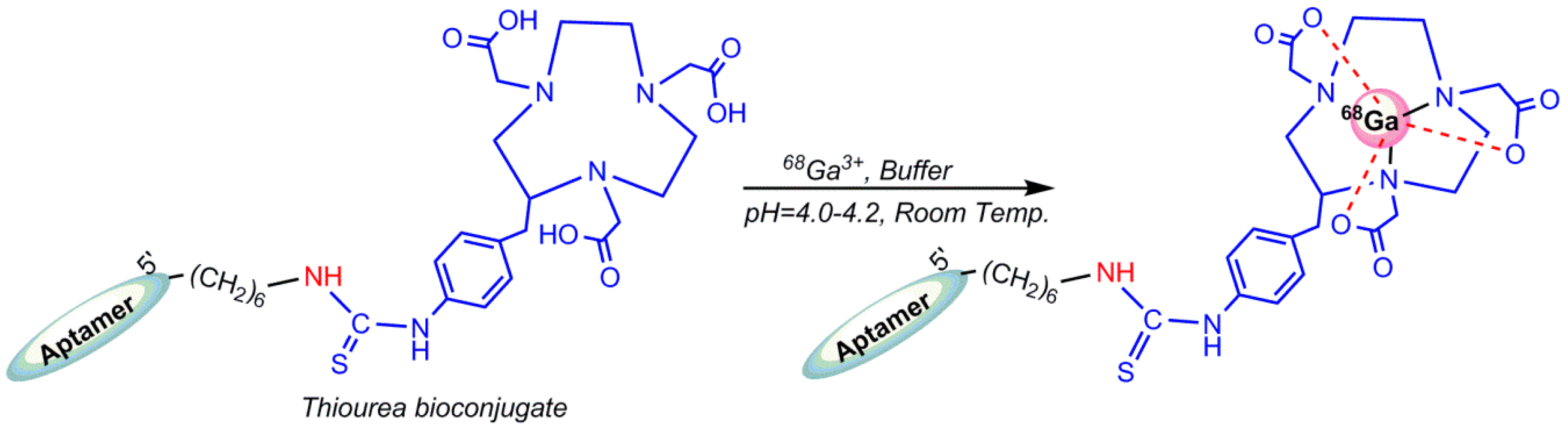
5.4. Gallium-68 (68Ga) Radiolabeling
6. Conclusions and Future Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Luker, G.D.; Piwnica-Worms, D. Molecular imaging in vivo with PET and SPECT. Acad. Radiol. 2001, 8, 4–14. [Google Scholar] [CrossRef]
- Pimlott, S.L.; Sutherland, A. Molecular tracers for the PET and SPECT imaging of disease. Chem. Soc. Rev. 2011, 40, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Thorsten, D.; Viktor, G. Molecular imaging in oncology using positron emission tomography. Medicine 2018, 115, 175–181. [Google Scholar]
- Saha, G.B. Fundamentals of Nuclear Pharmacy, 7th ed.; Springer: Basel, Switzerland, 2018; pp. 53–55, 70, 88, 93, 111–112, 148. [Google Scholar]
- Aerts, A.; Nren, I.; Gijs, M. Biological carrier molecules of radiopharmaceuticals for molecular cancer imaging and targeted cancer therapy. Curr. Pharm. Des. 2014, 20, 5218–5244. [Google Scholar] [CrossRef] [PubMed]
- Gijs, M.; Arets, A. Aptamers as radiopharmaceuticals for nuclear imaging and therapy. Nucl. Med. Biol. 2016, 43, 253–271. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G.; Carrillo, M.P. Development, screening, and analysis of DNA aptamer libraries potentially useful for diagnosis and passive immunity of arboviruses. BMC Res. Notes 2012, 5, 633. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G.; Kiel, J.L. In vitro selection of DNA aptamers to anthrax spores with electrochemiluminescence detection. Biosens. Bioelectron. 1999, 14, 457–464. [Google Scholar] [CrossRef]
- Ciesiolka, J.; Gorski, J. Selection of an RNA domain that binds zn2+. RNA 1995, 1, 538–550. [Google Scholar] [PubMed]
- Shangguan, D.; Li, Y. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef] [PubMed]
- Famulok, M. Molecular recognition of amino acids byRNA-aptamers: An l-citrulline binding RNA motif and its evolution into an l-arginine binder. J. Am. Chem. Soc. 1994, 116, 1698–1706. [Google Scholar] [CrossRef]
- Geiger, A.; Burgstaller, P. RNA aptamers that bind l-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 1996, 24, 1029–1036. [Google Scholar] [CrossRef] [PubMed]
- Jenison, R.D.; Gill, S.C. High-resolution molecular discrimination by RNA. Science 1994, 263, 1425–1429. [Google Scholar] [CrossRef] [PubMed]
- Sassanfar, M.; Szostak, J.W. An RNA motif that binds ATP. Nature 1993, 364, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Famulok, M.; Hartig, J.S. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 2007, 107, 3715–3743. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.E.; Wu, H.E. Improving the stability of aptamers by chemical modification. Curr. Med. Chem. 2011, 18, 4126–4138. [Google Scholar] [CrossRef] [PubMed]
- Lipi, F.; Chen, S. In vitro evolution of chemically-modified nucleic acid aptamers: Pros and cons, and comprehensive selection strategies. RNA Biol. 2016, 13, 1232–1245. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Cui, C. Bioapplications of cell-selex-generated aptamers in cancer diagnostics, therapeutics, theranostics and biomarker discovery: A comprehensive review. Cancers 2018, 10, 47. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Rossi, J. Targeted molecular imaging using aptamers in cancer. Pharmaceuticals 2018, 11, 71. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S. Radiopharmaceuticals for molecular imaging. Open Nucl. Med. J. 2010, 2, 178–185. [Google Scholar] [CrossRef]
- Eszter Borbas, K.B.; Catia, S.M.F. Design and synthesis of mono- and multimeric targeted radiopharmaceuticals based on novel cyclen ligands coupled to anti-muc1 aptamers for the diagnostic imaging and targeted radiotherapy of cancer. Bioconjug. Chem. 2007, 18, 1205–1212. [Google Scholar] [CrossRef] [PubMed]
- Barros, A.L.B.; Andrade, S.F. Radiolabeling of low molecular weight d-galactose-based glycodendrimer with technetium-99m and biodistribution studies. J. Radioanal. Nucl. Chem. 2013, 298, 605–609. [Google Scholar] [CrossRef]
- Cristiane, R.C.; Barros, A.L. Aptamers directly radiolabeled with technetium-99m as a potential agent capable of identifying carcinoembryonic antigen (CEA) in tumor cells T84. Bioorg. Med. Chem. Lett. 2014, 24, 1998–2001. [Google Scholar]
- Sousa, L.; Camila, M. (1→3)-β-d-glucan aptamers labeled with technetium-99 m: Biodistribution and imaging in experimental models of bacterial and fungal infection. Nucl. Med. Biol. 2017, 46, 19–24. [Google Scholar]
- Santos, S.R.; Rodrigues, C. Identification of staphylococcus aureus infection by aptamers directly radiolabeled with technetium-99m. Nucl. Med. Biol. 2015, 42, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liang, H. Cell-selex aptamer for highly specific radionuclide molecular imaging of glioblastoma in vivo. PLoS ONE 2014, 9, e90752. [Google Scholar] [CrossRef] [PubMed]
- Yuxia, L.; Guozheng, L. A brief review of chelators for radiolabeling oligomers. Materials 2010, 3, 3204–3217. [Google Scholar]
- Wang, Y.; Liu, G. Methods for MAG3 conjugation and 99mTc radiolabeling of biomolecules. Nat. Protoc. 2006, 1, 1477. [Google Scholar] [CrossRef] [PubMed]
- Abram, U.; Alberto, R. Technetium and rhenium: Coordination chemistry and nuclear medical applications. J. Braz. Chem. Soc. 2006, 17, 1486–1500. [Google Scholar] [CrossRef]
- Bormans, G.; Cleynhens, B. Comparison of benzyl, benzoyl and benzamidomethyl as protective groups for mercaptoacetyltriglycine (MAG3). J. Label. Compd. Radiopharm. 1989, 26, 50–52. [Google Scholar] [CrossRef]
- Winnard, P., Jr.; Chang, F. Preparation and use of NHS-MAG3 for technetium-99m labeling of DNA. Nucl. Med. Biol. 1997, 24, 425–432. [Google Scholar] [CrossRef]
- Duncan, R.J.; Weston, P.D. A new reagent which may be used to introduce sulfhydryl groups into proteins, and its use in the preparation of conjugates for immunoassay. Anal. Biochem. 1983, 132, 68–73. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, S. Improving the labeling of s-acetyl NHS−MAG3-conjugated morpholino oligomers. Bioconjug. Chem. 2002, 13, 893–897. [Google Scholar] [CrossRef] [PubMed]
- Rusckowski, M.; Qu, T. Inflammation and infection imaging with a (99m)Tc-neutrophil elastase inhibitor in monkeys. J. Nucl. Med. 2000, 41, 363–374. [Google Scholar] [PubMed]
- Yu-Min, Z.; Ning, L. Influence of different chelators (HYNIC, MAG3 and DTPA) on tumor cell accumulation and mouse biodistribution of technetium-99m labeled to antisense DNA. Eur. J. Nucl. Med. 2000, 27, 1700–1707. [Google Scholar]
- Kang, L.; Huo, Y. Noninvasive visualization of microRNA-155 in multiple kinds of tumors using a radiolabeled anti-miRNA oligonucleotide. Nucl. Med. Biol. 2016, 43, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Huo, Y. 99mTc radiolabeled anti-miR-155 oligonucleotide-phospholipids enveloped nanoparticles for cellular delivery and in vivo imaging. J. Nucl. Med. 2017, 58, 923. [Google Scholar]
- Liu, G.; Dou, S. Radiolabeling of MAG3-morpholino oligomers with 188Re at high labeling efficiency and specific radioactivity for tumor pre-targeting. Appl. Radiat. Isot. 2006, 64, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Yan-Rong, Z.; Yong-Xue, Z. Uptake kinetics of 99mTc-MAG3-antisense oligonucleotide to PCNA and effect on gene expression in vascular smooth muscle cells. J. Nucl. Med. 2005, 46, 1052–1058. [Google Scholar]
- Gomes, S.D.R. 99mTc-MAG3-aptamer for imaging human tumors associated with high level of matrix metalloprotease-9. Bioconjug. Chem. 2012, 23, 2192–2200. [Google Scholar] [CrossRef] [PubMed]
- Kryza, D.; Debordeaux, F. Ex vivo and in vivo imaging and biodistribution of aptamers targeting the human matrix metalloprotease-9 in melanomas. PLoS ONE 2016, 11, e0149387. [Google Scholar] [CrossRef] [PubMed]
- Childs, R.; Hnatowich, D. Optimum conditions for labeling of DTPA-coupled antibodies with technetium-99m. J. Nucl. Med. 1985, 26, 293–299. [Google Scholar] [PubMed]
- Hnatowich, D.; Layne, W. Radioactive labeling of antibody: A simple and efficient method. Science 1983, 220, 613–615. [Google Scholar] [CrossRef] [PubMed]
- Eckelman, W.C.; Karesh, S.M. New compounds: Fatty acid and long chain hydrocarbon derivatives containing a strong chelating agent. J. Pharm. Sci. 1975, 64, 704–706. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Cheng, D. Replacing 99m Tc with 111In improves MORF/cMORF pretargeting by reducing intestinal accumulation. Mol. Imaging Biol. 2009, 11, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Hnatowich, D.J.; Winnard, P., Jr. Technetium-99m labeling of DNA oligonucleotides. J. Nucl. Med. 1995, 36, 2306–2314. [Google Scholar] [PubMed]
- Calzada, V.; Moreno, M. Development of new PTK7-targeting aptamer-fluorescent and-radiolabelled probes for evaluation as molecular imaging agents: Lymphoma and melanoma in vivo proof of concept. Bioorg. Med. Chem. 2017, 25, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- Noaparast, Z.; Hosseinimehr, S. Tumor targeting with a (99m)Tc-labeled as 1411 aptamer in prostate tumor cells. J. Drug Target. 2015, 23, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, J.; Fischer, C. Radiosynthesis of new [90y]-DOTA-based maleimide reagents suitable for the prelabeling of thiol-bearing l-oligonucleotides and peptides. Bioconjug. Chem. 2009, 20, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Theobald, T. Sampson’s Textbook of Radiopharmacy; Pharmaceutical Press: London, UK, 2011; p. 96. [Google Scholar]
- Ren, B.X.; Yang, F. Magnetic resonance tumor targeting imaging using gadolinium labeled human telomerase reverse transcriptase antisense probes. Cancer Sci. 2012, 103, 1434–1439. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, D.W.; Su, H. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc. Natl. Acad. Sci. USA 2007, 104, 15549–15554. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Li, Z. PET/MRI dual-modality tumor imaging using arginine-glycine-aspartic (RGD)-conjugated radiolabeled iron oxide nanoparticles. J. Nucl. Med. 2008, 49, 1371–1379. [Google Scholar] [CrossRef] [PubMed]
- Sicco, E.; Baez, J. Derivatizations of sgc8-c aptamer to prepare metallic radiopharmaceuticals as imaging diagnostic agents: Syntheses, isolations, and physicochemical characterizations. Chem. Biol. Drug Des. 2018, 91, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Nunn, A.D. Radiopharmaceuticals: Chemistry and Pharmacology; CRC Press: Boca raton, FL, USA, 1992; Volume 55. [Google Scholar]
- de Vries, E.F.; Vroegh, J. Evaluation of fluorine-18-labeled alkylating agents as potential synthons for the labeling of oligonucleotides. Appl. Radiat. Isot. 2003, 58, 469–476. [Google Scholar] [CrossRef]
- Elisabeth, H.C.; Léngstroma’b, B. Synthesis of 4-([18f] fluoromethyl) phenyl isothiocyanate and its use in labelling oligonucleotides. Acta Chem. Scand. 1997, 51, 1236–1240. [Google Scholar]
- Gonda, J.; Kristian, P. Some nucleophilic reactions of 2-isothiocyanatobenzyl bromide. A new simple synthesis of 2-substituted 4H-benzo [d][1,3] thiazines. Collect. Czech. Chem. Commun. 1986, 51, 2802–2809. [Google Scholar] [CrossRef]
- Litak, P.T.; Kauffman, J.M. Syntheses of reactive fluorescent stains derived from 5 (2)-aryl-2 (5)-(4-pyridyl) oxazoles and bifunctionally reactive linkers. J. Hetrocycl. Chem. 1994, 31, 457–479. [Google Scholar] [CrossRef]
- Rotstein, B.H.; Stephenson, N.A.; Vasdev, N.; Liang, S.H. Spirocyclic hypervalent iodine (III)-mediated radiofluorination of non-activated and hindered aromatics. Nat. Commun. 2014, 5, 4365. [Google Scholar] [CrossRef] [PubMed]
- Orit Jacobson, I.D.W.; Lu Wang, Z.W. 18F-labeled single-stranded DNA aptamer for PET imaging of protein tyrosine kinase-7 expression. J. Nucl. Med. 2015, 56, 1780–1785. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Jacobson, O.; Zhu, G.; Chen, Z.; Liang, S.H.; Tian, R.; Yang, Z.; Niu, G.; Zhu, X.; Chen, X. PET imaging of EGFR expression using an (18)F-labeled RNA aptamer. Eur. J. Nucl. Med. Mol. Imaging 2018. [Google Scholar] [CrossRef]
- Zhu, G.; Zhang, H. Combinatorial screening of DNA aptamers for molecular imaging of HER2 in cancer. Bioconjug. Chem. 2017, 28, 1068–1075. [Google Scholar] [CrossRef] [PubMed]
- James, D.; Escudier, J.M. A ‘click chemistry’approach to the efficient synthesis of modified nucleosides and oligonucleotides for PET imaging. Tetrahedron Lett. 2010, 51, 1230–1232. [Google Scholar] [CrossRef]
- Lucas, R.; Zerrouki, R. A rapid efficient microwave-assisted synthesis of a 3′, 5′-pentathymidine by copper (I)-catalyzed [3 + 2] cycloaddition. Tetrahedron 2008, 64, 5467–5471. [Google Scholar] [CrossRef]
- Sommer, L.; Marans, N. A new reaction in organosilicon chemistry. J. Am. Chem. Soc. 1951, 73, 882. [Google Scholar] [CrossRef]
- Orit, J.; Xuefeng, Y. PET imaging of tenascin-c with a radiolabeled single-stranded DNA aptamer. J. Nucl. Med. 2015, 56, 616–621. [Google Scholar]
- Jacobson, O.; Weiss, I.D. PET of tumor CXCR4 expression with 4-18F-T140. J. Nucl. Med. 2010, 51, 1796–1804. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Park, J.Y.; Lee, T.S. Hybridization-based aptamer labeling using complementary oligonucleotide platform for PET and optical imaging. Biomaterials 2016, 100, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.J.; Ferdani, R. Copper-64 radiopharmaceuticals for PET imaging of cancer: Advances in preclinical and clinical research. Cancer Biother. Radiopharm. 2009, 24, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.J.; Wadas, T.J. Cross-bridged macrocyclic chelators for stable complexation of copper radionuclides for PET imaging. Q. J. Nucl. Med. Mol. Imaging 2008, 52, 185–193. [Google Scholar] [PubMed]
- Bass, L.A.; Wang, M. In vivo transchelation of copper-64 from TETA-octreotide to superoxide dismutase in rat liver. Bioconjug. Chem. 2000, 11, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Shokeen, M.; Anderson, C.J. Molecular imaging of cancer with copper-64 radiopharmaceuticals and positron emission tomography (PET). Acc. Chem. Res. 2009, 42, 832–841. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.V. Molecular imaging with copper-64. J. Inorg. Biochem. 2004, 98, 1874–1901. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wuest, M. Radiolabeling and in vivo behavior of copper-64-labeled cross-bridged cyclam ligands. J. Med. Chem. 2002, 45, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Li, Z. Evaluation of copper-64 labeled ambasar conjugated cyclic RGD peptide for improved microPET imaging of integrin αvβ3 expression. Bioconjug. Chem. 2010, 21, 1417–1424. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.L.; Yapp, D.T. Comparison of bifunctional chelates for 64cu antibody imaging. Eur. J. Nucl. Med. Mol. Imaging 2010, 37, 2117–2126. [Google Scholar] [CrossRef] [PubMed]
- William, M.; Rockey, B. Synthesis and radiolabeling of chelator–RNA aptamer bioconjugates with copper-64 for targeted molecular imaging. Bioorg. Med. Chem. 2011, 19, 4080–4090. [Google Scholar]
- Paudyal, B.; Zhang, K. Determining efficacy of breast cancer therapy by PET imaging of HER2 mRNA. Nucl. Med. Biol. 2013, 40, 994–999. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, H. Aptamer imaging with cu-64 labeled as 1411: Preliminary assessment in lung cancer. Nucl. Med. Biol. 2014, 41, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Abou, D.S.; Thorek, D.L. 89zr-labeled paramagnetic octreotide-liposomes for PET-MR imaging of cancer. Pharm. Res. 2013, 30, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Meijs, W.E.; Haisma, H.J. A facile method for the labeling of proteins with zirconium isotopes. Nucl. Med. Biol. 1996, 23, 439–448. [Google Scholar] [CrossRef]
- Meijs, W.E.; Herscheid, J.D. Evaluation of desferal as a bifunctional chelating agent for labeling antibodies with zr-89. Int. J. Rad. Appl. Instrum. A 1992, 43, 1443–1447. [Google Scholar] [CrossRef]
- Wadas, T.J.; Wong, E.H. Coordinating radiometals of copper, gallium, indium, yttrium, and zirconium for pet and spect imaging of disease. Chem. Rev. 2010, 110, 2858–2902. [Google Scholar] [CrossRef] [PubMed]
- Baroncelli, F.; Grossi, G. The complexing power of hydroxamic acids and its effect on the behaviour of organic extractants in the reprocessing of irradiated fuels—I. J. Inorg. Nucl. Chem. 1965, 27, 1085–1092. [Google Scholar] [CrossRef]
- Fuchs, A.V.; Tse, B.W. Evaluation of polymeric nanomedicines targeted to psma: Effect of ligand on targeting efficiency. Biomacromolecules 2015, 16, 3235–3247. [Google Scholar] [CrossRef] [PubMed]
- Pearce, A.K.; Rolfe, B.E. Development of a polymer theranostic for prostate cancer. Polym. Chem. 2014, 5, 6932–6942. [Google Scholar] [CrossRef]
- Rolfe, B.E.; Blakey, I. Multimodal polymer nanoparticles with combined 19F magnetic resonance and optical detection for tunable, targeted, multimodal imaging in vivo. J. Am. Chem. Soc. 2014, 136, 2413–2419. [Google Scholar] [CrossRef] [PubMed]
- England, R.M.; Rimmer, S. Hyper/highly-branched polymers by radical polymerisations. Polym. Chem. 2010, 1, 1533–1544. [Google Scholar] [CrossRef]
- Fletcher, N.L.; Houston, Z.H.; Simpson, J.D.; Veedu, R.N.; Thurecht, K.J. Designed multifunctional polymeric nanomedicines: Long-term biodistribution and tumour accumulation of aptamer-targeted nanomaterials. Chem. Commun. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hörsch, D.; Ezziddin, S. Peptide receptor radionuclide therapy for neuroendocrine tumors in germany: First results of a multi-institutional cancer registry. In Theranostics, Gallium-68, and Other Radionuclides; Baum, R., Rösch, F., Eds.; Springer: Berlin, Heidelberg, 2013; pp. 457–465. [Google Scholar]
- Zeglis, B.M.; Lewis, J.S. A practical guide to the construction of radiometallated bioconjugates for positron emission tomography. Dalton Trans. 2011, 40, 6168–6195. [Google Scholar] [CrossRef] [PubMed]
- Price, E.W.; Orvig, C. Matching chelators to radiometals for radiopharmaceuticals. Chem. Soc. Rev. 2014, 43, 260–290. [Google Scholar] [CrossRef] [PubMed]
- Gijs, M.; Dammicco, S. Gallium-68-labelled NOTA-oligonucleotides: An optimized method for their preparation. J. Label. Compd. Radiopharm. 2016, 59, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Baes, C.F.; Mesmer, R.S. The Hydrolysis of Cations; John Wiley & Sons: Hoboken, NJ, USA, 1976; pp. 318–319. [Google Scholar]
- Gijs, M.; Becker, G. Biodistribution of novel 68Ga-radiolabelled HER2 aptamers in mice. J. Nucl. Med. Radiat. Ther. 2016, 7, 300. [Google Scholar] [CrossRef]
- Kang, W.J.; Lee, J. Multimodal imaging probe for targeting cancer cells using umuc-1 aptamer. Colloids Surf. B Biointerfaces 2015, 136, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Veedu, R.N. Aptamers: Tools for Nanotherapy and Molecular Imaging; CRC Press: Boca raton, FL, USA, 2017; pp. 1–363. [Google Scholar]
- Kalia, J.; Raines, R.T. Advances in bioconjugation. Curr. Org. Chem. 2010, 14, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Kang, H. Bioimaging of nucleolin aptamer-containing 5-(n-benzylcarboxyamide)-2′-deoxyuridine more capable of specific binding to targets in cancer cells. J. Biomed. Biotech. 2010. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, X. Real-time imaging of protein internalization using aptamer conjugates. Anal. Chem. 2008, 80, 5002–5008. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; He, X. Locked nucleic acid/DNA chimeric aptamer probe for tumor diagnosis with improved serum stability and extended imaging window in vivo. Anal. Chim. Acta 2014, 812, 138–144. [Google Scholar] [CrossRef] [PubMed]



| Radioisotopes | Atomic Number | Physical Half-Life | Decay Mode (%) |
|---|---|---|---|
| 11C | 6 | 20.4 min | β+(100) |
| 13N | 7 | 9.96 min | β+(100) |
| 15O | 8 | 2.03 min | β+(100) |
| 18F | 9 | 109.8 min | β+(97) |
| 62Cu | 29 | 9.76 min | β+(97), EC(3) |
| 64Cu | 29 | 12.8 h | β+ Or β−, EC |
| 67Ga | 31 | 3.3 days | EC(100) |
| 68Ga | 31 | 68 min | β+(89), EC(11) |
| 82Rb | 37 | 75 s | β+(95), EC(5) |
| 89Zr | 40 | 78.1 h | β+(23), EC(77) |
| 94mTc | 43 | 52 min | β+(72), EC(28) |
| 99mTc | 43 | 6.0 h | IT(100) |
| 111In | 49 | 2.8 days | EC(100) |
| 123I | 53 | 13.2 h | EC(100) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassanzadeh, L.; Chen, S.; Veedu, R.N. Radiolabeling of Nucleic Acid Aptamers for Highly Sensitive Disease-Specific Molecular Imaging. Pharmaceuticals 2018, 11, 106. https://doi.org/10.3390/ph11040106
Hassanzadeh L, Chen S, Veedu RN. Radiolabeling of Nucleic Acid Aptamers for Highly Sensitive Disease-Specific Molecular Imaging. Pharmaceuticals. 2018; 11(4):106. https://doi.org/10.3390/ph11040106
Chicago/Turabian StyleHassanzadeh, Leila, Suxiang Chen, and Rakesh N. Veedu. 2018. "Radiolabeling of Nucleic Acid Aptamers for Highly Sensitive Disease-Specific Molecular Imaging" Pharmaceuticals 11, no. 4: 106. https://doi.org/10.3390/ph11040106
APA StyleHassanzadeh, L., Chen, S., & Veedu, R. N. (2018). Radiolabeling of Nucleic Acid Aptamers for Highly Sensitive Disease-Specific Molecular Imaging. Pharmaceuticals, 11(4), 106. https://doi.org/10.3390/ph11040106





