Active Opto-Magnetic Biosensing with Silicon Microring Resonators
Abstract
:1. Introduction
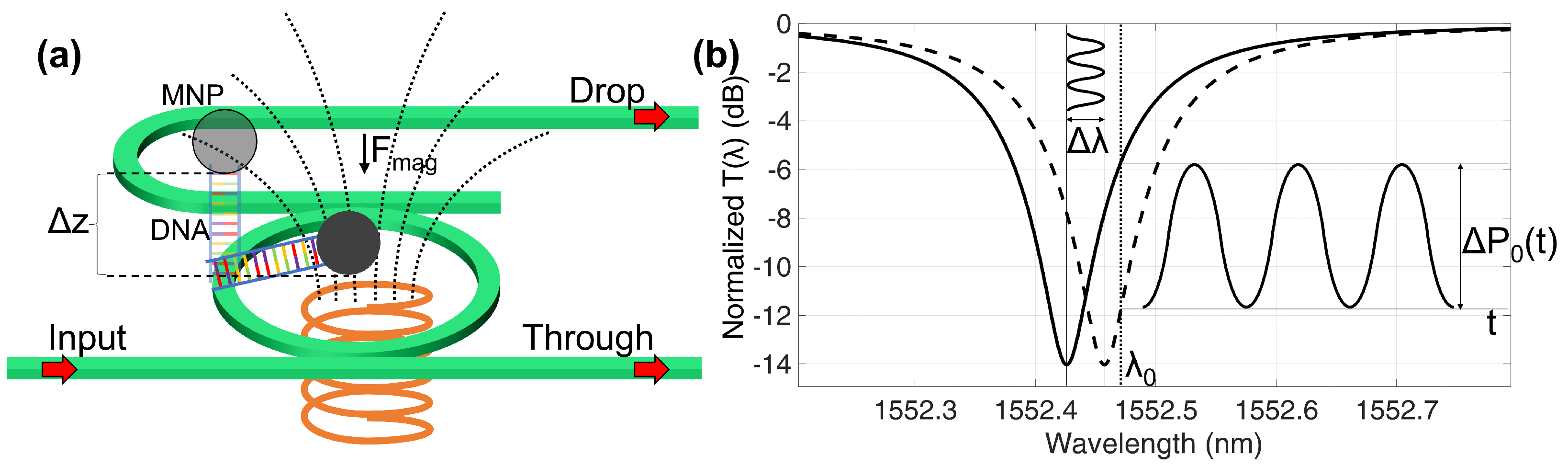
2. Opto-Magnetic Assay Concept
3. Photonic Circuit and Magnetic Labelling
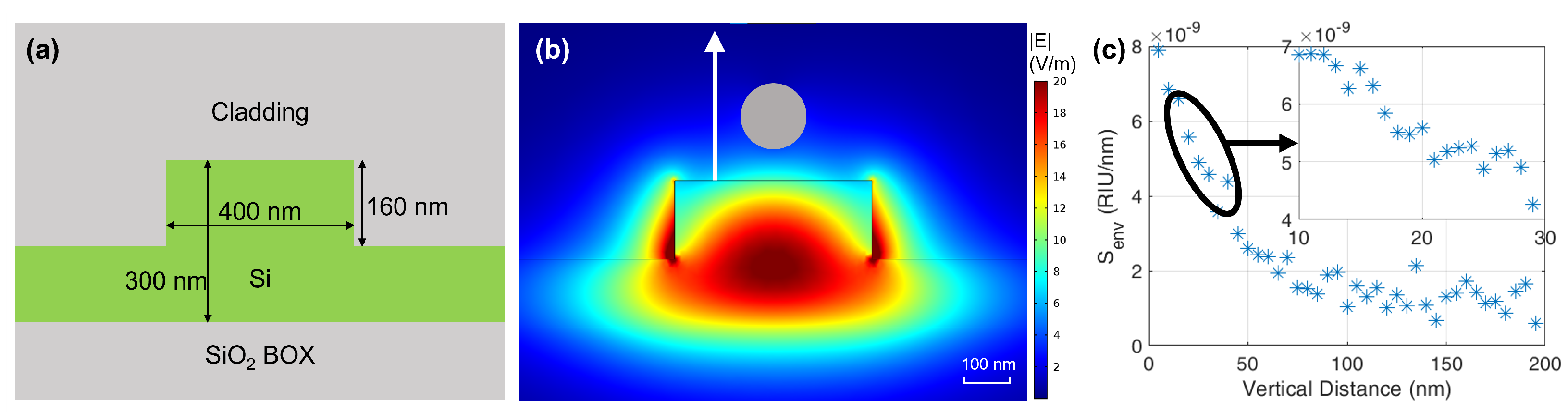
3.1. Optical Aspects
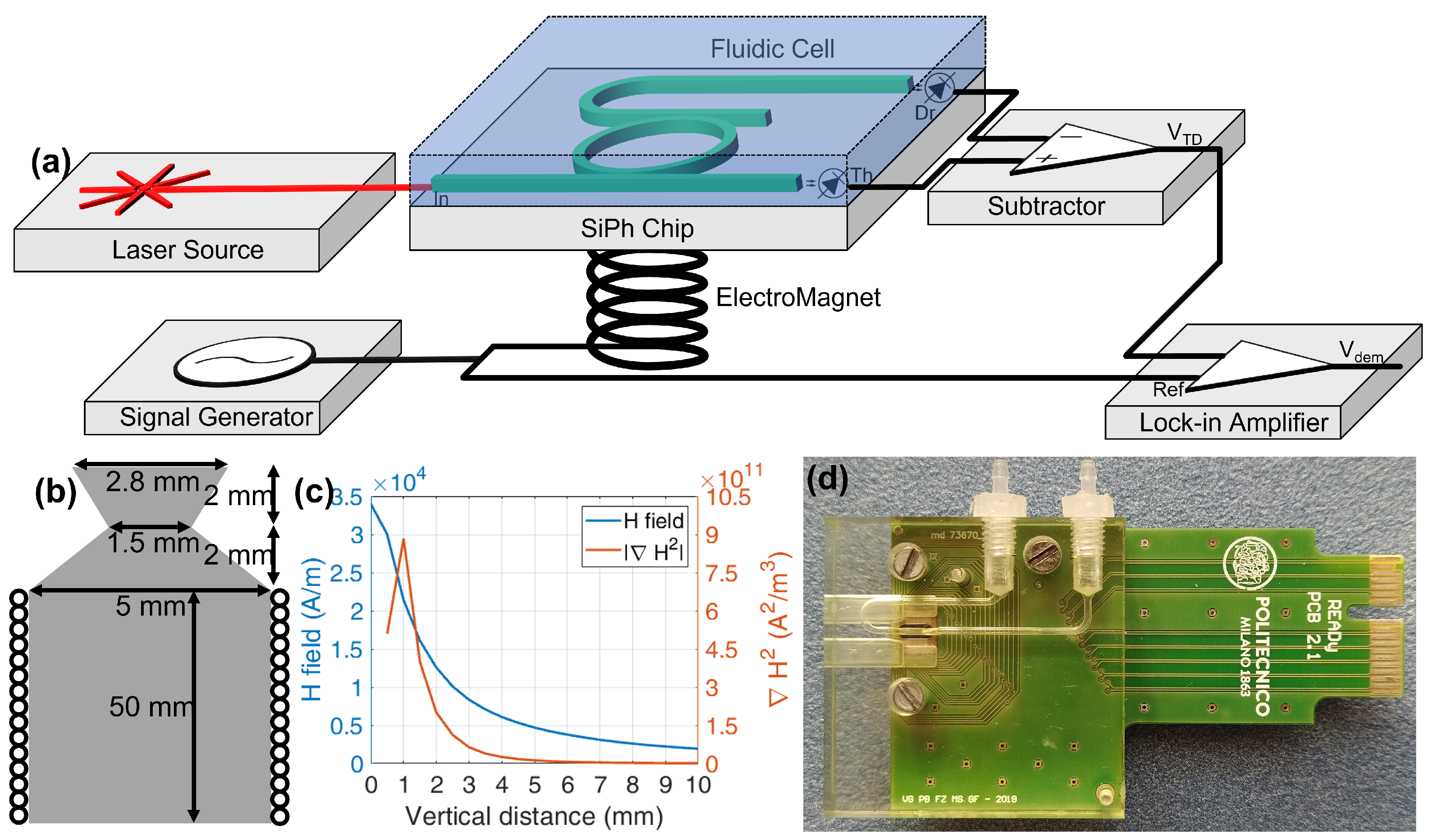
3.2. Magnetic Aspects
4. Chip Preparation
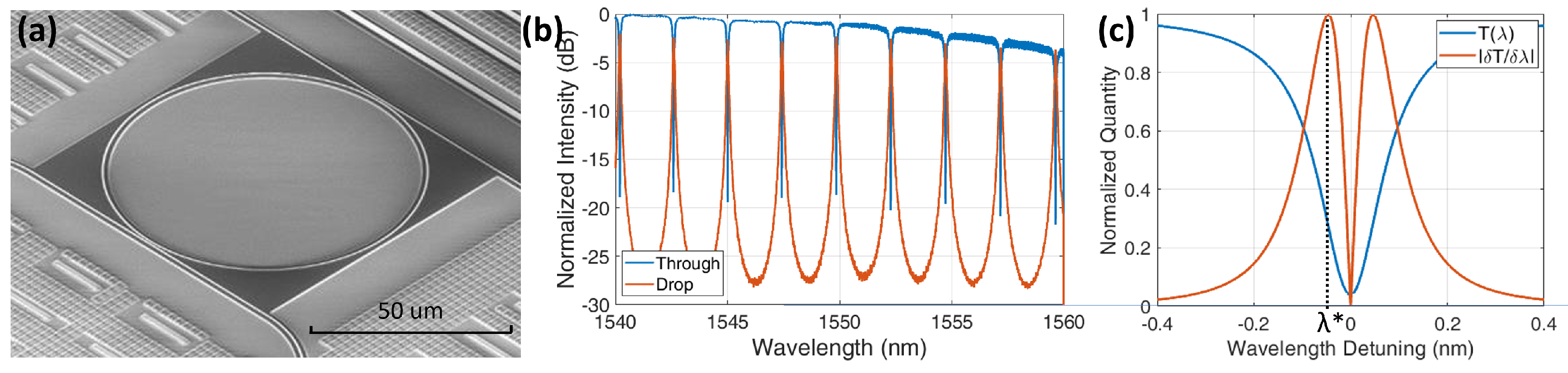
4.1. Silicon Photonic Chip Preparation
4.2. Chip Functionalization
4.3. DNA Hybridization
5. Experimental Setup
6. Measurements and Results
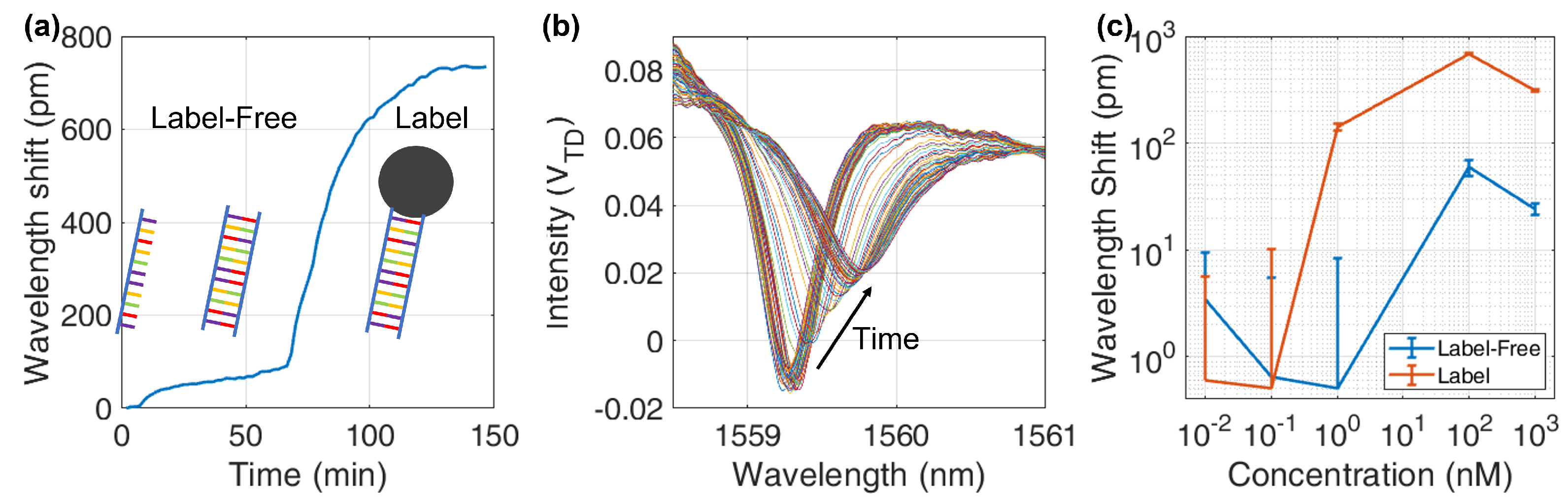
6.1. Hybridization
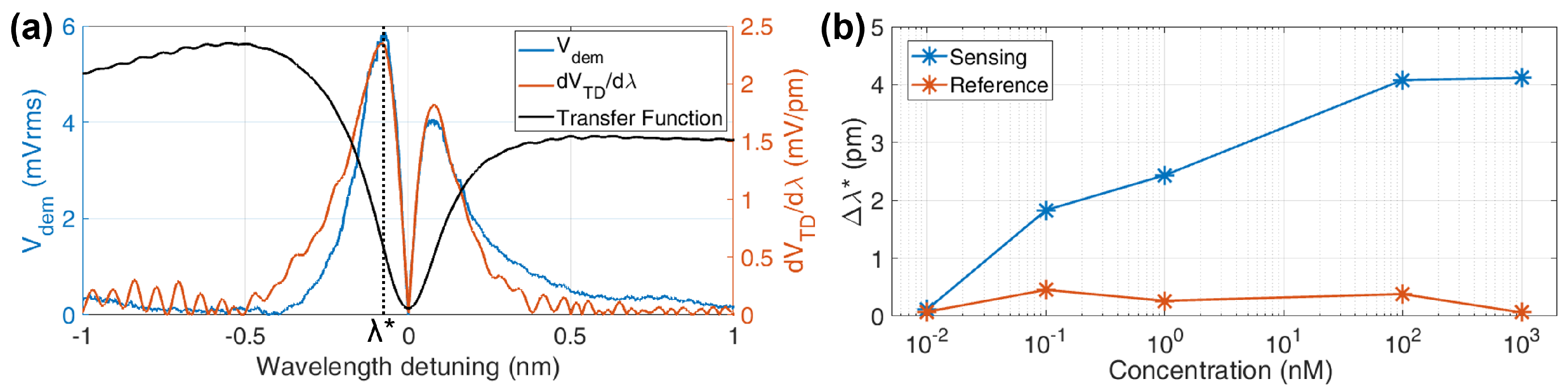
6.2. Opto-Magnetic Measurements
6.3. Opto-Magnetic Results
7. Conclusions
8. Patents
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, J.; Yang, Z.; Wilkinson, J.S.; Zhou, X. Optical biosensors based on refractometric sensing schemes: A review. Biosens. Bioelectron. 2019, 144, 111693. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, J. Optical biosensors: An exhaustive and comprehensive review. Analyst 2020, 145, 1605–1628. [Google Scholar] [CrossRef]
- Prabowo, B.A.; Purwidyantri, A.; Liu, K.C. Surface Plasmon Resonance Optical Sensor: A Review on Light Source Technology. Biosensors 2018, 8, 80. [Google Scholar] [CrossRef] [Green Version]
- Shi, Q.; Zhao, J.; Liang, L. Two dimensional photonic crystal slab biosensors using label free refractometric sensing schemes: A review. Prog. Quantum Electron. 2021, 77, 100298. [Google Scholar] [CrossRef]
- Pol, E.; Roos, H.; Markey, F.; Elwinger, F.; Shaw, A.; Karlsson, R. Evaluation of calibration-free concentration analysis provided by BiacoreTM systems. Anal. Biochem. 2016, 510, 88–97. [Google Scholar] [CrossRef]
- Kim, W.; Lee, S.H.; Ahn, Y.J.; Lee, S.H.; Ryu, J.; Choi, S.K.; Choi, S. A label-free cellulose SERS biosensor chip with improvement of nanoparticle-enhanced LSPR effects for early diagnosis of subarachnoid hemorrhage-induced complications. Biosens. Bioelectron. 2018, 111, 59–65. [Google Scholar] [CrossRef]
- Su, J.; Wang, D.; Nörbel, L.; Shen, J.; Zhao, Z.; Dou, Y.; Peng, T.; Shi, J.; Mathur, S.; Fan, C.; et al. Multicolor Gold-Silver Nano-Mushrooms as Ready-to-Use SERS Probes for Ultrasensitive and Multiplex DNA/miRNA Detection. Anal. Chem. 2017, 89, 2531–2538. [Google Scholar] [CrossRef]
- Scullion, M.G.; Krauss, T.F.; Di Falco, A. Slotted Photonic Crystal Sensors. Sensors 2013, 13, 3675–3710. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Wang, C.; Park, J. User-friendly point-of-care detection of influenza A (H1N1) virus using light guide in three-dimensional photonic crystal. RSC Adv. 2018, 8, 22991–22997. [Google Scholar] [CrossRef] [Green Version]
- Fahrenkopf, N.M.; McDonough, C.; Leake, G.L.; Su, Z.; Timurdogan, E.; Coolbaugh, D.D. The AIM Photonics MPW: A Highly Accessible Cutting Edge Technology for Rapid Prototyping of Photonic Integrated Circuits. IEEE J. Sel. Top. Quantum Electron. 2019, 25, 1–6. [Google Scholar] [CrossRef]
- Murib, M.S.; Martens, D.; Bienstman, P. Label-free real-time optical monitoring of DNA hybridization using SiN Mach–Zehnder interferometer-based integrated biosensing platform. J. Biomed. Opt. 2018, 23, 127002. [Google Scholar] [CrossRef] [PubMed]
- Steglich, P.; Hülsemann, M.; Dietzel, B.; Mai, A. Optical biosensors based on silicon-on-insulator ring resonators: A review. Molecules 2019, 24, 519. [Google Scholar] [CrossRef] [Green Version]
- Su, J. Label-free single molecule detection using microtoroid optical resonators. J. Vis. Exp. 2015, 2015, e53180. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, S.; Flueckiger, J.; Wu, W.; Grist, S.M.; Talebi Fard, S.; Donzella, V.; Khumwan, P.; Thompson, E.R.; Wang, Q.; Kulik, P.; et al. Improving the performance of silicon photonic rings, disks, and Bragg gratings for use in label-free biosensing. Biosens. Nanomed. VII 2014, 9166, 91660M. [Google Scholar] [CrossRef]
- Dhakal, A.; Peyskens, F.; Clemmen, S.; Raza, A.; Wuytens, P.; Zhao, H.; Le Thomas, N.; Baets, R. Single mode waveguide platform for spontaneous and surface-enhanced on-chip Raman spectroscopy. Interface Focus 2016, 6, 20160015. [Google Scholar] [CrossRef] [PubMed]
- Ettabib, M.A.; Marti, A.; Liu, Z.; Bowden, B.M.; Zervas, M.N.; Bartlett, P.N.; Wilkinson, J.S. Waveguide Enhanced Raman Spectroscopy for Biosensing: A Review. ACS Sens. 2021, 6, 2025–2045. [Google Scholar] [CrossRef]
- Botsialas, A.; Salapatas, A.; Makarona, E.; Jobst, G.; Raptis, I.; Misiakos, K.; Hoekman, M.; Stoffer, R. Broad-band Mach-Zehnder interferometers as high performance refractive index sensors: Theory and monolithic implementation. Opt. Express 2014, 22, 8856–8870. [Google Scholar] [CrossRef]
- Schmitt, K.; Schirmer, B.; Hoffmann, C.; Brandenburg, A.; Meyrueis, P. Interferometric biosensor based on planar optical waveguide sensor chips for label-free detection of surface bound bioreactions. Biosens. Bioelectron. 2007, 22, 2591–2597. [Google Scholar] [CrossRef]
- Zamora, V.; Díez, A.; Andrés, M.V.; Gimeno, B. Cylindrical optical microcavities: Basic properties and sensor applications. Photonics Nanostruct.—Fundam. Appl. 2011, 9, 149–158. [Google Scholar] [CrossRef]
- Su, J.; Goldberg, A.F.F.F.; Stoltz, B.M. Label-free detection of single nanoparticles and biological molecules using microtoroid optical resonators. Light Sci. Appl. 2016, 5, 2–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.J.; Zou, J.; Ye, J.; Li, M.; Jiang, X. Cascaded silicon-on-insulator double-ring sensors operating in high-sensitivity transverse-magnetic mode. Opt. Lett. 2013, 38, 1349–1351. [Google Scholar] [CrossRef]
- Henneke, D.; Khodadad, I.; Khorasaninejad, M.; Clarke, N.; Saini, S.S. Optimization of multiple-slot waveguides for biochemical sensing. Appl. Opt. 2014, 53, 5169–5178. [Google Scholar] [CrossRef]
- Khan, M.A.; Sun, J.; Li, B.; Przybysz, A.; Kosel, J. Magnetic sensors-A review and recent technologies. Eng. Res. Express 2021, 3, 022005. [Google Scholar] [CrossRef]
- Lin, G.; Makarov, D.; Schmidt, O.G. Magnetic sensing platform technologies for biomedical applications. Lab Chip 2017, 17, 1884–1912. [Google Scholar] [CrossRef]
- Graham, D.L.; Ferreira, H.A.; Freitas, P.P. Magnetoresistive-based biosensors and biochips. Trends Biotechnol. 2004, 22, 455–462. [Google Scholar] [CrossRef]
- Freitas, P.P.; Ferreira, R.; Cardoso, S.; Cardoso, F. Magnetoresistive sensors. J. Phys. Condens. Matter 2007, 19, 165221. [Google Scholar] [CrossRef]
- Besse, P.A.; Boero, G.; Demierre, M.; Pott, V.; Popovic, R. Detection of a single magnetic microbead using a miniaturized silicon Hall sensor. Appl. Phys. Lett. 2002, 80, 4199. [Google Scholar] [CrossRef]
- Sharma, P.P.; Albisetti, E.; Massetti, M.; Scolari, M.; La Torre, C.; Monticelli, M.; Leone, M.; Damin, F.; Gervasoni, G.; Ferrari, G.; et al. Integrated platform for detecting pathogenic DNA via magnetic tunneling junction-based biosensors. Sens. Actuators B Chem. 2017, 242, 280–287. [Google Scholar] [CrossRef] [Green Version]
- Matschegewski, C.; Kowalski, A.; Müller, K.; Teller, H.; Grabow, N.; Großmann, S.; Schmitz, K.P.; Siewert, S. Biocompatibility of magnetic iron oxide nanoparticles for biomedical applications. Curr. Dir. Biomed. Eng. 2019, 5, 573–576. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Su, D.; Liu, J.; Saha, R.; Wang, J.P. Magnetic nanoparticles in nanomedicine: A review of recent advances. Nanotechnology 2019, 30, 502003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Reenen, A.; De Jong, A.M.; Den Toonder, J.M.; Prins, M.W.; Reenen, A.; Jong, A.M.; Toonder, J.M.J.; Prins, M.W.J.; Van Reenen, A.; De Jong, A.M.; et al. Integrated lab-on-chip biosensing systems based on magnetic particle actuation-a comprehensive review. Lab Chip 2014, 14, 1966–1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moerland, C.P.; Van IJzendoorn, L.J.; Prins, M.W. Rotating magnetic particles for lab-on-chip applications-a comprehensive review. Lab Chip 2019, 19, 919–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Donolato, M.; Pinto, A.; Bosco, F.G.; Hwu, E.T.; Chen, C.H.; Alstrøm, T.S.; Lee, G.H.; Schäfer, T.; Vavassori, P.; et al. Blu-ray based optomagnetic aptasensor for detection of small molecules. Biosens. Bioelectron. 2016, 75, 396–403. [Google Scholar] [CrossRef] [Green Version]
- Tian, B.; Wetterskog, E.; Qiu, Z.; Zardán Gómez de la Torre, T.; Donolato, M.; Fougt Hansen, M.; Svedlindh, P.; Strömberg, M. Shape anisotropy enhanced optomagnetic measurement for prostate-specific antigen detection via magnetic chain formation. Biosens. Bioelectron. 2017, 98, 285–291. [Google Scholar] [CrossRef]
- Uddin, R.; Burger, R.; Donolato, M.; Fock, J.; Creagh, M.; Hansen, M.F.; Boisen, A. Lab-on-a-disc agglutination assay for protein detection by optomagnetic readout and optical imaging using nano- and micro-sized magnetic beads. Biosens. Bioelectron. 2016, 85, 351–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Munir, A.; Zhu, Z.; Zhou, H.S. Magnetic Nanoparticle Enhanced Surface Plasmon Resonance Sensing and Its Application for the Ultrasensitive Detection of Magnetic Nanoparticle-Enriched Small Molecules. Anal. Chem. 2010, 82, 6782–6789. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dostalek, J.; Knoll, W. Magnetic Nanoparticle-Enhanced Biosensor Based on Grating-Coupled Surface Plasmon Resonance. Anal. Chem. 2011, 83, 6202–6207. [Google Scholar] [CrossRef]
- Valera, E.; McClellan, M.S.; Bailey, R.C. Magnetically-actuated, bead-enhanced silicon photonic immunosensor. Anal. Methods 2015, 7, 8539–8544. [Google Scholar] [CrossRef] [Green Version]
- Prabhathan, P.; Murukeshan, V.M.; Jing, Z.; Ramana, P.V.; Prieto, F.; Sepulveda, B.; Calle, A.; Llobera, A.; Dominguez, C.; Abad, A.; et al. Compact SOI nanowire refractive index sensor using phase shifted Bragg grating. Opt. Express 2009, 17, 15330–15341. [Google Scholar] [CrossRef]
- Milvich, J.; Kohler, D.; Freude, W.; Koos, C. Integrated phase-sensitive photonic sensors: A system design tutorial. Adv. Opt. Photonics 2021, 13, 584–642. [Google Scholar] [CrossRef]
- Bogaerts, W.; de Heyn, P.; van Vaerenbergh, T.; de Vos, K.; Kumar Selvaraja, S.; Claes, T.; Dumon, P.; Bienstman, P.; van Thourhout, D.; Baets, R. Silicon microring resonators. Laser Photonics Rev. 2012, 6, 47–73. [Google Scholar] [CrossRef]
- Luan, E.; Shoman, H.; Ratner, D.M.; Cheung, K.C.; Chrostowski, L. Silicon photonic biosensors using label-free detection. Sensors 2018, 18, 3519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piedimonte, P.; Sola, L.; Cretich, M.; Gori, A.; Chiari, M.; Marchisio, E.; Borga, P.; Bertacco, R.; Melloni, A.; Ferrari, G.; et al. Differential Impedance Sensing platform for high selectivity antibody detection down to few counts: A case study on Dengue Virus. Biosens. Bioelectron. 2022, 202, 113996. [Google Scholar] [CrossRef]
- Rahim, A.; Goyvaerts, J.; Szelag, B.; Fedeli, J.M.; Absil, P.; Aalto, T.; Harjanne, M.; Littlejohns, C.; Reed, G.; Winzer, G.; et al. Open-access silicon photonics platforms in Europe. IEEE J. Sel. Top. Quantum Electron. 2019, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Chu, K.Y.; Thompson, A.R. Densities and Refractive Indices of Alcohol-Water Solutions of n-Propyl, Isopropyl, and Methyl Alcohols. J. Chem. Eng. Data 1962, 7, 358–360. [Google Scholar] [CrossRef]
- Biradar, U.V.; Dongarge, S.M. Refractive Index of Salt (NaCl) from Aquous Solution. Int. J. Comput. Math. Sci. 2015, 4, 33–39. [Google Scholar]
- Bedanta, S.; Kleemann, W. Supermagnetism. J. Phys. D Appl. Phys. 2008, 42, 013001. [Google Scholar] [CrossRef]
- Pankhurst, Q.A.; Connolly, J.; Jones, S.K.; Dobson, J. Applications of magnetic nanoparticles in biomedicine. J. Phys. D Appl. Phys. 2003, 36, R167. [Google Scholar] [CrossRef] [Green Version]
- Bahadorimehr, A.; Alvankarian, J.; Majlis, B.Y. Magnetic force on a magnetic bead. In Proceedings of the 2012 10th IEEE International Conference on Semiconductor Electronics, ICSE 2012, Kuala Lumpur, Malaysia, 19–21 September 2012; pp. 280–284. [Google Scholar] [CrossRef]
- Boeuf, F.; Cremer, S.; Vulliet, N.; Pinguet, T.; Mekis, A.; Masini, G.; Verslegers, L.; Sun, P.; Ayazi, A.; Hon, N.K.; et al. A multi-wavelength 3D-compatible silicon photonics platform on 300mm SOI wafers for 25Gb/s applications. In Proceedings of the Technical Digest—International Electron Devices Meeting, IEDM, Washington, DC, USA, 9–11 December 2013. [Google Scholar] [CrossRef]
- Cretich, M.; Pirri, G.; Damin, F.; Solinas, I.; Chiari, M. A new polymeric coating for protein microarrays. Anal. Biochem. 2004, 332, 67–74. [Google Scholar] [CrossRef]
- Özkumur, E.; Yalçin, A.; Cretich, M.; Lopez, C.A.; Bergstein, D.A.; Goldberg, B.B.; Chiari, M.; Ünlü, M.S. Quantification of DNA and protein adsorption by optical phase shift. Biosens. Bioelectron. 2009, 25, 167–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirri, G.; Damin, F.; Chiari, M.; Bontempi, E.; Depero, L.E. Characterization of A Polymeric Adsorbed Coating for DNA Microarray Glass Slides. Anal. Chem. 2004, 76, 1352–1358. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Chen, H.; Lou, X.W.; Archer, L.A. DNA bending stiffness on small length scales. Phys. Rev. Lett. 2008, 100, 1–4. [Google Scholar] [CrossRef]
- Yeom, E.; Kang, Y.J.; Lee, S.J. Changes in velocity profile according to blood viscosity in a microchannel. Biomicrofluidics 2014, 8, 034110. [Google Scholar] [CrossRef] [Green Version]
- Cardenosa-Rubio, M.C.; Robison, H.M.; Bailey, R.C. Recent advances in environmental and clinical analysis using microring resonator–based sensors. Curr. Opin. Environ. Sci. Health 2019, 10, 38–46. [Google Scholar] [CrossRef]
- Rho, D.; Breaux, C.; Kim, S. Label-Free Optical Resonator-Based Biosensors. Sensors 2020, 20, 5901. [Google Scholar] [CrossRef]

| Time | RF | ICP | Pressure | DC | |||
|---|---|---|---|---|---|---|---|
| s | sccm | sccm | W | W | mBar | V | |
| Clamp | 20 | / | / | / | / | / | / |
| Cooling | 5 | / | / | / | / | 5 | / |
| Repeat | |||||||
| Stabilization 1 | 5 | 80 | / | / | / | 5 | / |
| Etching | 12 | 80 | / | 50 (0) | 1500 (5) | 5 | 26 |
| Pump Down 1 | 10 | / | / | / | / | 5 | / |
| Stabilization 2 | 2 | / | 24 | / | / | 5 | / |
| Passivation | 5 | / | 24 | 50 (0) | 1500 (3) | 5 | 25 |
| Pump Down 2 | 10 | / | / | / | / | 5 | / |
| Loop | |||||||
| Final | 60 | 80 | / | 50 (0) | 1500 (5) | 5 | 26 |
| ssDNA | Sequence | Molecular Weight (kDa) |
|---|---|---|
| Probe | 5-NH- TCA TCG GTC AGG TGC AAC AAA TTG ATA AGC | 18 |
| AAT GCT TTT TTG GCC CTA TCT TCT AAC AGC-3 | ||
| Target | 5-Biotin- GCT GTT AGA AGA TAG GGC CAA AAA AGC ATT | 18 |
| GCT TAT CAA TTT GTT GCA CCT GAC CGA TGA-3 |
| Solution | Components |
|---|---|
| Washing | 2xSSC (Saline-Sodium Citrate) |
| PBS | 1xPBS (Phosphate Buffered Saline Solution) |
| DNA | target ssDNA in 2xSSC, concentrations from 10 m to 1 m |
| MNP | streptavidin-coated MNP in PBS, ∼ particles/ |
| Step | Solution | Repetitions | Volume/Repetition L | Flow L/min | Step Pause min |
|---|---|---|---|---|---|
| 1 | Washing | 1 | >500 | 100 | |
| 2 | DNA | 1 | 200 | 100 | |
| 3 | DNA | 1 | 300 | 20 | >5 |
| 4 | Washing | 1 | 300 | 100 | >5 |
| 5 | PBS | 1 | 300 | 100 | >5 |
| 6 | MNP | 1 | 200 | 100 | |
| 7 | MNP | 4 | 25 | 20 | >4 |
| 8 | PBS | 1 | >500 | 20 | |
| Total time: | >100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borga, P.; Milesi, F.; Peserico, N.; Groppi, C.; Damin, F.; Sola, L.; Piedimonte, P.; Fincato, A.; Sampietro, M.; Chiari, M.; et al. Active Opto-Magnetic Biosensing with Silicon Microring Resonators. Sensors 2022, 22, 3292. https://doi.org/10.3390/s22093292
Borga P, Milesi F, Peserico N, Groppi C, Damin F, Sola L, Piedimonte P, Fincato A, Sampietro M, Chiari M, et al. Active Opto-Magnetic Biosensing with Silicon Microring Resonators. Sensors. 2022; 22(9):3292. https://doi.org/10.3390/s22093292
Chicago/Turabian StyleBorga, Piero, Francesca Milesi, Nicola Peserico, Chiara Groppi, Francesco Damin, Laura Sola, Paola Piedimonte, Antonio Fincato, Marco Sampietro, Marcella Chiari, and et al. 2022. "Active Opto-Magnetic Biosensing with Silicon Microring Resonators" Sensors 22, no. 9: 3292. https://doi.org/10.3390/s22093292
APA StyleBorga, P., Milesi, F., Peserico, N., Groppi, C., Damin, F., Sola, L., Piedimonte, P., Fincato, A., Sampietro, M., Chiari, M., Melloni, A., & Bertacco, R. (2022). Active Opto-Magnetic Biosensing with Silicon Microring Resonators. Sensors, 22(9), 3292. https://doi.org/10.3390/s22093292









