Evaluating Synthetic Medical Images Using Artificial Intelligence with the GAN Algorithm
Abstract
1. Introduction
- Quantitative evaluation: the assessment of the quality (noise, similarity) of the images.
- Qualitative evaluation: the assessment of the reliability of the images.
2. Related Work
| Name of Method/Metric | Showed High Accuracy Only In | Advantage | Disadvantage |
|---|---|---|---|
| MAE/MSE/SSIM [30,31,32] | Synthetic PET/CT/MRI images | Has high accuracy in assessing noise in images | Require reference image for each synthetic image, Cannot be used for assessing echocardiogram synthetic image |
| NIQE [36] | Image quality assessment | Do not require reference image for each synthetic image | It can only correctly evaluate noisy synthetic images. Cannot evaluate better quality synthetic images with high accuracy. |
| IS [37] | Natural images assessment | Do not require reference image for each synthetic image. | It can only evaluate the distribution of generated images. Adapted to the evaluation of natural images; |
| FID [15] | Natural images assessment | It can estimate the distance between the distribution of generated image set and that of real image set. | Long calculation time; Adapted to the evaluation of natural images; |
| FastFID [16] | Natural images assessment | Fast calculation time | Adapted to the evaluation of natural images; |
| DQA [38] | MRI images | Higher evaluation accuracy | Adapted to the evaluation of MRI images; |
| HYPE [39] | Medical and natural images | Has highest accuracy; Used as a gold standard; | Costly and time consuming |
| Proposed Method | Echocardiogram images | Fast and reliable | Combination of two methods |
3. Image Generation Processes
3.1. The Working Principle of the GAN
3.2. GAN Architecture and Parameters
3.3. Dataset
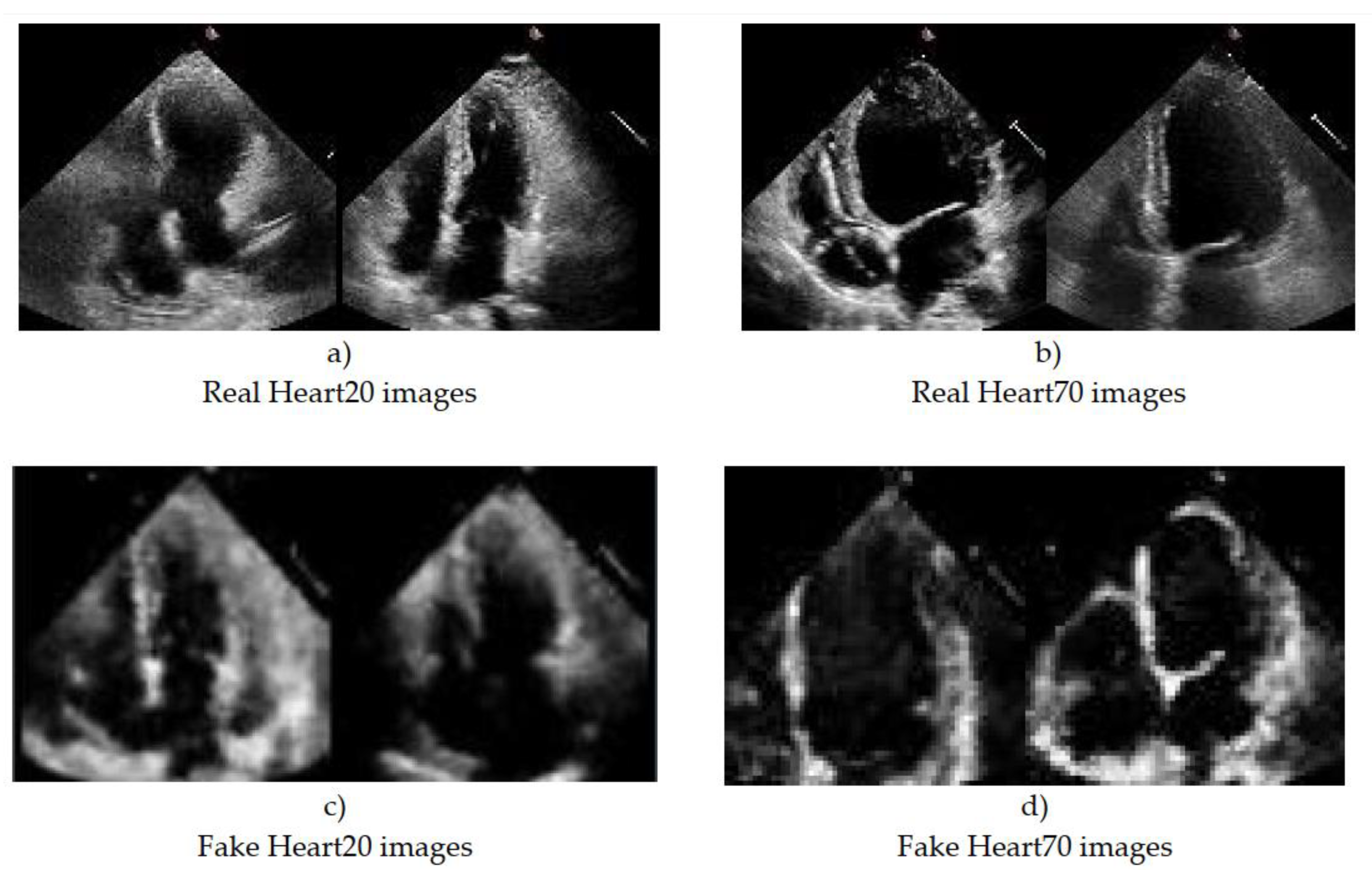
3.4. Training the GAN Network and the Results
4. Proposed Evaluation Method
4.1. Problem Statement
- -
- They are noisier and blurrier than ordinary images;
- -
- The edges are not clearly defined;
- -
- Usually generated synthetic echo images are of gray-scale quality, i.e., mostly single-channel.
4.2. Method Description
4.3. Experimental Results
4.4. New CNN-Based Evaluation Method
- Less than 40—Very bad. Low quality or the same images, mode collapse, or overfitting occurs in GAN.
- 30–50—Bad. The diversity is very low. Most images are unrealistic and of poor quality.
- 50–70—Satisfactory. The diversity is low. Some images are unrealistic and of low quality.
- 70–80—Good-quality images. The diversity is high, but there are still some disturbances in some images.
- More than 80—Much like the real images, the diversity is very high.
5. Discussion
6. Future Direction
7. Conclusions
- It can evaluate the quality of synthetic images belonging to two or more classes at the same time.
- It is possible to evaluate the diversity of the generated Images and the presence of the same images.
- It can estimate how close the distribution distance of synthetic images is to that of real images of the same class and how far it is from that of other classes.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nasimova, N.; Muminov, B.; Nasimov, R.; Abdurashidova, K.; Abdullaev, M. November. Comparative Analysis of the Results of Algorithms for Dilated Cardiomyopathy and Hypertrophic Cardiomyopathy Using Deep Learning. In Proceedings of the 2021 International Conference on Information Science and Communications Technologies (ICISCT), Tashkent, Uzbekistan, 3–5 November 2021; pp. 1–5. [Google Scholar]
- Muminov, B.; Nasimov, R.; Mirzahalilov, S.; Sayfullaeva, N.; Gadoyboyeva, N. Localization and Classification of Myocardial Infarction Based on Artificial Neural Network. In Proceedings of the 2020 Information Communication Technologies Conference (ICTC), Nanjing, China, 29–31 May 2020; pp. 245–249. [Google Scholar]
- He, X.; Lei, Y.; Liu, Y.; Tian, Z.; Wang, T.; Curran, W.J.; Liu, T.; Yang, X. Deep attentional GAN-based high-resolution ultrasound imaging. In Proceedings of the SPIE 11319, Medical Imaging 2020: Ultrasonic Imaging and Tomography, 113190B, Bellingham, WA, USA, 16 March 2020. [Google Scholar]
- Peng, B.; Huang, X.; Wang, S.; Jiang, J. A Real-Time Medical Ultrasound Simulator Based on a Generative Adversarial Network Model. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; pp. 4629–4633. [Google Scholar]
- Teng, L.; Fu, Z.; Yao, Y. Interactive Translation in Echocardiography Training System with Enhanced Cycle-GAN. IEEE Access 2020, 8, 106147–106156. [Google Scholar] [CrossRef]
- Devi, M.K.A.; Suganthi, K. Review of Medical Image Synthesis using GAN Techniques. ITM Web Conf. 2021, 37, 1005. [Google Scholar] [CrossRef]
- Wang, S.; Cao, G.; Wang, Y.L.; Liao, S.; Wang, Q.; Shi, J.; Li, C.; Shen, D. Review and Prospect: Artificial Intelligence in Advanced Medical Imaging. Front. Radiol. 2021, 1, 781868. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhao, J.; Long, X.; Luo, Q.; Wang, R.; Ding, X.; Shen, C. AUE-Net: Automated Generation of Ultrasound Elastography Using Generative Adversarial Network. Diagnostics 2022, 12, 253. [Google Scholar] [CrossRef]
- Singh, N.K.; Raza, K. Medical Image Generation Using Generative Adversarial Networks: A Review. In Health Informatics: A Computational Perspective in Healthcare. Studies in Computational Intelligence; Patgiri, R., Biswas, A., Roy, P., Eds.; Springer: Singapore, 2021; Volume 932. [Google Scholar]
- Abdi, A.H.; Tsang, T.S.; Abolmaesumi, P. GAN-enhanced Conditional Echocardiogram Generation. arXiv 2019, arXiv:1911.02121. [Google Scholar]
- Fabiani, I.; Pugliese, N.R.; Santini, V.; Conte, L.; Di Bello, V. Speckle-Tracking Imaging, Principles and Clinical Applications: A Review for Clinical Cardiologists. Echocardiogr. Heart Fail. Card. Electrophysiol. 2016, 2016, 85–114. [Google Scholar] [CrossRef]
- Morra, L.; Piano, L.; Lamberti, F.; Tommasi, T. Bridging the gap between Natural and Medical Images through Deep Colorization. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 835–842. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Al-Amidie, M.; Al-Asadi, A.; Humaidi, A.J.; Al-Shamma, O.; Fadhel, M.A.; Zhang, J.; Santamaría, J.; Duan, Y. Novel Transfer Learning Approach for Medical Imaging with Limited Labeled Data. Cancers 2021, 13, 1590. [Google Scholar] [CrossRef]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. GANs Trained by a Two Time-Scale Update Rule Converge to a Local Nash Equilibrium. Adv. Neural Inf. Process. Syst. 2017, arXiv:1706.08500. [Google Scholar]
- Coyner, A.S.; Chen, J.S.; Chang, K.; Singh, P.; Ostmo, S.; Chan, R.P.; Imaging and Informatics in Retinopathy of Prematurity Consortium. Synthetic Medical Images for Robust, Privacy-Preserving Training of Artificial Intelligence: Application to Retinopathy of Prematurity Diagnosis. Ophthalmol. Sci. 2022, 2, 100126. [Google Scholar] [CrossRef]
- Khan, F.; Tarimer, I.; Alwageed, H.S.; Karadağ, B.C.; Fayaz, M.; Abdusalomov, A.B.; Cho, Y.-I. Effect of Feature Selection on the Accuracy of Music Popularity Classification Using Machine Learning Algorithms. Electronics 2022, 11, 3518. [Google Scholar] [CrossRef]
- Available online: https://cardiologytasmania.com.au/for-referring-doctors/understanding-your-echo-report/ (accessed on 25 January 2023).
- Siddani, B.; Balachandar, S.; Moore, W.C.; Yang, Y.; Fang, R. Machine learning for physics-informed generation of dispersed multiphase flow using generative adversarial networks. Theor. Comput. Fluid Dyn. 2021, 35, 807–830. [Google Scholar] [CrossRef]
- Coutinho-Almeida, J.; Rodrigues, P.P.; Cruz-Correia, R.J. GANs for Tabular Healthcare Data Generation: A Review on Utility and Privacy. In Discovery Science; Soares, C., Torgo, L., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 282–291. [Google Scholar]
- Lee, H.; Kang, S.; Chung, K. Robust Data Augmentation Generative Adversarial Network for Object Detection. Sensors 2023, 23, 157. [Google Scholar] [CrossRef]
- Figueira, A.; Vaz, B. Survey on Synthetic Data Generation, Evaluation Methods and GANs. Mathematics 2022, 10, 2733. [Google Scholar] [CrossRef]
- Li, X.; Jiang, Y.; Rodriguez-Andina, J.J.; Luo, H.; Yin, S.; Kaynak, O. When medical images meet generative adversarial network: Recent development and research opportunities. Discov. Artif. Intell. 2021, 1, 5. [Google Scholar] [CrossRef]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef]
- Khatun, A.; Denman, S.; Sridharan, S.; Fookes, C. Pose-driven attention-guided image generation for person re-Identification. Pattern Recognit. 2023, 137, 109246. [Google Scholar] [CrossRef]
- Mendes, J.; Pereira, T.; Silva, F.; Frade, J.; Morgado, J.; Freitas, C.; Oliveira, H.P. Lung CT image synthesis using GANs. Expert Syst. Appl. 2023, 215, 119350. [Google Scholar] [CrossRef]
- Dirvanauskas, D.; Maskeliūnas, R.; Raudonis, V.; Damaševičius, R.; Scherer, R. HEMIGEN: Human Embryo Image Generator Based on Generative Adversarial Networks. Sensors 2019, 19, 3578. [Google Scholar] [CrossRef] [PubMed]
- Borji, A. Pros and cons of gan evaluation measures. Comput. Vis. Image Underst. 2019, 179, 41–65. [Google Scholar] [CrossRef]
- Lucic, M.; Kurach, K.; Michalski, M.; Gelly, S.; Bousquet, O. Are GANs Created Equal? A Large-Scale Study. In Proceedings of the 32nd Conference on Neural Information Processing Systems (NIPS 2018), Montréal, Canada, 3–8 December 2018. [Google Scholar]
- Tereza, N.; Ninon, B.; David, S. Chapter 25—Validation and evaluation metrics for medical and biomedical image synthesis. In Biomedical Image Synthesis and Simulation; Academic Press: Cambridge, MA, USA, 2020; pp. 573–600. ISBN 9780128243497. [Google Scholar] [CrossRef]
- Middel, L.; Palm, C.; Erdt, M. Synthesis of Medical Images Using GANs. In Uncertainty for Safe Utilization of Machine Learning in Medical Imaging and Clinical Image-Based Procedures; Springer International Publishing: Cham, Switzerland, 2019; pp. 125–134. [Google Scholar] [CrossRef]
- Nodirov, J.; Abdusalomov, A.B.; Whangbo, T.K. Attention 3D U-Net with Multiple Skip Connections for Segmentation of Brain Tumor Images. Sensors 2022, 22, 6501. [Google Scholar] [CrossRef]
- Bauer, D.F.; Russ, T.; Waldkirch, B.I.; Tönnes, C.; Segars, W.P.; Schad, L.R.; Golla, A.K. Generation of annotated multimodal ground truth datasets for abdominal medical image registration. Int. J. CARS 2021, 16, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Dar, S.U.; Yurt, M.; Karacan, L.; Erdem, A.; Erdem, E.; Cukur, T. Image Synthesis in Multi-Contrast MRI With Conditional Generative Adversarial Networks. IEEE Trans. Med. Imaging 2019, 38, 2375–2388. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Lei, Y.; Fu, Y.; Wynne, J.F.; Curran, W.J.; Liu, T.; Yang, X. A review on medical imaging synthesis using deep learning and its clinical applications. J. Appl. Clin. Med. Phys. 2021, 22, 11–36. [Google Scholar] [CrossRef] [PubMed]
- Kuldoshbay, A.; Abdusalomov, A.; Mukhiddinov, M.; Baratov, N.; Makhmudov, F.; Cho, Y.I. An improvement for the automatic classification method for ultrasound images used on CNN. Int. J. Wavelets Multiresolution Inf. Process. 2022, 20, 2150054. [Google Scholar]
- Treder, M.S.; Codrai, R.; Tsvetanov, K.A. Quality assessment of anatomical MRI images from generative adversarial networks: Human assessment and image quality metrics. J. Neurosci. Methods 2022, 374, 109579. [Google Scholar] [CrossRef]
- Thambawita, V.; Salehi, P.; Sheshkal, S.A.; Hicks, S.A.; Hammer, H.L.; Parasa, S.; Riegler, M.A. SinGAN-Seg: Synthetic training data generation for medical image segmentation. PLoS ONE 2022, 17, e0267976. [Google Scholar] [CrossRef]
- Salimans, T.; Goodfellow, I.J.; Zaremba, W.; Cheung, V.; Radford, A.; Chen, X. Improved techniques for training GANs. In Proceedings of the Advances in Neural Information Processing Systems 29 (NIPS 2016), Barcelona, Spain, 5–10 December 2016; Volume 29. [Google Scholar]
- Sharon, Z.; Mitchell, G.; Ranjay, K.; Austin, N.; Durim, M.; Michael, B. HYPE: Human eYe Perceptual Evaluation of Generative Models. arXiv 2019, arXiv:1904.01121v4. [Google Scholar] [CrossRef]
- Zhang, R.; Isola, P.; Efros, A.A.; Shechtman, E.; Wang, O. The Unreasonable Effectiveness of Deep Features as a Perceptual Metric. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar] [CrossRef]
- Woodland, M.; Wood, J.; Anderson, B.M.; Kundu, S.; Lin, E.; Koay, E.; Brock, K.K. Evaluating the Performance of StyleGAN2-ADA on Medical Images. In Simulation and Synthesis in Medical Imaging. SASHIMI 2022. Lecture Notes in Computer Science; Zhao, C., Svoboda, D., Wolterink, J.M., Escobar, M., Eds.; Springer International Publishing: Cham, Switzerland, 2022; Volume 13570. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Proceedings of the Advances in Neural Information Processing Systems 27 (NIPS 2014), Montreal, QC, Canada, 8–13 December 2014; Volume 27. [Google Scholar]
- Anuja, N.; Raj, J.; Noel, A.; Ruban, N.; Zhemin, Z.; Murugappan, M. RDA-UNET-WGAN: An Accurate Breast Ultrasound Lesion Sigmentation Using Wasserstein Generative Adversarial Networks. Arab. J. Sci. Eng. 2020, 45, 6399–6410. [Google Scholar]
- Moghadam, A.Z.; Azarnoush, H.; Seyyedsalehi, S.A. Multi WGAN-GP loss for pathological stain transformation using GAN. In Proceedings of the 2021 29th Iranian Conference on Electrical Engineering (ICEE), Tehran, Iran, 18–20 May 2021; pp. 927–933. [Google Scholar] [CrossRef]
- Armanious, K.; Jiang, C.; Fischer, M.; Küstner, T.; Hepp, T.; Nikolaou, K.; Gatidis, S.; Yang, B. MedGAN: Medical image translation using GANs. Comput. Med. Imaging Graph. 2020, 79, 101684. [Google Scholar] [CrossRef]
- Yusuke, S.; Sayaka, S.; Hitoshi, K. Checkerboard artifacts free convolutional neural networks. APSIPA Trans. Signal Inf. Process. 2019, 8, e9. [Google Scholar] [CrossRef]
- Ouyang, D.; He, B.; Ghorbani, A.; Lungren, M.P.; Ashley, E.A.; Liang, D.H.; Zou, J.Y. Echonet-dynamic: A large new cardiac motion video data resource for medical machine learning. In Proceedings of the NeurIPS ML4H Workshop, Vancouver, BC, Canada, 13 December 2019. [Google Scholar]
- Mathiasen, A.; Hvilshoj, F. Backpropagating through Fréchet Inception Distance. arXiv 2020, arXiv:2009.14075. [Google Scholar]
- Unterthiner, T.; Steenkiste, S.V.; Kurach, K.; Marinier, R.; Michalski, M.; Gelly, S. FVD: A new Metric for Video Generation. In Proceedings of the ICLR 2019, New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]
- Jakhongir, N.; Abdusalomov, A.; Whangbo, T.K. 3D Volume Reconstruction from MRI Slices based on VTK. In Proceedings of the 2021 International Conference on Information and Communication Technology Convergence (ICTC), Jeju Island, Korea, 19–21 October 2021; pp. 689–692. [Google Scholar] [CrossRef]
- Aljohani, A.; Alharbe, N. Generating Synthetic Images for Healthcare with Novel Deep Pix2Pix GAN. Electronics 2022, 11, 3470. [Google Scholar] [CrossRef]
- Dimitriadis, A.; Trivizakis, E.; Papanikolaou, N.; Tsiknakis, M.; Marias, K. Enhancing cancer differentiation with synthetic MRI examinations via generative models: A systematic review. Insights Imaging 2022, 13, 188. [Google Scholar] [CrossRef] [PubMed]
- Umirzakova, S.; Abdusalomov, A.; Whangbo, T.K. Fully Automatic Stroke Symptom Detection Method Based on Facial Features and Moving Hand Differences. In Proceedings of the 2019 International Symposium on Multimedia and Communication Technology (ISMAC), Quezon City, Philippines, 19–21 August 2019; pp. 1–5. [Google Scholar]
- Chen, R.J.; Lu, M.Y.; Chen, T.Y.; Williamson, D.F.K.; Mahmood, F. Synthetic data in machine learning for medicine and healthcare. Nat. Biomed. Eng. 2021, 5, 493–497. [Google Scholar] [CrossRef]
- Emam, K.; Mosquera, L.; Hoptroff, R. Chapter 1: Introducing Synthetic Data Generation. In Practical Synthetic Data Generation: Balancing Privacy and the Broad Availability of Data; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2020; pp. 1–22. [Google Scholar]
- Abdusalomov, A.B.; Safarov, F.; Rakhimov, M.; Turaev, B.; Whangbo, T.K. Improved Feature Parameter Extraction from Speech Signals Using Machine Learning Algorithm. Sensors 2022, 22, 8122. [Google Scholar] [CrossRef]
- Islam, Z.; Abdel-Aty, M.; Cai, Q.; Yuan, J. Crash data augmentation using variational autoencoder. Accid. Anal. Prev. 2021, 151, 105950. [Google Scholar] [CrossRef]
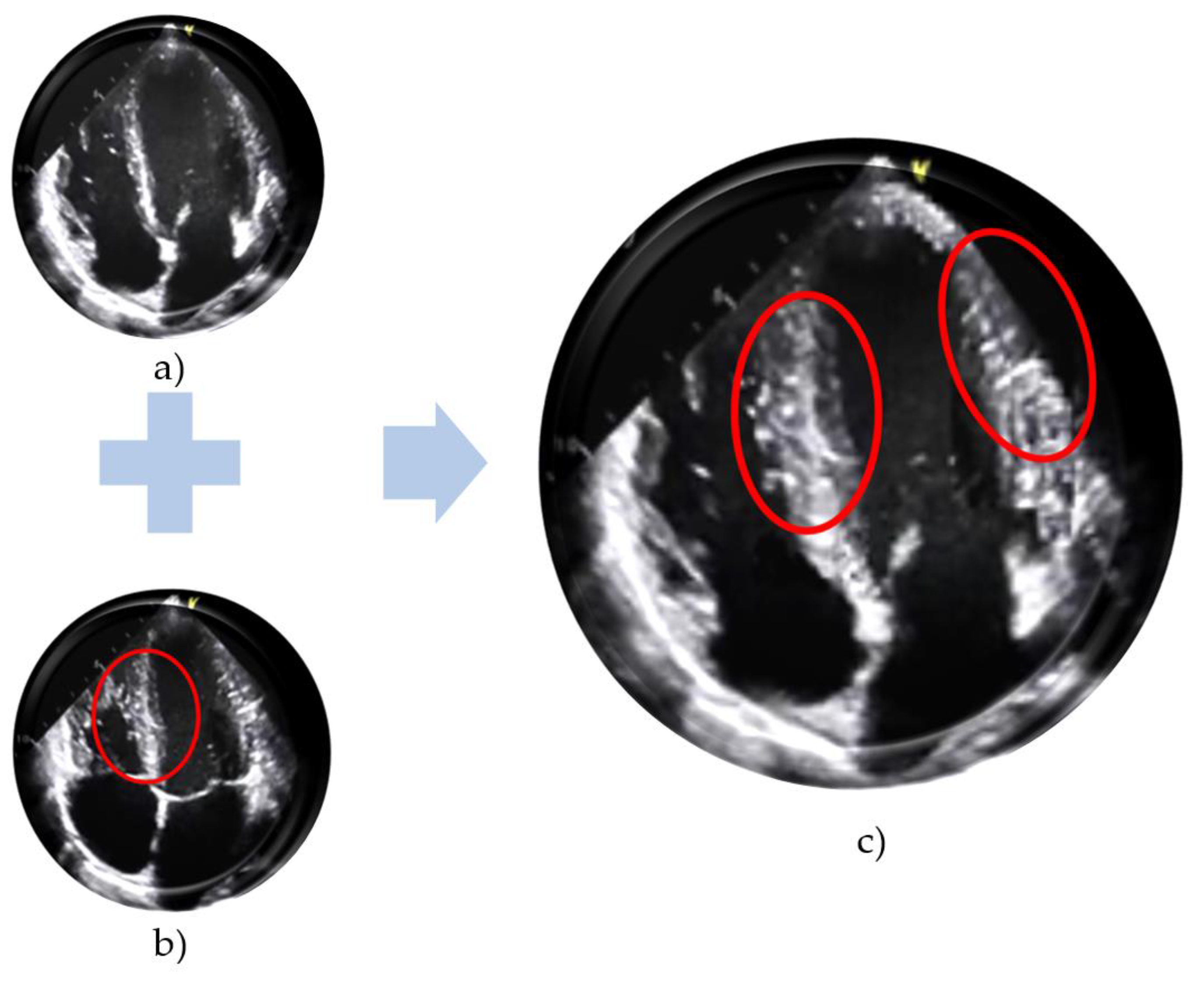
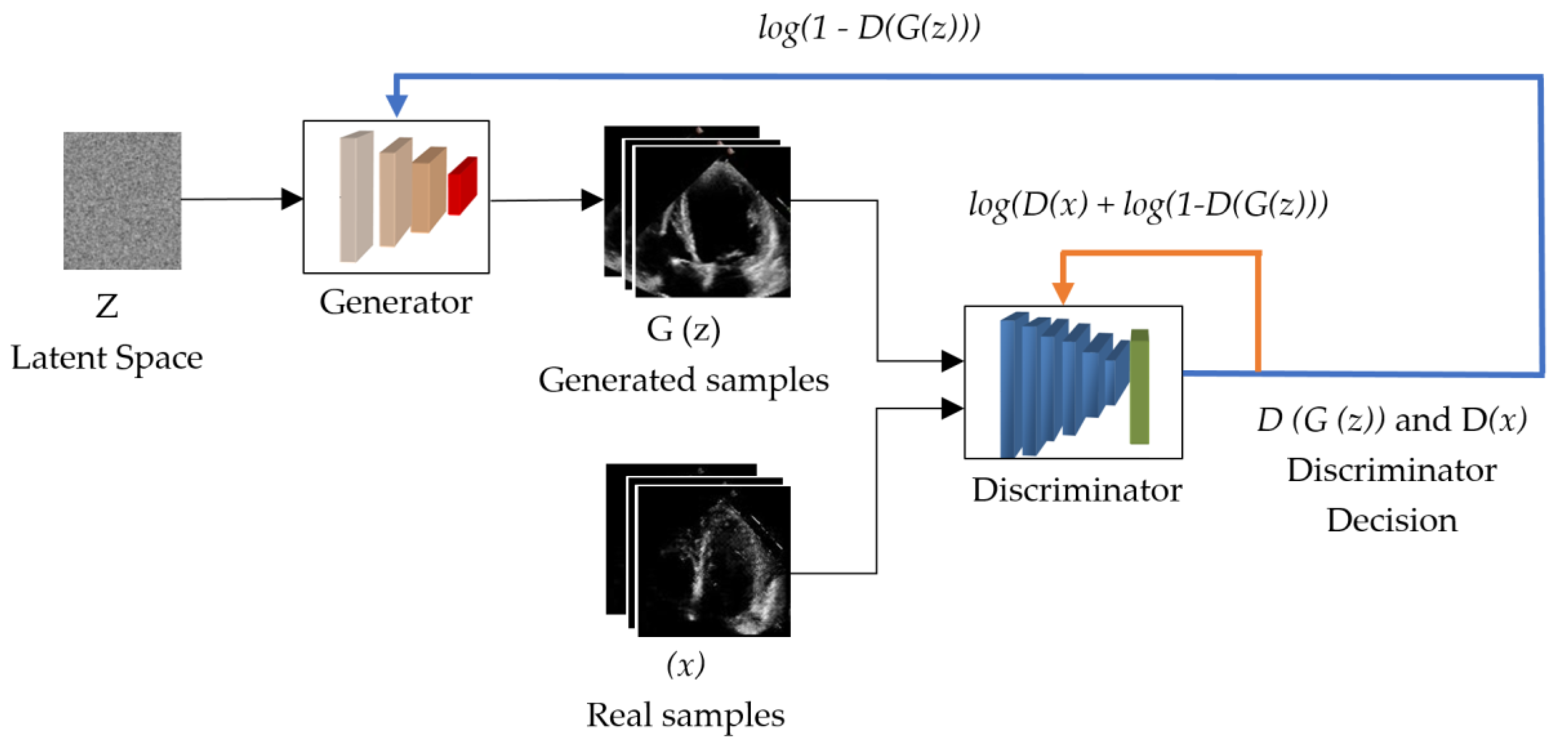
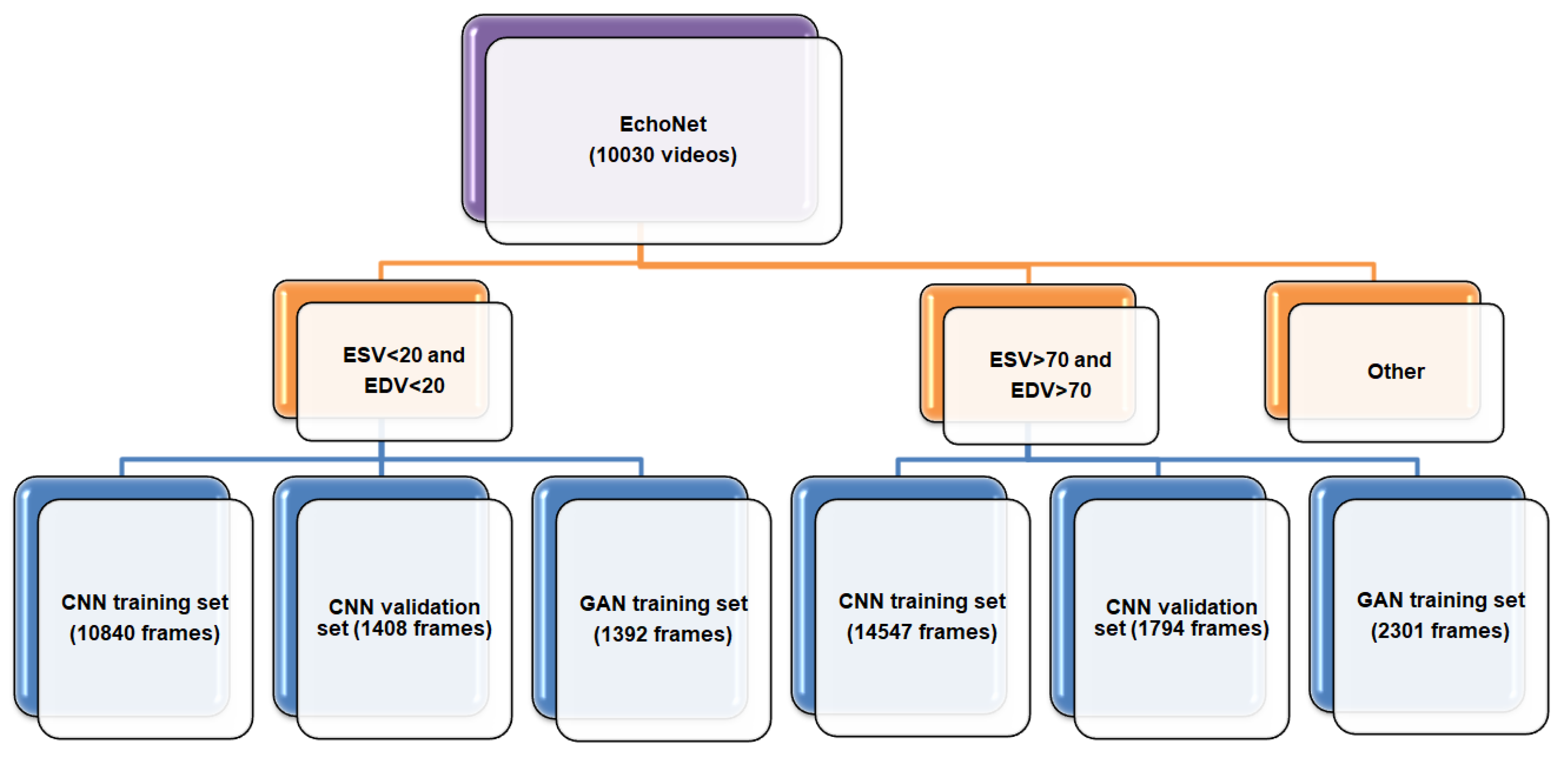
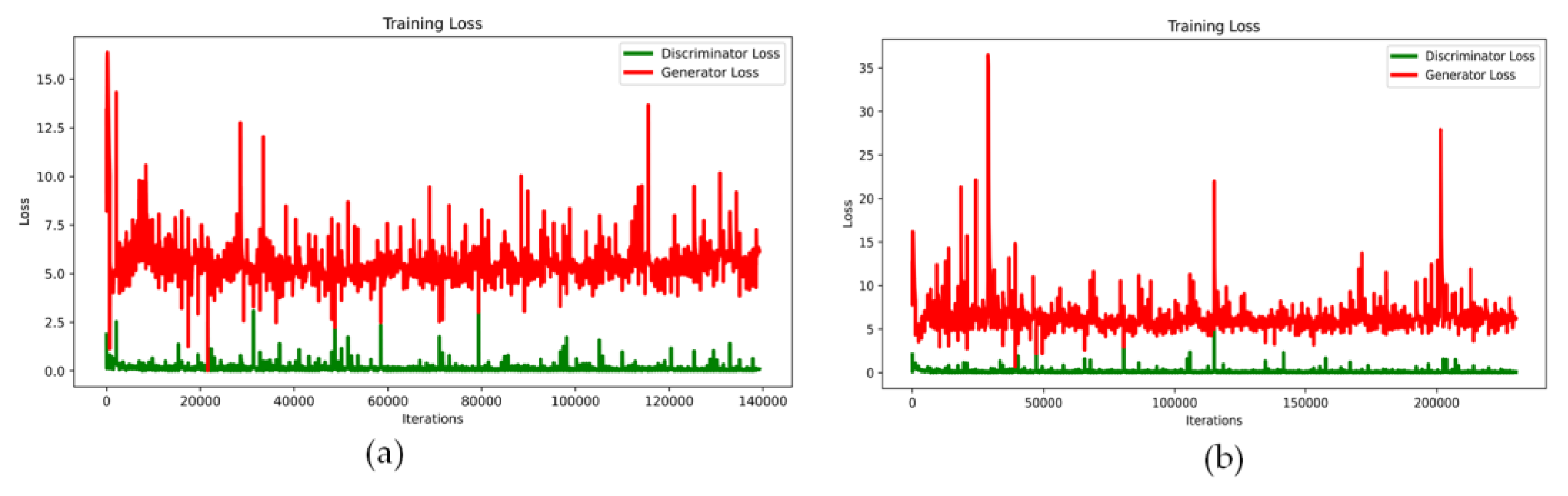


| No. | Names of the Layers | Number of Convolutional Layer Filters | Convolutional Layer Filter Size/Stride/Padding |
|---|---|---|---|
| 1. | Input + Reshape | ||
| 2. | ConvTranspose2d + BatchNorm + ReLU | 512 | 4/1/0 |
| 3. | ConvTranspose2d + BatchNorm + ReLU | 256 | 4/2/1 |
| 4. | ConvTranspose2d + BatchNorm + ReLU | 128 | 4/2/1 |
| 5. | ConvTranspose2d + BatchNorm + ReLU | 64 | 4/2/1 |
| 6. | ConvTranspose2d + Tanh | 1 | 4/2/1 |
| No. | Names of the Layers | Number of Convolutional Layer Filters | Convolutional Layer Filter Size/Stride/Padding |
|---|---|---|---|
| 7. | Input | ||
| 8. | Conv2d + BatchNorm + LeakyReLU(0.2) | 64 | 4/2/1 |
| 9. | Conv2d + BatchNorm + LeakyReLU(0.2) | 128 | 4/2/1 |
| 10. | Conv2d + BatchNorm + LeakyReLU(0.2) | 256 | 4/2/1 |
| 11. | Conv2d + BatchNorm + LeakyReLU(0.2) | 512 | 4/2/1 |
| 12. | Conv2d + Sigmoid + Flatten | 1 | 4/1/0 |
| No. | Names of the Layers | Number of Convolutional Layer Filters | Convolutional Layer Filter Size/Stride/Padding | Dropout (%) |
|---|---|---|---|---|
| 1. | Input | |||
| 2. | Conv2d + BatchNorm + ReLU + Dropout | 256 | 4/2/1 | 20 |
| 3. | Conv2d + BatchNorm + ReLU + Dropout | 2 | 4/2/1 | 20 |
| 4. | Conv2d + BatchNorm + ReLU + Dropout | 128 | 4/2/1 | 20 |
| 5. | Conv2d + BatchNorm + ReLU + Dropout | 16 | 2/1/0 | 20 |
| 6. | Output |
| Batch Size | FID | FMD | Real Datasets Name | ||
|---|---|---|---|---|---|
| Time | Value | Time | Value | ||
| Heart20 fake dataset | |||||
| 1392 | 253.372 | 34.41 | 1.813 | 16.62 | GAN training set |
| 1408 | 313.716 | 42.56 | 2.456 | 29.45 | CNN validation set |
| Heart70 fake dataset | |||||
| 1794 | 308.225 | 62.81 | 1.930 | 138.38 | GAN training set |
| 2301 | 326.37 | 61.04 | 2.120 | 129.23 | CNN validation Set |
| Batch | FMD Time | FID Time | FID/FMD Ratio |
|---|---|---|---|
| 8 | 17.43 ms | 1.51 s | 86,632.24 |
| 16 | 37.559 ms | 3.468 s | 92,334.73 |
| 32 | 62.775 ms | 6.226 s | 99,179.61 |
| 64 | 112.565 ms | 12.143 s | 107,875.4 |
| 128 | 244.611 ms | 24.371 s | 99,631.66 |
| Labeled Name | Predicted | Confusion Matrix |
|---|---|---|
| Positive | Positive | TP |
| Positive | Negative | FN |
| Negative | Positive | FP |
| Negative | Negative | TP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdusalomov, A.B.; Nasimov, R.; Nasimova, N.; Muminov, B.; Whangbo, T.K. Evaluating Synthetic Medical Images Using Artificial Intelligence with the GAN Algorithm. Sensors 2023, 23, 3440. https://doi.org/10.3390/s23073440
Abdusalomov AB, Nasimov R, Nasimova N, Muminov B, Whangbo TK. Evaluating Synthetic Medical Images Using Artificial Intelligence with the GAN Algorithm. Sensors. 2023; 23(7):3440. https://doi.org/10.3390/s23073440
Chicago/Turabian StyleAbdusalomov, Akmalbek Bobomirzaevich, Rashid Nasimov, Nigorakhon Nasimova, Bahodir Muminov, and Taeg Keun Whangbo. 2023. "Evaluating Synthetic Medical Images Using Artificial Intelligence with the GAN Algorithm" Sensors 23, no. 7: 3440. https://doi.org/10.3390/s23073440
APA StyleAbdusalomov, A. B., Nasimov, R., Nasimova, N., Muminov, B., & Whangbo, T. K. (2023). Evaluating Synthetic Medical Images Using Artificial Intelligence with the GAN Algorithm. Sensors, 23(7), 3440. https://doi.org/10.3390/s23073440







