Adaptive Time–Frequency Segment Optimization for Motor Imagery Classification
Abstract
1. Introduction
- A new approach to optimize the time–frequency segment is defined, aiming to provide a subject-specific time–frequency segment.
- We develop a novel framework for MI decoding. Rather than employing a non-customized time–frequency segment, our approach adaptively explores the optimal time–frequency segment without constraints on limited optional segments.
- To verify the effectiveness of the proposed algorithm, three MI-EEG datasets are included for classification experiments, and the proposed algorithm shows competitive and robust performance.
2. Related Works
3. Materials and Methods
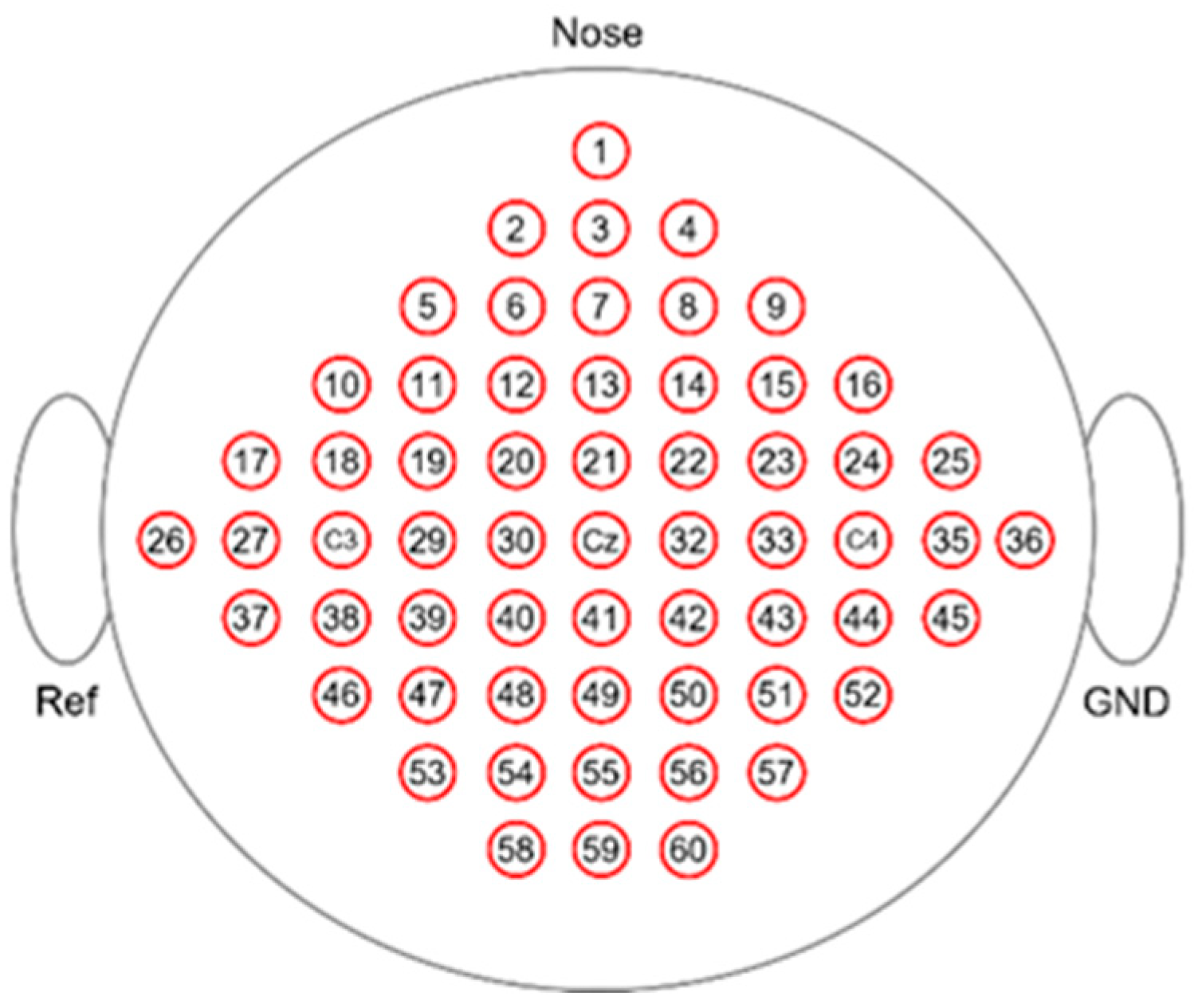
3.1. Datasets
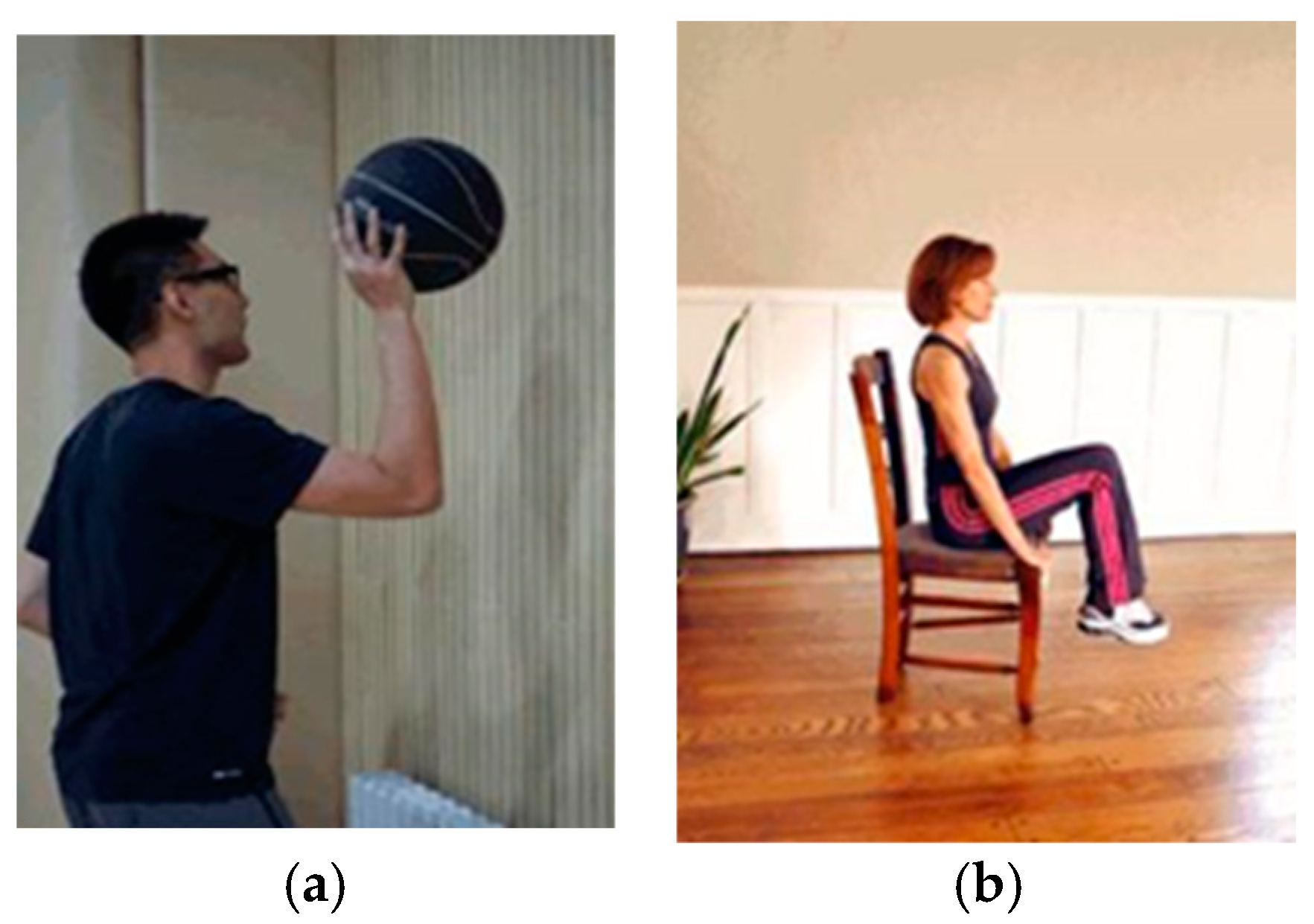
3.1.1. BCI Competition III Dataset IIIa (DS1)
3.1.2. Chinese Academy of Medical Sciences Autonomous Dataset (DS2)
3.1.3. BCI Competition IV Dataset 1 (DS3)
3.2. Method
3.2.1. Sparrow Search Algorithm-Based Time–Frequency Segment Optimization
- (1)
- Producer, responsible for providing appropriate exploration areas or directions for all sparrows seeking to optimize the time–frequency segments of EEG signals;
- (2)
- Scrounger, following the sparrows that can indicate the direction of better time–frequency segments;
- (3)
- Scout, aware of potential shortcomings in the current process of optimizing the time–frequency segment range of EEG signals, and seeking better time–frequency segments by escaping from the current position.
3.2.2. Correlation-Based Channel Selection Algorithm
3.2.3. Feature Extraction Based on Regularized Common Spatial Pattern
3.2.4. Feature Classification Based on Support Vector Machine
4. Results
4.1. Data Preprocessing
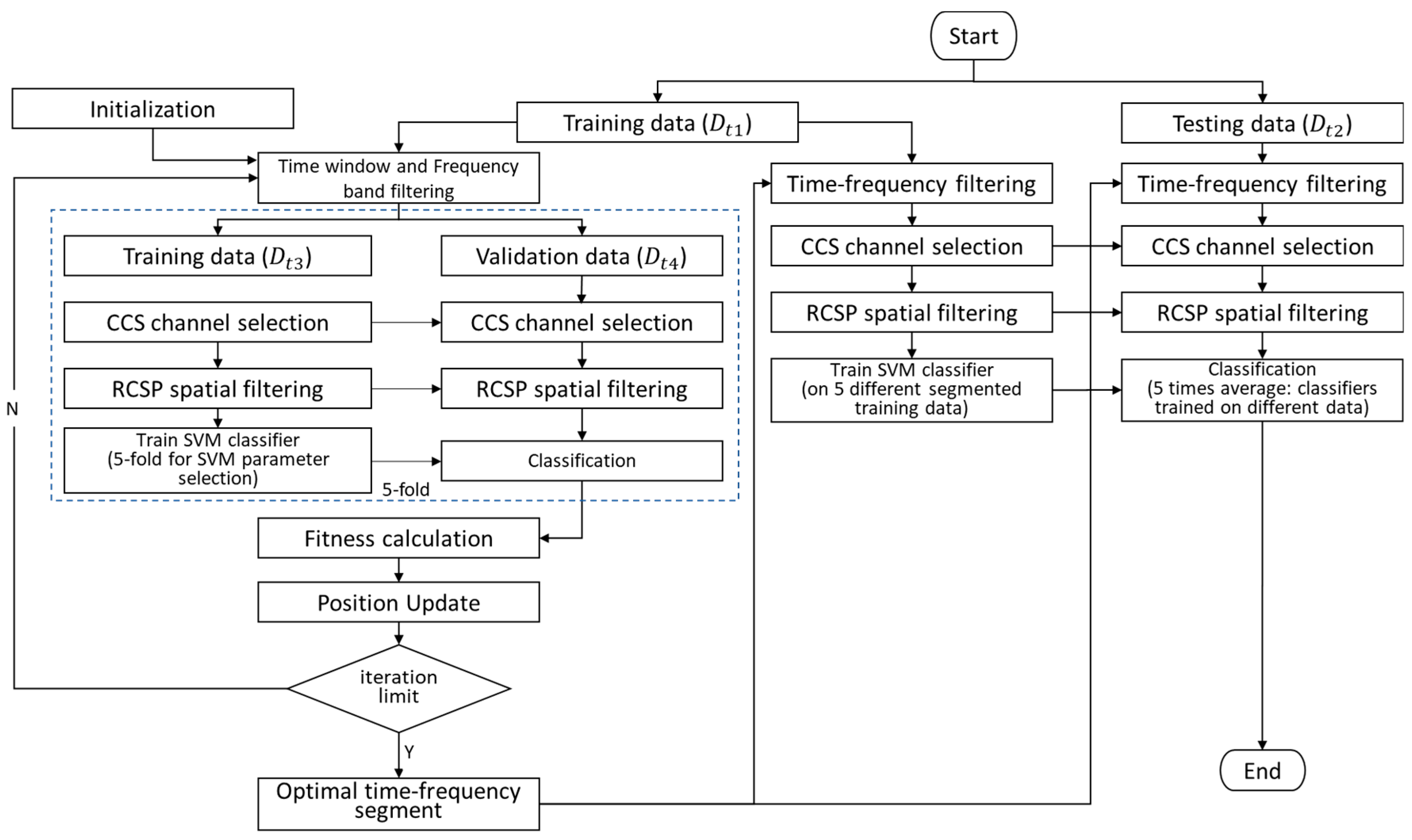
4.2. Whole Framework
- (1)
- Initialize the positions of n (n = 10) sparrows, corresponding to time–frequency segments;
- (2)
- Split the experimental data into a training set and a test set in a 7:3 ratio;
- (3)
- Apply time–frequency filtering to based on the current time–frequency segment;
- (4)
- Utilize a five-fold cross-validation method, where is split into a training set and a validation set in each iteration (in a 4:1 ratio);
- (5)
- Calculate the optimal channels for using the CCS algorithm and remove irrelevant channel data from both and ;
- (6)
- Compute RCSP spatial filters on and extract features from both and ;
- (7)
- Train an SVM classifier on and evaluate the accuracy (fitness) on the test set ;
- (8)
- Update the time–frequency segments for each sparrow based on the fitness;
- (9)
- Repeat steps (3) to (8) until the predefined iteration limit is reached; in this experiment, the iteration limit was set to 20. The current optimal result obtained is the best time–frequency segment;
- (10)
- Use as the training set and as the test set. The ratios are determined according to the specifications of the datasets providers: 1:1 for DS1, 3:1 for DS2, and 3:7 for DS3. Based on the best time–frequency segment obtained, calculate the accuracy of the current model on the test set according to steps (5) to (7).
4.3. Experimental Results
4.4. Analysis of Variance for Results
5. Discussion
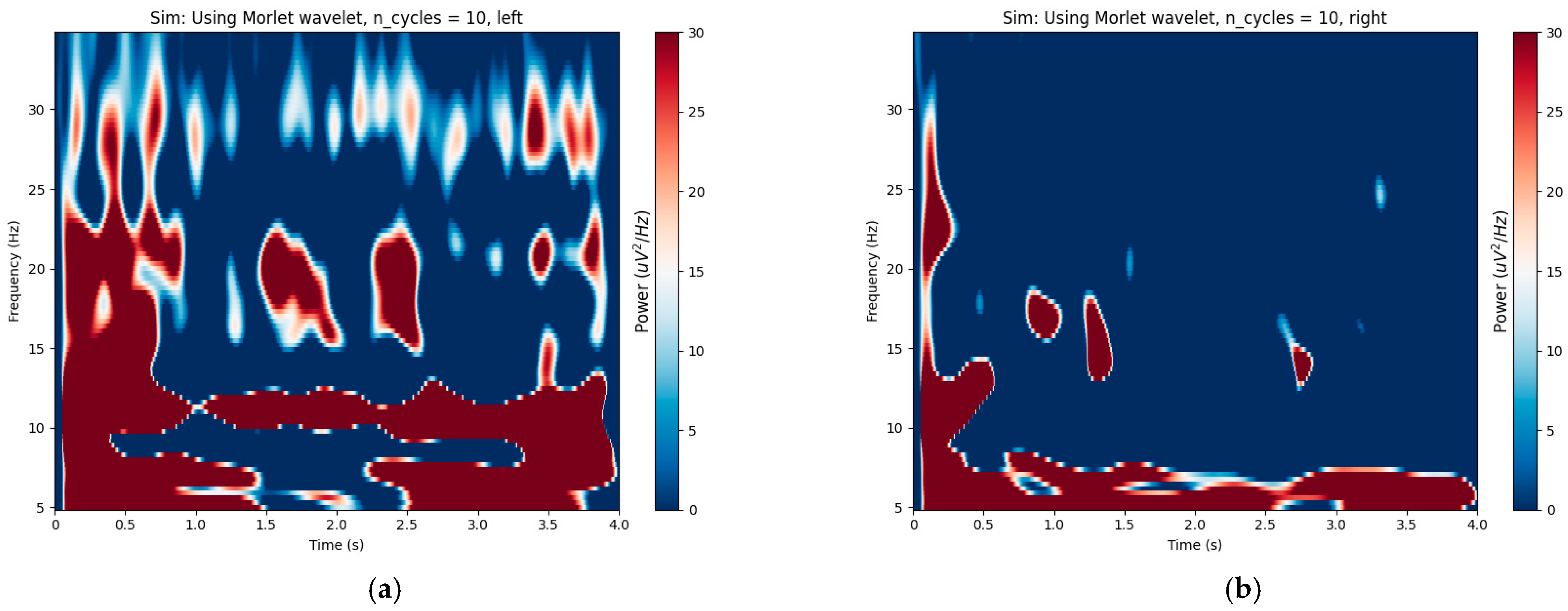
5.1. Optimal Time–Frequency Segment Distributions
5.2. Effect of SSA on Channel Selection
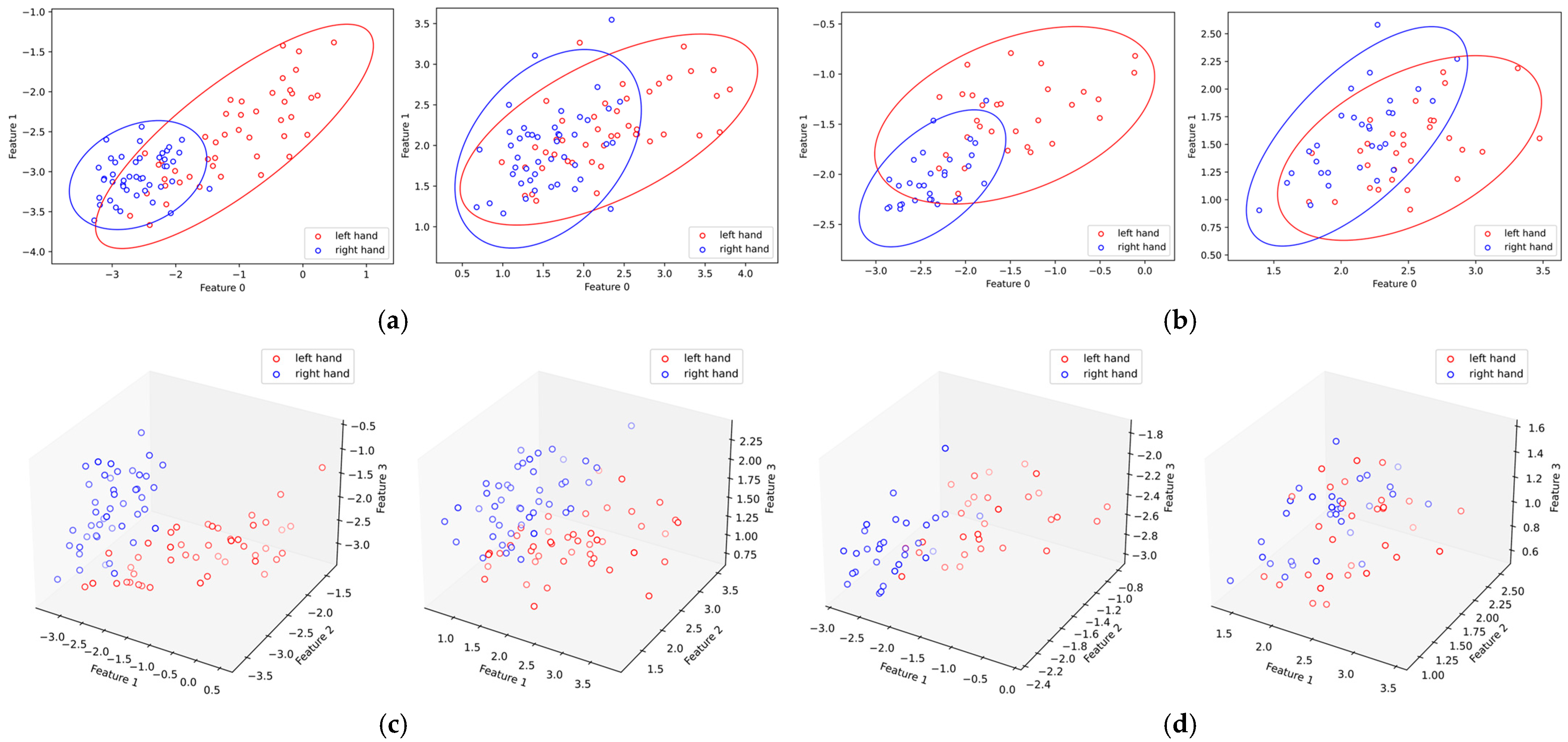
5.3. Effect of SSA on RCSP Feature Distribution
5.4. Effects of CCS and SSA on Improving Classification
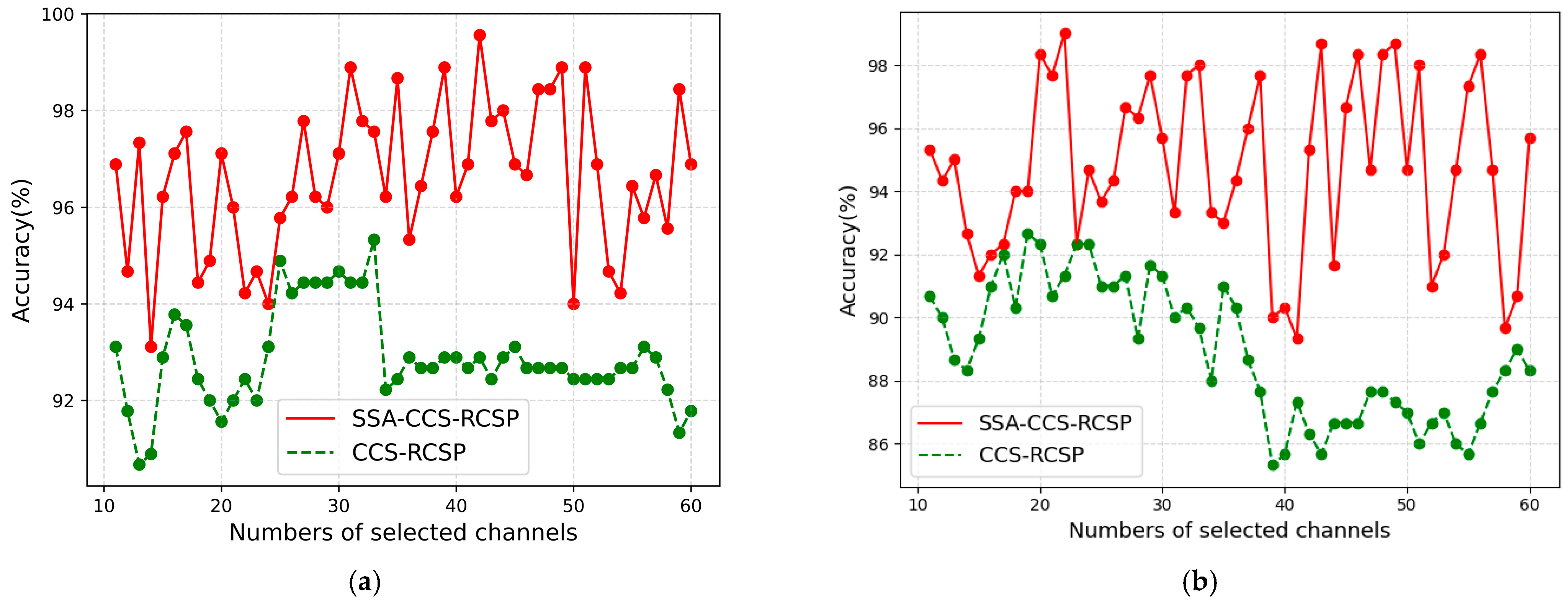
5.5. Performance of SSA with Different Channel Numbers
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolpaw, J.R.; Birbaumer, N.; Mcfarland, D.J.; Pfurtscheller, G.; Vaughan, T.M. Brain-computer interfaces for communication and control. Suppl. Clin. Neurophysiol. 2002, 113, 767–791. [Google Scholar] [CrossRef] [PubMed]
- Pfurtscheller, G.; Neuper, C.; Schlogl, A.; Lugger, K. Separability of EEG signals recorded during right and left motor imagery using adaptive autoregressive parameters. IEEE Trans. Rehabil. Eng. 1998, 6, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Jasper, H.; Penfield, W. Electrocorticograms in man: Effect of voluntary movement upon the electrical activity of the precentral gyrus. Arch. Psychiatr. Nervenkrankh. 1949, 183, 163–174. [Google Scholar] [CrossRef]
- Koles, Z.J.; Lazar, M.S.; Zhou, S.Z. Spatial patterns underlying population differences in the background EEG. Brain Topogr. 1990, 2, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Miao, Y.; Daly, I.; Zuo, C.; Cichocki, A. Correlation-based channel selection and regularized feature optimization for MI-based BCI. Neural Netw. 2019, 118, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Lemm, S.; Blankertz, B.; Curio, G.; Muller, K.-R. Spatio-spectral filters for improving the classification of single trial EEG. IEEE Trans. Biomed. Eng. 2005, 52, 1541–1548. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Eng, H.-L.; Guan, C.; Plataniotis, K.N.; Venetsanopoulos, A.N. Regularized Common Spatial Pattern with Aggregation for EEG Classification in Small-Sample Setting. IEEE Trans. Biomed. Eng. 2010, 57, 2936–2946. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, X.; Wu, Z.; Sun, B.; Li, T. Motor imagery recognition with automatic EEG channel selection and deep learning. J. Neural Eng. 2020, 18, 016004. [Google Scholar] [CrossRef]
- Quadrianto, N.; Cuntai, G.; Dat, T.H.; Xue, P. Sub-band Common Spatial Pattern (SBCSP) for Brain-Computer Interface. In Proceedings of the International IEEE/EMBS Conference on Neural Engineering, Kohala Coast, HI, USA, 2–5 May 2007. [Google Scholar]
- Ang, K.K.; Chin, Z.Y.; Zhang, H. Cuntai Guan Filter Bank Common Spatial Pattern (FBCSP) in Brain-Computer Interface. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks, Hong Kong, China, 1–8 June 2008; pp. 2390–2397. [Google Scholar]
- Kirar, J.S.; Agrawal, R.K. Relevant Feature Selection from a Combination of Spectral-Temporal and Spatial Features for Classification of Motor Imagery EEG. J. Med. Syst. 2018, 42, 78. [Google Scholar] [CrossRef]
- Varone, G.; Boulila, W.; Driss, M.; Kumari, S.; Khan, M.K.; Gadekallu, T.R.; Hussain, A. Finger pinching and imagination classification: A fusion of CNN architectures for IoMT-enabled BCI applications. Inf. Fusion 2024, 101, 102006. [Google Scholar] [CrossRef]
- Yu, X.; Aziz, M.Z.; Sadiq, M.T.; Jia, K.; Fan, Z.; Xiao, G. Computerized Multidomain EEG Classification System: A New Paradigm. IEEE J. Biomed. Health Inform. 2022, 26, 3626–3637. [Google Scholar] [CrossRef]
- Yu, X.; Aziz, M.Z.; Sadiq, M.T.; Fan, Z.; Xiao, G. A New Framework for Automatic Detection of Motor and Mental Imagery EEG Signals for Robust BCI Systems. IEEE Trans. Instrum. Meas. 2021, 70, 1–12. [Google Scholar] [CrossRef]
- Peterson, V.; Wyser, D.; Lambercy, O.; Spies, R.; Gassert, R. A penalized time–frequency band feature selection and classification procedure for improved motor intention decoding in multichannel EEG. J. Neural Eng. 2019, 16, 016019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Nam, C.S.; Zhou, G.; Jin, J.; Wang, X.; Cichocki, A. Temporally Constrained Sparse Group Spatial Patterns for Motor Imagery BCI. IEEE Trans. Cybern. 2019, 49, 3322–3332. [Google Scholar] [CrossRef] [PubMed]
- Gaur, P.; McCreadie, K.; Pachori, R.B.; Wang, H.; Prasad, G. An automatic subject specific channel selection method for enhancing motor imagery classification in EEG-BCI using correlation. Biomed. Signal Process. Control 2021, 68, 102574. [Google Scholar] [CrossRef]
- Shan, H.; Xu, H.; Zhu, S.; He, B. A novel channel selection method for optimal classification in different motor imagery BCI paradigms. Biomed. Eng. Online 2015, 14, 93. [Google Scholar] [CrossRef]
- Strypsteen, T.; Bertrand, A. End-to-end learnable EEG channel selection for deep neural networks with Gumbel-softmax. J. Neural Eng. 2021, 18, 0460a9. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, G.; Wang, Y.; Yang, L.; Liang, Z.; Cong, F. One-Dimensional Convolutional Neural Networks Combined with Channel Selection Strategy for Seizure Prediction Using Long-Term Intracranial EEG. Int. J. Neural Syst. 2022, 32, 2150048. [Google Scholar] [CrossRef]
- Sun, B.; Liu, Z.; Wu, Z.; Mu, C.; Li, T. Graph Convolution Neural Network based End-to-end Channel Selection and Classification for Motor Imagery Brain-computer Interfaces. IEEE Trans. Ind. Inform. 2022, 19, 9314–9324. [Google Scholar] [CrossRef]
- Sun, B.; Zhang, H.; Wu, Z.; Zhang, Y.; Li, T. Adaptive Spatiotemporal Graph Convolutional Networks for Motor Imagery Classification. IEEE Signal Process. Lett. 2021, 28, 219–223. [Google Scholar] [CrossRef]
- Sun, B.; Wu, Z.; Hu, Y.; Li, T. Golden subject is everyone: A subject transfer neural network for motor imagery-based brain computer interfaces. Neural Netw. 2022, 151, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Zhao, X.; Zhang, H.; Bai, R.; Li, T. EEG Motor Imagery Classification with Sparse Spectrotemporal Decomposition and Deep Learning. IEEE Trans. Autom. Sci. Eng. 2021, 18, 541–551. [Google Scholar] [CrossRef]
- Idowu, O.P.; Adelopo, O.; Ilesanmi, A.E.; Li, X.; Samuel, O.W.; Fang, P.; Li, G. Neuro-evolutionary approach for optimal selection of EEG channels in motor imagery based BCI application. Biomed. Signal Process. Control 2021, 68, 102621. [Google Scholar] [CrossRef]
- Blankertz, B.; Muller, K.-R.; Krusienski, D.J.; Schalk, G.; Wolpaw, J.R.; Schlogl, A.; Pfurtscheller, G.; Millan, J.R.; Schroder, M.; Birbaumer, N. The BCI competition III: Validating alternative approaches to actual BCI problems. IEEE Trans. Neural Syst. Rehabil. Eng. 2006, 14, 153–159. [Google Scholar] [CrossRef] [PubMed]
- BCI Competition: Download Area. Available online: https://bbci.de/competition/iii/#data_set_iiia (accessed on 27 December 2023).
- Blankertz, B.; Dornhege, G.; Krauledat, M.; Müller, K.-R.; Curio, G. The non-invasive Berlin Brain–Computer Interface: Fast acquisition of effective performance in untrained subjects. NeuroImage 2007, 37, 539–550. [Google Scholar] [CrossRef] [PubMed]
- BCI Competition: Download Area. Available online: https://www.bbci.de/competition/iv/ (accessed on 20 December 2023).
- Yang, Y.; Chevallier, S.; Wiart, J.; Bloch, I. Subject-specific time–frequency selection for multi-class motor imagery-based BCIs using few Laplacian EEG channels. Biomed. Signal Process. Control 2017, 38, 302–311. [Google Scholar] [CrossRef]
- Xue, J.; Shen, B. A novel swarm intelligence optimization approach: Sparrow search algorithm. Syst. Sci. Control Eng. Open Access J. 2020, 8, 22–34. [Google Scholar] [CrossRef]
- Feng, J.; Yin, E.; Jin, J.; Saab, R.; Daly, I.; Wang, X.; Hu, D.; Cichocki, A. Towards correlation-based time window selection method for motor imagery BCIs. Neural Netw. 2018, 102, 87–95. [Google Scholar] [CrossRef]
- Wang, L. Support Vector Machines: Theory and Applications; Springer Science & Business Media: Dordrecht, The Netherlands, 2005; Volume 177. [Google Scholar]
- Delorme, A.; Makeig, S. EEGLAB: An open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods 2004, 134, 9–21. [Google Scholar] [CrossRef]
- Pfurtscheller, G.; Neuper, C. Motor imagery and direct brain-computer communication. Proc. IEEE 2001, 89, 1123–1134. [Google Scholar] [CrossRef]







| Subjects | Accuracy (%) | ||||||
|---|---|---|---|---|---|---|---|
| CSP | CCS-CSP | RCSP | CCS-RCSP | SSA-RCSP | FBCSP | SSA-CCS-RCSP | |
| k3b | 92.22 | 94.89 | 92.22 | 95.33 | 98.00 | 79.38 | 99.56 |
| l1b | 91.33 | 94.33 | 88.33 | 92.67 | 93.00 | 53.93 | 98.67 |
| mean | 91.78 | 94.61 | 90.28 | 94.00 | 95.50 | 66.66 | 99.11 |
| A01 | 73.25 | 73.25 | 76.00 | 79.25 | 76.25 | 84.00 | 87.75 |
| A02 | 91.50 | 92.75 | 91.50 | 93.00 | 94.25 | 95.75 | 97.25 |
| A03 | 82.75 | 86.75 | 88.75 | 89.25 | 89.75 | 83.50 | 92.25 |
| A04 | 88.75 | 90.25 | 90.50 | 91.50 | 94.75 | 90.50 | 96.75 |
| A05 | 70.00 | 70.50 | 67.75 | 73.25 | 73.75 | 72.50 | 76.75 |
| A06 | 96.00 | 96.25 | 95.00 | 96.25 | 97.00 | 93.75 | 98.50 |
| A07 | 59.00 | 65.50 | 63.50 | 65.50 | 72.50 | 58.75 | 72.50 |
| A08 | 83.50 | 86.50 | 80.00 | 84.50 | 88.00 | 90.00 | 91.00 |
| A09 | 73.25 | 73.25 | 65.25 | 65.25 | 67.25 | 72.75 | 78.75 |
| mean | 79.78 | 81.67 | 79.81 | 81.97 | 83.72 | 82.39 | 87.94 |
| a | 63.67 | 86.67 | 59.33 | 91.00 | 86.00 | 81.60 | 94.00 |
| b | 72.67 | 76.67 | 71.33 | 76.67 | 74.33 | 62.13 | 82.67 |
| f | 75.17 | 86.00 | 88.33 | 89.33 | 88.00 | 81.80 | 91.33 |
| g | 75.17 | 91.33 | 89.00 | 92.00 | 90.33 | 84.53 | 94.00 |
| mean | 71.67 | 85.17 | 77.00 | 87.25 | 84.67 | 77.52 | 90.50 |
| Subjects | Degree of Freedom | Sum of Squares | Mean Square | F | PR (>F) |
|---|---|---|---|---|---|
| dataset | 2.0 | 0.077 | 0.039 | 3.517 | 0.034 * |
| algorithm | 6.0 | 0.198 | 0.033 | 3.002 | 0.010 * |
| dataset * algorithm | 12.0 | 0.141 | 0.012 | 1.070 | 0.396 |
| Residual | 84.0 | 0.924 | 0.011 |
| Comparison | Mean Difference | Standard Error | t-Value | p-Value | |
|---|---|---|---|---|---|
| CCS-CSP | CCS-RCSP | −0.007 | 0.038 | −0.171 | 0.865 |
| CCS-CSP | CSP | 0.051 | 0.038 | 1.328 | 0.187 |
| CCS-CSP | FBCSP | 0.053 | 0.038 | 1.387 | 0.169 |
| CCS-CSP | RCSP | 0.039 | 0.038 | 1.007 | 0.317 |
| CCS-CSP | SSA-CCS-RCSP | −0.058 | 0.038 | −1.505 | 0.136 |
| CCS-CSP | SSA-RCSP | −0.012 | 0.038 | −0.317 | 0.752 |
| CCS-RCSP | CSP | 0.058 | 0.038 | 1.499 | 0.137 |
| CCS-RCSP | FBCSP | 0.060 | 0.038 | 1.557 | 0.123 |
| CCS-RCSP | RCSP | 0.045 | 0.038 | 1.178 | 0.242 |
| CCS-RCSP | SSA-CCS-RCSP | −0.051 | 0.038 | −1.334 | 0.185 |
| CCS-RCSP | SSA-RCSP | −0.006 | 0.038 | −0.146 | 0.884 |
| CSP | FBCSP | 0.002 | 0.038 | 0.058 | 0.954 |
| CSP | RCSP | −0.012 | 0.038 | −0.322 | 0.748 |
| CSP | SSA-CCS-RCSP | −0.109 | 0.038 | −2.833 | 0.006 |
| CSP | SSA-RCSP | −0.063 | 0.038 | −1.645 | 0.103 |
| FBCSP | RCSP | −0.015 | 0.038 | −0.380 | 0.705 |
| FBCSP | SSA-CCS-RCSP | −0.111 | 0.038 | −2.891 | 0.005 |
| FBCSP | SSA-RCSP | −0.066 | 0.038 | −1.703 | 0.092 |
| RCSP | SSA-CCS-RCSP | −0.097 | 0.038 | −2.512 | 0.014 |
| RCSP | SSA-RCSP | −0.051 | 0.038 | −1.323 | 0.189 |
| SSA-CCS-RCSP | SSA-RCSP | 0.046 | 0.038 | 1.188 | 0.238 |
| Algorithm | Algorithm | DS1 | DS2 | DS3 | |||
|---|---|---|---|---|---|---|---|
| Accuracy (%) | NC | Accuracy (%) | NC | Accuracy (%) | NC | ||
| Spatial Pattern | CSP [4,28,35] | 91.78 | 60 | 79.78 | 26 | 71.67 | 59 |
| RCSP [5,7] | 90.28 | 60 | 79.81 | 26 | 77.00 | 59 | |
| SSA Improved | SSA-RCSP | 95.50 | 60 | 83.72 | 26 | 84.67 | 59 |
| Channel Selection | CCS-CSP | 94.61 | 19 | 81.67 | 20.4 | 85.17 | 25.25 |
| CCS-RCSP | 94.00 | 35 | 81.97 | 19.7 | 87.25 | 35.25 | |
| SSA Improved (This study) | SSA-CCS-RCSP | 99.11 | 42.5 | 87.94 | 21.4 | 90.50 | 25.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Li, G.; Zhang, Q.; Yu, Q.; Li, T. Adaptive Time–Frequency Segment Optimization for Motor Imagery Classification. Sensors 2024, 24, 1678. https://doi.org/10.3390/s24051678
Huang J, Li G, Zhang Q, Yu Q, Li T. Adaptive Time–Frequency Segment Optimization for Motor Imagery Classification. Sensors. 2024; 24(5):1678. https://doi.org/10.3390/s24051678
Chicago/Turabian StyleHuang, Junjie, Guorui Li, Qian Zhang, Qingmin Yu, and Ting Li. 2024. "Adaptive Time–Frequency Segment Optimization for Motor Imagery Classification" Sensors 24, no. 5: 1678. https://doi.org/10.3390/s24051678
APA StyleHuang, J., Li, G., Zhang, Q., Yu, Q., & Li, T. (2024). Adaptive Time–Frequency Segment Optimization for Motor Imagery Classification. Sensors, 24(5), 1678. https://doi.org/10.3390/s24051678







