Mammogram Image Enhancement Techniques for Online Breast Cancer Detection and Diagnosis
Abstract
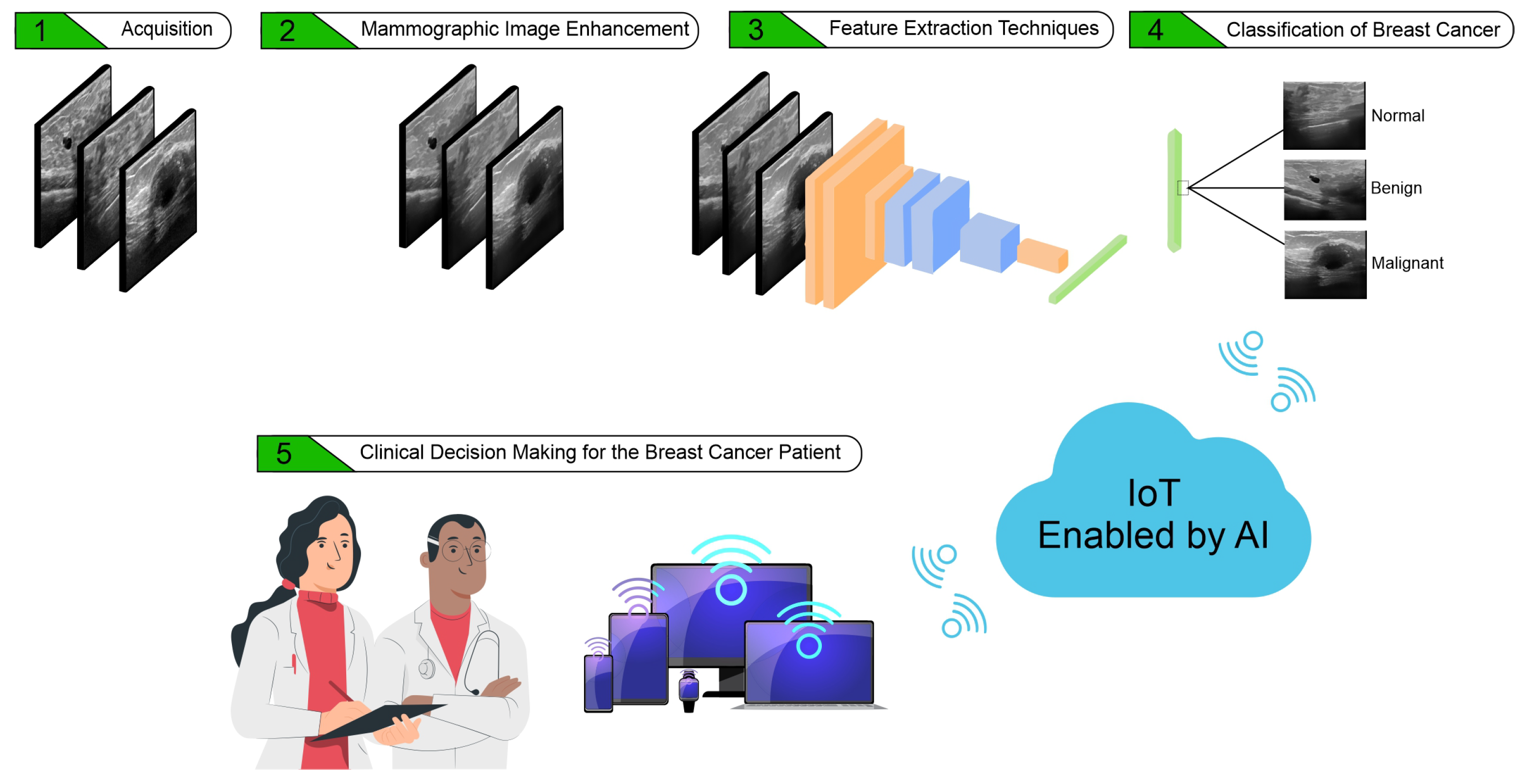
:1. Introduction
- Development of a comparative analysis on the evaluation of breast cancer image enhancement methods to improve the accuracy in the detection of malignant tumors;
- Comparing different image enhancement techniques and classification techniques focused on breast tumor;
- Validating the results through statistical evaluations and estimating a better strategy for pre-screnning of tumors;
- Providing an online processing tool for breast cancer detection for early diagnosis and treatment.
2. Materials and Methods
2.1. Database Description
Data Augmentation
2.2. Image Enhancement Techniques
2.2.1. Bilateral
2.2.2. Histogram Equalization
2.2.3. Total Variance
2.2.4. Low-Light Image Enhancement via Illumination Map Estimation
2.2.5. Exposure Fusion
2.2.6. Gamma Correction
2.2.7. Light-DehazeNet
2.2.8. Zero-Reference Deep Curve Estimation
2.2.9. Low-Light Image Enhancement with Normalizing Flow
2.3. Data Extraction
2.4. Classification Methods
2.4.1. Multi-Layer Perceptron
2.4.2. Support Vector Machine
2.4.3. k-Nearest Neighbor
2.5. Statistical Metrics
2.5.1. Quality Metrics
- RMSE: The Root Mean Square Error (RMSE) is a metric that considers the number of errors between two sets of data. In this metric, the closer to zero, the more accurate the observed forecast results [59]. Thus, a comparison will be made with the two breast ultrasound images, the original and the image after applying the improvement algorithms.where is the actual value of the data, is the predicted value, n is the number of data and is the total number of values.
- CNR: The contrast-to-noise ratio (CNR) is a metric to measure the contrast of images. It enables us to analyze the difference in contrast between the nodules and the other regions in the breast ultrasound images [60].where,which are the variation of signal strength inside and outside the target area, respectively.
- AMBE: The Absolute Mean Brightness Error (AMBE) is a metric that evaluates the difference between the average intensity level of the enhanced ultrasound image and the average intensity level of the original image [61].where is the average intensity level of the enhanced image and is the average intensity level of the original image.
- AG: The Average Gradient (AG) is a metric that represents the clarity of the breast ultrasound image, reflecting the image’s ability to express contrast details between the nodule and the other regions [62].where M and N are the width and height of the image, and refers to the horizontal and vertical gradients.
- PSNR: Peak Signal-to-Noise Ratio (PSNR) is a metric that evaluates the relationship between the maximum value of the measured signal and the amount of noise that affects the signal of breast ultrasound images [59].where is the maximum value and is the result of the RMSE.
- SSIM: The Structural Similarity Index (SSIM) is one of the quality assessment metric used to measure the visual changes and similarity between two images, by performing quality assessment and comparing the structural characteristics, which is described through the structural similarities [59]. In this way, it helps to analyze the similarity between the original breast ultrasound image and the image after applying the image improvement algorithm.where, with an image and being the average value for x or luminance x, being the average value for y or luminance y, the contrast value for y, the contrast value for x, and being two variables used to stabilize the division if the divisor is 0.
2.5.2. Rank Metrics
- True Positive class Benign (VB): VB occurs when in the actual dataset, class Benign was correctly predicted as class Benign.
- True Positive Malignant class (VM): The VM occurs when in the actual dataset, the Malignant class was correctly predicted as the Malignant class.
- True Positive Normal class (VN): The VN occurs when in the actual dataset, the Normal class was correctly predicted as the Normal class.
- False Negative (FN): FN occurs when in the actual data set, the class we are trying to predict was predicted incorrectly. That is, when it was supposed to be cancer and was diagnosed as non-cancer.
- False Positive (FP): FP occurs when in the actual dataset, the class we are trying to predict was predicted incorrectly. That is, when it was supposed to be non-cancer and was diagnosed as cancer.
- Accuracy: This is the general probability of success, which shows the global success rate considering the analyzed classes. Thus, it takes into account the hits of the three classes under all hits and misses.
- F1-score: It is the harmonic average between precision and recall. It is a commonly used metric to assess unbalanced data.
- Hit rate of benign class (Benign): This is the probability that a patient who has a positive diagnosis for benign actually has a benign nodule.
- Hit rate of Malignant class: This is the probability that a patient who has a positive diagnosis for malignant actually has a malignant nodule.
- Hit rate of the Normal class (Normal): This is the probability that a patient who has a negative diagnosis for nodules actually does not have nodules.
2.6. Experimental Configuration
3. Results and Discussion
3.1. Image Enhancement
3.2. Classification
3.3. Online System/Web Interfaces
4. Conclusions and Future Work
Future Work
- Developing novel light image enhancement strategies specific for breast cancer, considering the applied generic enhancement algorithms;
- Embedding such new enhancement approaches in specific hardware modules with the possibility of interacting with the cloud;
- Building 3D reconstruction models to perform the volumetric quantification of the nodules;
- Building a web dashboard to analyze the experiments as well as the 3D reconstruction with better visualizations.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, M. Research on the Detection Method of Breast Cancer Deep Convolutional Neural Network Based on Computer Aid. In Proceedings of the 2021 IEEE Asia-Pacific Conference on Image Processing, Electronics and Computers (IPEC), IEEE, Dalian, China, 14–16 April 2021; pp. 536–540. [Google Scholar]
- Matic, Z.; Kadry, S. Tumor Segmentation in Breast MRI Using Deep Learning. In Proceedings of the 2022 Fifth International Conference of Women in Data Science at Prince Sultan University (WiDS PSU), Riyadh, Saudi Arabia, 28–29 March 2022; pp. 49–51. [Google Scholar] [CrossRef]
- Ahmed, M.; Islam, M.R. Breast Cancer Classification from Histopathological Images using Convolutional Neural Network. In Proceedings of the 2021 International Conference on Computer, Communication, Chemical, Materials and Electronic Engineering (IC4ME2), Rajshahi, Bangladesh, 26–27 December 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Khumdee, M.; Assawaroongsakul, P.; Phasukkit, P.; Houngkamhang, N. Breast Cancer Detection using IR-UWB with Deep Learning. In Proceedings of the 2021 16th International Joint Symposium on Artificial Intelligence and Natural Language Processing (iSAI-NLP), Ayutthaya, Thailand, 21–23 December 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Afaq, S.; Jain, A. MAMMO-Net: An Approach for Classification of Breast Cancer using CNN with Gabor Filter in Mammographic Images. In Proceedings of the 2022 International Conference on Computational Intelligence and Sustainable Engineering Solutions (CISES), Greater Noida, India, 20–21 May 2022; pp. 177–182. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, N.; Xu, M.; Yu, J.; Qin, C.; Luo, X.; Yang, X.; Wang, T.; Li, A.; Ni, D. Deeply-Supervised Networks With Threshold Loss for Cancer Detection in Automated Breast Ultrasound. IEEE Trans. Med. Imaging 2020, 39, 866–876. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Huo, Y.; Pan, Y.; Xu, Z.; Huang, R.; Xie, Y.; Han, C.; Liu, Z.; Wang, Y. Learning Pre- and Post-contrast Representation for Breast Cancer Segmentation in DCE-MRI. In Proceedings of the 2022 IEEE 35th International Symposium on Computer-Based Medical Systems (CBMS), Shenzhen, China, 21–23 July 2022; pp. 355–359. [Google Scholar] [CrossRef]
- Huang, C.; Song, P.; Gong, P.; Trzasko, J.D.; Manduca, A.; Chen, S. Debiasing-Based Noise Suppression for Ultrafast Ultrasound Microvessel Imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2019, 66, 1281–1291. [Google Scholar] [CrossRef] [PubMed]
- Moshrefi, A.; Nabki, F. An Efficient Method to Enhance the Quality of Ultrasound Medical Images. In Proceedings of the 2021 IEEE International Midwest Symposium on Circuits and Systems (MWSCAS), Lansing, MI, USA, 9–11 August 2021; pp. 267–270. [Google Scholar] [CrossRef]
- de Souza, R.W.; Silva, D.S.; Passos, L.A.; Roder, M.; Santana, M.C.; Pinheiro, P.R.; de Albuquerque, V.H.C. Computer-assisted Parkinson’s disease diagnosis using fuzzy optimum- path forest and Restricted Boltzmann Machines. Comput. Biol. Med. 2021, 131, 104260. [Google Scholar] [CrossRef] [PubMed]
- Qadri, S.F.; Shen, L.; Ahmad, M.; Qadri, S.; Zareen, S.S.; Akbar, M.A. SVseg: Stacked sparse autoencoder-based patch classification modeling for vertebrae segmentation. Mathematics 2022, 10, 796. [Google Scholar] [CrossRef]
- Afonso, L.C.; Rosa, G.H.; Pereira, C.R.; Weber, S.A.; Hook, C.; Albuquerque, V.H.C.; Papa, J.P. A recurrence plot-based approach for Parkinson’s disease identification. Future Gener. Comput. Syst. 2019, 94, 282–292. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, X.; Sun, S.; Wu, D.; Bai, J.; Yin, Y.; Liu, X.; Zhang, H.; de Albuquerque, V.H.C. Learning physical properties in complex visual scenes: An intelligent machine for perceiving blood flow dynamics from static CT angiography imaging. Neural Netw. 2020, 123, 82–93. [Google Scholar] [CrossRef]
- Khan, M.A.; Muhammad, K.; Sharif, M.; Akram, T.; Albuquerque, V.H.C.d. Multi-Class Skin Lesion Detection and Classification via Teledermatology. IEEE J. Biomed. Health Inform. 2021, 25, 4267–4275. [Google Scholar] [CrossRef]
- Cao, L.; Wang, W.; Huang, C.; Xu, Z.; Wang, H.; Jia, J.; Chen, S.; Dong, Y.; Fan, C.; de Albuquerque, V.H.C. An Effective Fusing Approach by Combining Connectivity Network Pattern and Temporal-Spatial Analysis for EEG-Based BCI Rehabilitation. IEEE Trans. Neural Syst. Rehabil. Eng. 2022, 30, 2264–2274. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, Y.; Liang, Y.; Zhan, Z.; Jiang, M.; Zhang, X.; Daniel, S.d.S.; Wu, W.; Albuquerque, V.H.C.d. Edge2Analysis: A Novel AIoT Platform for Atrial Fibrillation Recognition and Detection. IEEE J. Biomed. Health Inform. 2022. [Google Scholar] [CrossRef]
- de Mesquita, V.A.; Cortez, P.C.; Ribeiro, A.B.; de Albuquerque, V.H.C. A novel method for lung nodule detection in computed tomography scans based on Boolean equations and vector of filters techniques. Comput. Electr. Eng. 2022, 100, 107911. [Google Scholar] [CrossRef]
- Huang, C.; Zhang, G.; Chen, S.; de Albuquerque, V.H.C. An Intelligent Multisampling Tensor Model for Oral Cancer Classification. IEEE Trans. Ind. Inform. 2022, 18, 7853–7861. [Google Scholar] [CrossRef]
- Mohamed, A.; Wahba, A.A.; Sayed, A.M.; Haggag, M.A.; El-Adawy, M.I. Enhancement of Ultrasound Images Quality Using a New Matching Material. In Proceedings of the 2019 International Conference on Innovative Trends in Computer Engineering (ITCE), Aswan, Egypt, 2–4 February 2019; pp. 47–51. [Google Scholar] [CrossRef]
- Singh, P.; Mukundan, R.; Ryke, R.d. Feature Enhancement in Medical Ultrasound Videos Using Multifractal and Contrast Adaptive Histogram Equalization Techniques. In Proceedings of the 2019 IEEE Conference on Multimedia Information Processing and Retrieval (MIPR), San Jose, CA, USA, 28–30 March 2019; pp. 240–245. [Google Scholar] [CrossRef] [Green Version]
- Latif, G.; Butt, M.; Al Anezi, F.Y.; Alghazo, J. Remoção de manchas de imagem por ultrassom e detecção de câncer de mama usando Deep CNN. In Proceedings of the 2020 RIVF International Conference on Computing and Communication Technologies (RIVF), Ho Chi Minh City, Vietnam, 14–15 October 2020. [Google Scholar] [CrossRef]
- Zhao, H.; Niu, J.; Meng, H.; Wang, Y.; Li, Q.; Yu, Z. Focal U-Net: A Focal Self-attention based U-Net for Breast Lesion Segmentation in Ultrasound Images. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; pp. 1506–1511. [Google Scholar] [CrossRef]
- Jahwar, A.F.; Mohsin Abdulazeez, A. Segmentation and Classification for Breast Cancer Ultrasound Images Using Deep Learning Techniques: A Review. In Proceedings of the 2022 IEEE 18th International Colloquium on Signal Processing & Applications (CSPA), Selangor, Malaysia, 12 May 2022; pp. 225–230. [Google Scholar] [CrossRef]
- Dabass, J.; Arora, S.; Vig, R.; Hanmandlu, M. Segmentation Techniques for Breast Cancer Imaging Modalities-A Review. In Proceedings of the 2019 9th International Conference on Cloud Computing, Data Science & Engineering (Confluence), Noida, India, 10–11 January 2019; pp. 658–663. [Google Scholar] [CrossRef]
- Chen, C.; Wang, Y.; Niu, J.; Liu, X.; Li, Q.; Gong, X. Domain Knowledge Powered Deep Learning for Breast Cancer Diagnosis Based on Contrast-Enhanced Ultrasound Videos. IEEE Trans. Med. Imaging 2021, 40, 2439–2451. [Google Scholar] [CrossRef] [PubMed]
- Badawy, S.M.; Mohamed, A.E.N.A.; Hefnawy, A.A.; Zidan, H.E.; GadAllah, M.T.; El-Banby, G.M. Classification of Breast Ultrasound Images Based on Convolutional Neural Networks—A Comparative Study. In Proceedings of the 2021 International Telecommunications Conference (ITC-Egypt), Alexandria, Egypt, 13–15 July 2021; pp. 1–8. [Google Scholar] [CrossRef]
- Wang, Y.; Lei, B.; Elazab, A.; Tan, E.L.; Wang, W.; Huang, F.; Gong, X.; Wang, T. Breast Cancer Image Classification via Multi-Network Features and Dual-Network Orthogonal Low-Rank Learning. IEEE Access 2020, 8, 27779–27792. [Google Scholar] [CrossRef]
- Yaganteeswarudu, A. Multi Disease Prediction Model by using Machine Learning and Flask API. In Proceedings of the 2020 5th International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 10–12 June 2020; pp. 1242–1246. [Google Scholar] [CrossRef]
- Mufid, M.R.; Basofi, A.; Al Rasyid, M.U.H.; Rochimansyah, I.F.; rokhim, A. Design an MVC Model using Python for Flask Framework Development. In Proceedings of the 2019 International Electronics Symposium (IES), Surabaya, Indonesia, 27–28 September 2019; pp. 214–219. [Google Scholar] [CrossRef]
- Ahmed, I.; Ahmad, A.; Jeon, G. An IoT-Based Deep Learning Framework for Early Assessment of Covid-19. IEEE Internet Things J. 2021, 8, 15855–15862. [Google Scholar] [CrossRef]
- Munadi, K.; Muchtar, K.; Maulina, N.; Pradhan, B. Image Enhancement for Tuberculosis Detection Using Deep Learning. IEEE Access 2020, 8, 217897–217907. [Google Scholar] [CrossRef]
- Al-Dhabyani, W.; Gomaa, M.; Khaled, H.; Fahmy, A. Dataset of breast ultrasound images. Data Brief 2020, 28, 104863. [Google Scholar] [CrossRef]
- Tomasi, C.; Manduchi, R. Bilateral filtering for gray and color images. In Proceedings of the Sixth International Conference on Computer Vision (IEEE Cat. No.98CH36271), Bombay, India, 7 January 1998; pp. 839–846. [Google Scholar] [CrossRef]
- Zhihong, W.; Xiaohong, X. Study on Histogram Equalization. In Proceedings of the 2011 2nd International Symposium on Intelligence Information Processing and Trusted Computing, Wuhan, China, 22–23 October 2011; pp. 177–179. [Google Scholar] [CrossRef]
- Chambolle, A. An algorithm for total variation minimization and applications. J. Math. Imaging Vis. 2004, 20, 89–97. [Google Scholar]
- Guo, X.; Li, Y.; Ling, H. LIME: Low-Light Image Enhancement via Illumination Map Estimation. IEEE Trans. Image Process. 2017, 26, 982–993. [Google Scholar] [CrossRef]
- Ying, Z.; Li, G.; Ren, Y.; Wang, R.; Wang, W. A New Image Contrast Enhancement Algorithm Using Exposure Fusion Framework. In Proceedings of the Computer Analysis of Images and Patterns, Ystad, Sweden, 22–24 August 2017; Felsberg, M., Heyden, A., Krüger, N., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 36–46. [Google Scholar]
- Cao, G.; Huang, L.; Tian, H.; Huang, X.; Wang, Y.; Zhi, R. Contrast enhancement of brightness-distorted images by improved adaptive gamma correction. Comput. Electr. Eng. 2018, 66, 569–582. [Google Scholar] [CrossRef] [Green Version]
- Ullah, H.; Muhammad, K.; Irfan, M.; Anwar, S.; Sajjad, M.; Imran, A.S.; de Albuquerque, V.H.C. Light-DehazeNet: A Novel Lightweight CNN Architecture for Single Image Dehazing. IEEE Trans. Image Process. 2021, 30, 8968–8982. [Google Scholar] [CrossRef]
- Guo, C.; Li, C.; Guo, J.; Loy, C.C.; Hou, J.; Kwong, S.; Cong, R. Zero-Reference Deep Curve Estimation for Low-Light Image Enhancement. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 1777–1786. [Google Scholar] [CrossRef]
- Wang, Y.; Wan, R.; Yang, W.; Li, H.; Chau, L.P.; Kot, A.C. Low-Light Image Enhancement with Normalizing Flow. arXiv 2021. [Google Scholar] [CrossRef]
- Muhammad, K.; Hussain, T.; Tanveer, M.; Sannino, G.; de Albuquerque, V.H.C. Cost-Effective Video Summarization Using Deep CNN With Hierarchical Weighted Fusion for IoT Surveillance Networks. IEEE Internet Things J. 2020, 7, 4455–4463. [Google Scholar] [CrossRef]
- Khan, S.; Muhammad, K.; Mumtaz, S.; Baik, S.W.; de Albuquerque, V.H.C. Energy-Efficient Deep CNN for Smoke Detection in Foggy IoT Environment. IEEE Internet Things J. 2019, 6, 9237–9245. [Google Scholar] [CrossRef]
- Hussain, T.; Muhammad, K.; Ullah, A.; Cao, Z.; Baik, S.W.; de Albuquerque, V.H.C. Cloud-Assisted Multiview Video Summarization Using CNN and Bidirectional LSTM. IEEE Trans. Ind. Inform. 2020, 16, 77–86. [Google Scholar] [CrossRef]
- Muhammad, K.; Mustaqeem; Ullah, A.; Imran, A.S.; Sajjad, M.; Kiran, M.S.; Sannino, G.; de Albuquerque, V.H.C. Human action recognition using attention based LSTM network with dilated CNN features. Future Gener. Comput. Syst. 2021, 125, 820–830. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Wu, Z.; Tang, C. Modulation Recognition Algorithm Based on ResNet50 Multi-feature Fusion. In Proceedings of the 2021 International Conference on Intelligent Transportation, Big Data & Smart City (ICITBS), Xi’an, China, 27–28 March 2021; pp. 677–680. [Google Scholar] [CrossRef]
- Yang, X.; Yang, D.; Huang, C. An interactive prediction system of breast cancer based on ResNet50, chatbot and PyQt. In Proceedings of the 2021 2nd International Seminar on Artificial Intelligence, Networking and Information Technology (AINIT), Shanghai, China, 15–17 October 2021; pp. 309–316. [Google Scholar] [CrossRef]
- Wan, Z.; Gu, T. A Bootstrapped Transfer Learning Model Based on ResNet50 and Xception to Classify Buildings Post Hurricane. In Proceedings of the 2021 2nd International Conference on Big Data & Artificial Intelligence & Software Engineering (ICBASE), Zhuhai, China, 24–26 September 2021; pp. 265–270. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Z.; He, J.; Zhu, Y.; Wei, X. A method of vehicle flow training and detection based on ResNet50 with CenterNet method. In Proceedings of the 2021 International Conference on Communications, Information System and Computer Engineering (CISCE), Beijing, China, 14–16 May 2021; pp. 335–339. [Google Scholar] [CrossRef]
- Dutta, J.; Chanda, D. Music Emotion Recognition in Assamese Songs using MFCC Features and MLP Classifier. In Proceedings of the 2021 International Conference on Intelligent Technologies (CONIT), Hubli, India, 25–17 June 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Ahil, M.N.; Vanitha, V.; Rajathi, N. Apple and Grape Leaf Disease Classification using MLP and CNN. In Proceedings of the 2021 International Conference on Advancements in Electrical, Electronics, Communication, Computing and Automation (ICAECA), Coimbatore, India, 8–9 October 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Wang, H.; Wang, J. Short Term Wind Speed Forecasting Based on Feature Extraction by CNN and MLP. In Proceedings of the 2021 2nd International Symposium on Computer Engineering and Intelligent Communications (ISCEIC), Nanjing, China, 6–8 August 2021; pp. 191–197. [Google Scholar] [CrossRef]
- Liu, H.; Xiao, X.; Li, Y.; Mi, Q.; Yang, Z. Effective Data Classification via Combining Neural Networks and SVM. In Proceedings of the 2019 Chinese Control And Decision Conference (CCDC), Nanchang, China, 3–5 June 2019; pp. 4006–4009. [Google Scholar] [CrossRef]
- Kr, K.; Kv, A.R.; Pillai, A. An Improved Feature Selection and Classification of Gene Expression Profile using SVM. In Proceedings of the 2019 2nd International Conference on Intelligent Computing, Instrumentation and Control Technologies (ICICICT), Kannur, India, 5–6 June 2019; Volume 1, pp. 1033–1037. [Google Scholar] [CrossRef]
- Turesson, H.K.; Ribeiro, S.; Pereira, D.R.; Papa, J.P.; de Albuquerque, V.H.C. Machine learning algorithms for automatic classification of marmoset vocalizations. PLoS ONE 2016, 11, e0163041. [Google Scholar] [CrossRef] [Green Version]
- Salim, A.P.; Laksitowening, K.A.; Asror, I. Time Series Prediction on College Graduation Using KNN Algorithm. In Proceedings of the 2020 8th International Conference on Information and Communication Technology (ICoICT), Yogyakarta, Indonesia, 24–26 June 2020; pp. 1–4. [Google Scholar] [CrossRef]
- Ma, J.; Li, J.; Wang, W. Application of Wavelet Entropy and KNN in Motor Fault Diagnosis. In Proceedings of the 2022 4th International Conference on Communications, Information System and Computer Engineering (CISCE), Shenzhen, China, 27–29 May 2022; pp. 221–226. [Google Scholar] [CrossRef]
- Sabilla, I.A.; Meirisdiana, M.; Sunaryono, D.; Husni, M. Best Ratio Size of Image in Steganography using Portable Document Format with Evaluation RMSE, PSNR, and SSIM. In Proceedings of the 2021 4th International Conference of Computer and Informatics Engineering (IC2IE), Depok, Indonesia, 14–15 September 2021; pp. 289–294. [Google Scholar] [CrossRef]
- Rodriguez-Molares, A.; Hoel Rindal, O.M.; D’hooge, J.; Måsøy, S.E.; Austeng, A.; Torp, H. The Generalized Contrast-to-Noise Ratio. In Proceedings of the 2018 IEEE International Ultrasonics Symposium (IUS), Kobe, Japan, 22–25 October 2018; pp. 1–4. [Google Scholar] [CrossRef]
- Harun, N.H.; Bakar, J.A.; Wahab, Z.A.; Osman, M.K.; Harun, H. Color Image Enhancement of Acute Leukemia Cells in Blood Microscopic Image for Leukemia Detection Sample. In Proceedings of the 2020 IEEE 10th Symposium on Computer Applications & Industrial Electronics (ISCAIE), Penang, Malaysia, 18–19 April 2020; pp. 24–29. [Google Scholar] [CrossRef]
- AbdAlRahman, A.; Ismail, S.M.; Said, L.A.; Radwan, A.G. Double Fractional-order Masks Image Enhancement. In Proceedings of the 2021 3rd Novel Intelligent and Leading Emerging Sciences Conference (NILES), Giza, Egypt, 23–25 October 2021; pp. 261–264. [Google Scholar] [CrossRef]









| Information | Amount | Percent |
|---|---|---|
| Normal Images | 133 | 17.05 |
| Benign Images | 437 | 56.03 |
| Malignant Images | 210 | 26.92 |
| Algorithms | Processing Time (minutes) |
|---|---|
| Bilateral | 1344.5736 |
| Gamma correction | 2.8820 |
| HE | 7.1598 |
| LDNet | 6.7305 |
| LIME | 651.8196 |
| LLFLOW | 9.2683 |
| TV | 17.3219 |
| Ying | 8.8536 |
| ZDCE | 9.5254 |
| Algorithms | RMSE | CNR | AMBE | AG | PSNR | SSIM |
|---|---|---|---|---|---|---|
| Bilateral | 0.0329 | 0.0317 | 0.0063 | 0.0 | 77.8054 | 0.8726 |
| Gamma correction | 0.0936 | 0.4060 | 0.0763 | 0.0363 | 36.7292 | 0.8924 |
| HE | 0.2391 | 1.1135 | 0.2161 | 0.0364 | 61.2157 | 0.6347 |
| LDNet | 0.2360 | 0.9508 | 0.1941 | 0.0361 | 60.7778 | 0.3705 |
| LIME | 0.2801 | 1.0946 | 0.2272 | 0.0 | 59.4688 | 0.3778 |
| LLFLOW | 0.1342 | 0.5578 | 0.1069 | 0.0184 | 66.3159 | 0.8116 |
| TV | 0.0227 | 0.0088 | 0.0017 | 3.86e-05 | 81.0792 | 0.8569 |
| Ying | 0.0691 | 0.3125 | 0.0621 | 0.1676 | 71.6003 | 0.9394 |
| ZDCE | 0.1334 | 0.6059 | 0.1189 | 0.0362 | 65.6991 | 0.7858 |
| Algorithms | Times (seconds) | MLP | kNN | SVM |
|---|---|---|---|---|
| Original | Training | 161.605 | 0.105 | 109.754 |
| Test | 0.036 | 1.119 | 5.582 | |
| Bilateral | Training | 0.1076 | 0.107 | 114,538 |
| Test | 0.034 | 1.105 | 5.741 | |
| Gamma correction | Training | 142.177 | 0.106 | 113.221 |
| Test | 0.039 | 1.231 | 5.801 | |
| HE | Training | 164.593 | 0.104 | 117.841 |
| Test | 0.033 | 1.176 | 5.906 | |
| LDNET | Training | 197.522 | 0.106 | 119.007 |
| Test | 0.040 | 1.143 | 6.101 | |
| LIME | Training | 158.675 | 0.105 | 110.196 |
| Test | 0.032 | 1.193 | 5.622 | |
| LLFLOW | Training | 120.524 | 0.106 | 114.953 |
| Test | 0.032 | 1.205 | 5.803 | |
| TV | Training | 141.409 | 0.106 | 115.173 |
| Test | 0.031 | 1.214 | 5.900 | |
| Ying | Training | 156.628 | 0.105 | 115.086 |
| Test | 0.033 | 1.194 | 5.874 | |
| Z-DCE | Training | 169.422 | 0.105 | 123.348 |
| Test | 0.037 | 1.122 | 6.170 |
| Algorithms | Metrics | MLP | kNN | SVM |
|---|---|---|---|---|
| Original | ACC Global | 94.99 ± 1.65 | 77.85 ± 3.30 | 96.50 ± 1.82 |
| Benign | 97.08 ± 1.50 | 76.44 ± 4.98 | 97.42 ± 1.44 | |
| Malignant | 92.50 ± 3.21 | 71.90 ± 5.90 | 94.40 ± 2.94 | |
| Normal | 92.15 ± 7.50 | 91.91 ± 5.87 | 96.80 ± 3.99 | |
| F1-score | 94.96 ± 1.69 | 78.57 ± 3.20 | 96.49 ± 1.56 | |
| Bilateral | ACC Global | 95.54 ± 1.36 | 79.07 ± 2.45 | 96.69 ± 1.56 |
| Benign | 96.68 ± 1.91 | 78.78 ± 3.06 | 97.31 ± 1.94 | |
| Malignant | 92.61 ± 3.44 | 73.21 ± 5.92 | 95.11 ± 2.86 | |
| Normal | 96.41 ± 4.41 | 89.31 ± 5.65 | 97.18 ± 3.54 | |
| F1-score | 95.53 ± 1.37 | 79.70 ± 2.41 | 96.69 ± 1.56 | |
| Gamma correction | ACC Global | 94.93 ± 1.59 | 77.91 ± 2.66 | 96.47 ± 1.33 |
| Benign | 96.74 ± 1.57 | 77.17 ± 4.08 | 97.54 ± 1.31 | |
| Malignant | 91.90 ± 3.86 | 70.83 ± 5.52 | 93.92 ± 3.32 | |
| Normal | 93.83 ± 5.90 | 91.54 ± 6.45 | 97.00 ± 3.66 | |
| F1-score | 94.91 ± 1.61 | 78.72 ± 2.65 | 96.46 ± 1.35 | |
| HE | ACC Global | 95.28 ± 1.71 | 78.07 ± 2.62 | 96.31 ± 1.38 |
| Benign | 96.45 ± 2.01 | 78.20 ± 4.21 | 97.25 ± 1.50 | |
| Malignant | 92.97 ± 3.87 | 75.23 ± 5.80 | 94.40 ± 2.84 | |
| Normal | 95.15 ± 4.55 | 82.15 ± 7.88 | 96.26 ± 3.51 | |
| F1-score | 95.27 ± 1.71 | 78.45 ± 2.54 | 96.30 ± 1.39 | |
| LDNET | ACC Global | 94.96 ± 1.96 | 83.23 ± 2.54 | 96.31 ± 1.52 |
| Benign | 96.90 ± 1.81 | 84.61 ± 3.07 | 97.19 ± 1.59 | |
| Malignant | 93.09 ± 4.63 | 76.90 ± 5.48 | 94.40 ± 3.55 | |
| Normal | 91.50 ± 7.48 | 88.73 ± 6.95 | 96.45 ± 4.60 | |
| F1-score | 94.93 ± 1.99 | 83.44 ± 2.46 | 96.30 ± 1.53 | |
| LIME | ACC Global | 95.51 ± 1.46 | 81.57 ± 2.57 | 96.47 ± 1.24 |
| Benign | 96.91 ± 1.72 | 82.21 ± 3.45 | 97.25 ± 1.36 | |
| Malignant | 92.38 ± 3.49 | 75.00 ± 5.90 | 94.16 ± 3.14 | |
| Normal | 95.88 ± 3.71 | 89.87 ± 6.47 | 97.56 ± 2.45 | |
| F1-score | 95.50 ± 1.46 | 82.00 ± 2.55 | 96.46 ± 1.24 | |
| LLFLOW | ACC Global | 95.06 ± 1.66 | 75.96 ± 3.40 | 96.34 ± 1.07 |
| Benign | 96.23 ± 2.25 | 75.74 ± 3.73 | 97.25 ± 1.32 | |
| Malignant | 92.49 ± 3.47 | 68.09 ± 6.79 | 93.69 ± 2.94 | |
| Normal | 95.34 ± 6.82 | 89.10 ± 6.33 | 97.57 ± 3.77 | |
| F1-score | 95.04 ± 1.66 | 76.75 ± 3.25 | 96.33 ± 1.07 | |
| TV | ACC Global | 95.64 ± 1.41 | 77.33 ± 3.21 | 96.66 ± 1.66 |
| Benign | 96.97 ± 1.57 | 73.97 ± 5.25 | 97.42 ± 1.57 | |
| Malignant | 93.09 ± 3.18 | 76.30 ± 5.02 | 94.88 ± 2.84 | |
| Normal | 95.31 ± 4.38 | 90.05 ± 5.44 | 97.00 ± 3.86 | |
| F1-score | 95.63 ± 1.42 | 78.08 ± 3.06 | 96.66 ± 1.67 | |
| Ying | ACC Global | 95.22 ± 1.56 | 78.14 ± 3.35 | 96.57 ± 1.39 |
| Benign | 96.22 ± 1.76 | 76.90 ± 5.01 | 97.37 ± 1.44 | |
| Malignant | 93.80 ± 3.94 | 73.21 ± 7.25 | 94.64 ± 3.18 | |
| Normal | 94.18 ± 6.42 | 90.02 ± 5.97 | 96.99 ± 3.69 | |
| F1-score | 95.21 ± 1.58 | 78.93 ± 3.16 | 96.56 ± 1.39 | |
| Z-DCE | ACC Global | 94.23 ± 2.18 | 76.89 ± 2.96 | 95.80 ± 1.35 |
| Benign | 96.68 ± 1.56 | 78.56 ± 5.18 | 97.14 ± 1.46 | |
| Malignant | 92.14 ± 4.06 | 69.40 ± 5.54 | 93.09 ± 2.90 | |
| Normal | 89.52 ± 9.41 | 83.31 ± 7.36 | 95.70 ± 3.74 | |
| F1-score | 94.17 ± 2.25 | 77.25 ± 2.94 | 95.79 ± 1.35 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Silva, D.S.; Nascimento, C.S.; Jagatheesaperumal, S.K.; Albuquerque, V.H.C.d. Mammogram Image Enhancement Techniques for Online Breast Cancer Detection and Diagnosis. Sensors 2022, 22, 8818. https://doi.org/10.3390/s22228818
da Silva DS, Nascimento CS, Jagatheesaperumal SK, Albuquerque VHCd. Mammogram Image Enhancement Techniques for Online Breast Cancer Detection and Diagnosis. Sensors. 2022; 22(22):8818. https://doi.org/10.3390/s22228818
Chicago/Turabian Styleda Silva, Daniel S., Caio S. Nascimento, Senthil K. Jagatheesaperumal, and Victor Hugo C. de Albuquerque. 2022. "Mammogram Image Enhancement Techniques for Online Breast Cancer Detection and Diagnosis" Sensors 22, no. 22: 8818. https://doi.org/10.3390/s22228818
APA Styleda Silva, D. S., Nascimento, C. S., Jagatheesaperumal, S. K., & Albuquerque, V. H. C. d. (2022). Mammogram Image Enhancement Techniques for Online Breast Cancer Detection and Diagnosis. Sensors, 22(22), 8818. https://doi.org/10.3390/s22228818











