An Ensemble Approach for the Prediction of Diabetes Mellitus Using a Soft Voting Classifier with an Explainable AI
Abstract
:1. Introduction
1.1. Diabetes-Facts and Figures
1.2. Problem Statement
1.3. Artificial Intelligence (AI) Research Challenges in a Diabetes Diagnosis
1.4. Research Motivation
1.5. Aim, Contribution, and Paper Organization
- Several machine learning algorithms were applied and using the two best classification methods, an ensemble method was developed to diagnose diabetes.
- The model’s inside explainability was provided to make the model more reliable and to produce a good balance between the accuracy and interpretability, which will be convenient for doctors or clinicians to understand and apply the model.
- SHAP plots were created to provide physicians with some insights into the main driving factors affecting the disease prediction from various perspectives, including visualization, feature importance, and each attribute’s contribution to making a decision.
2. Literature Review
3. Methodology
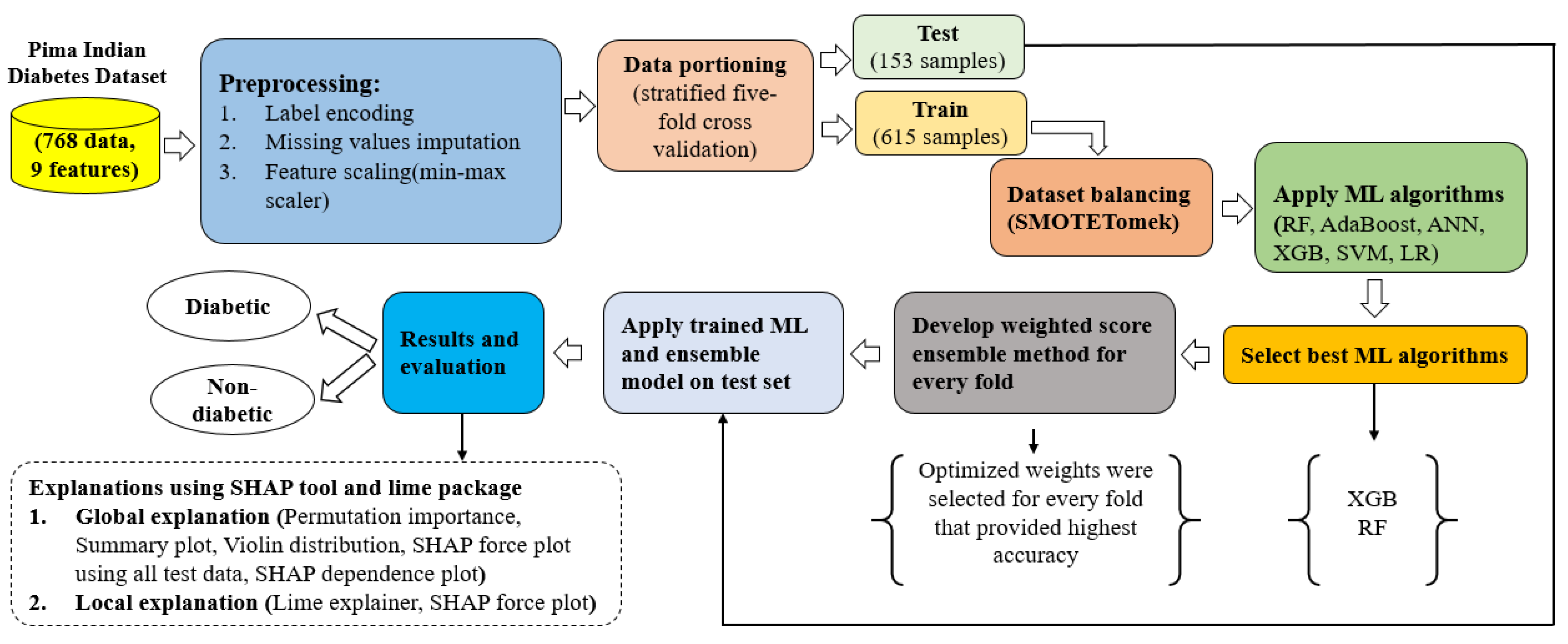
3.1. Proposed Approach
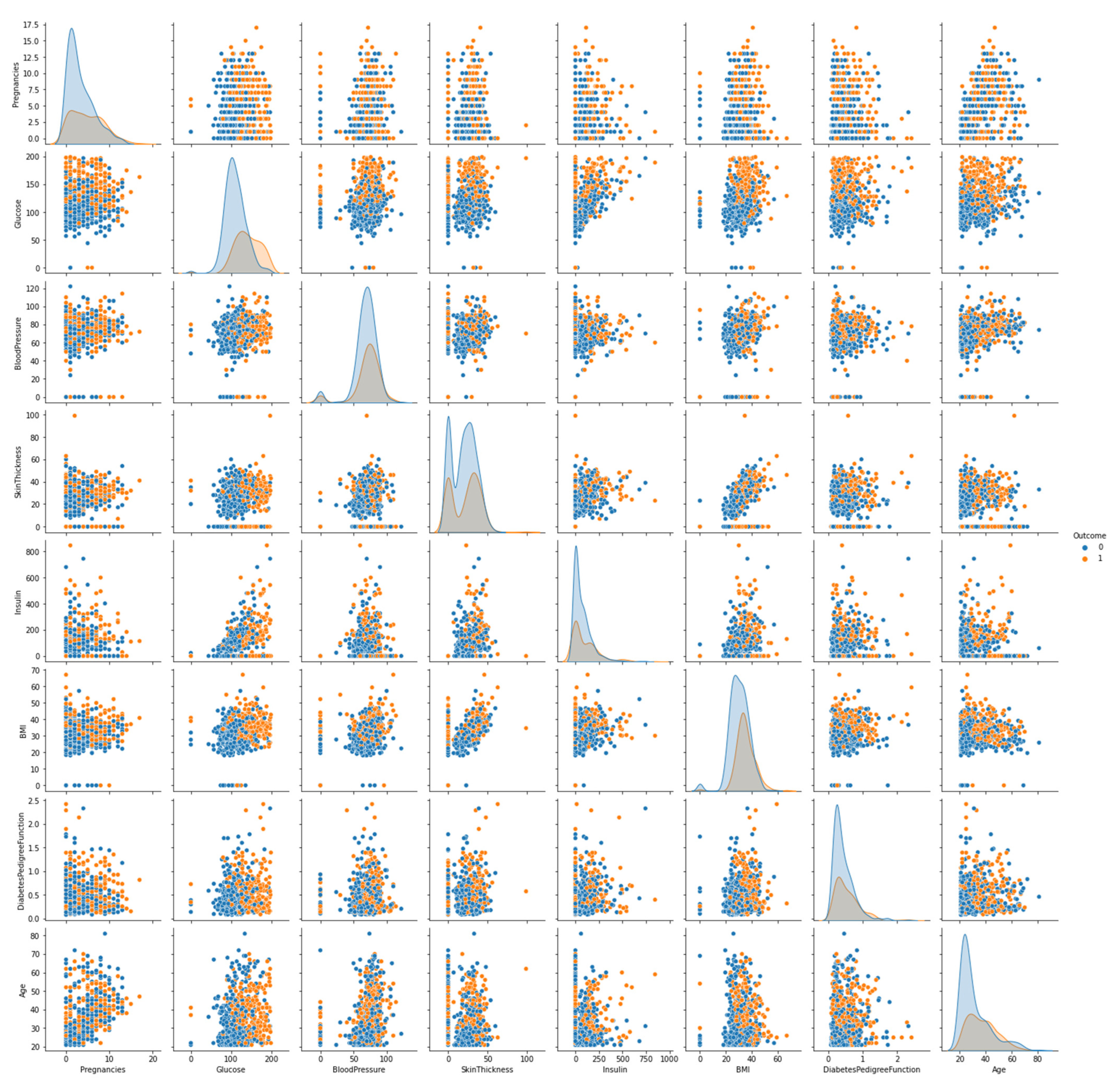
3.2. Dataset Description
3.3. Preprocessing
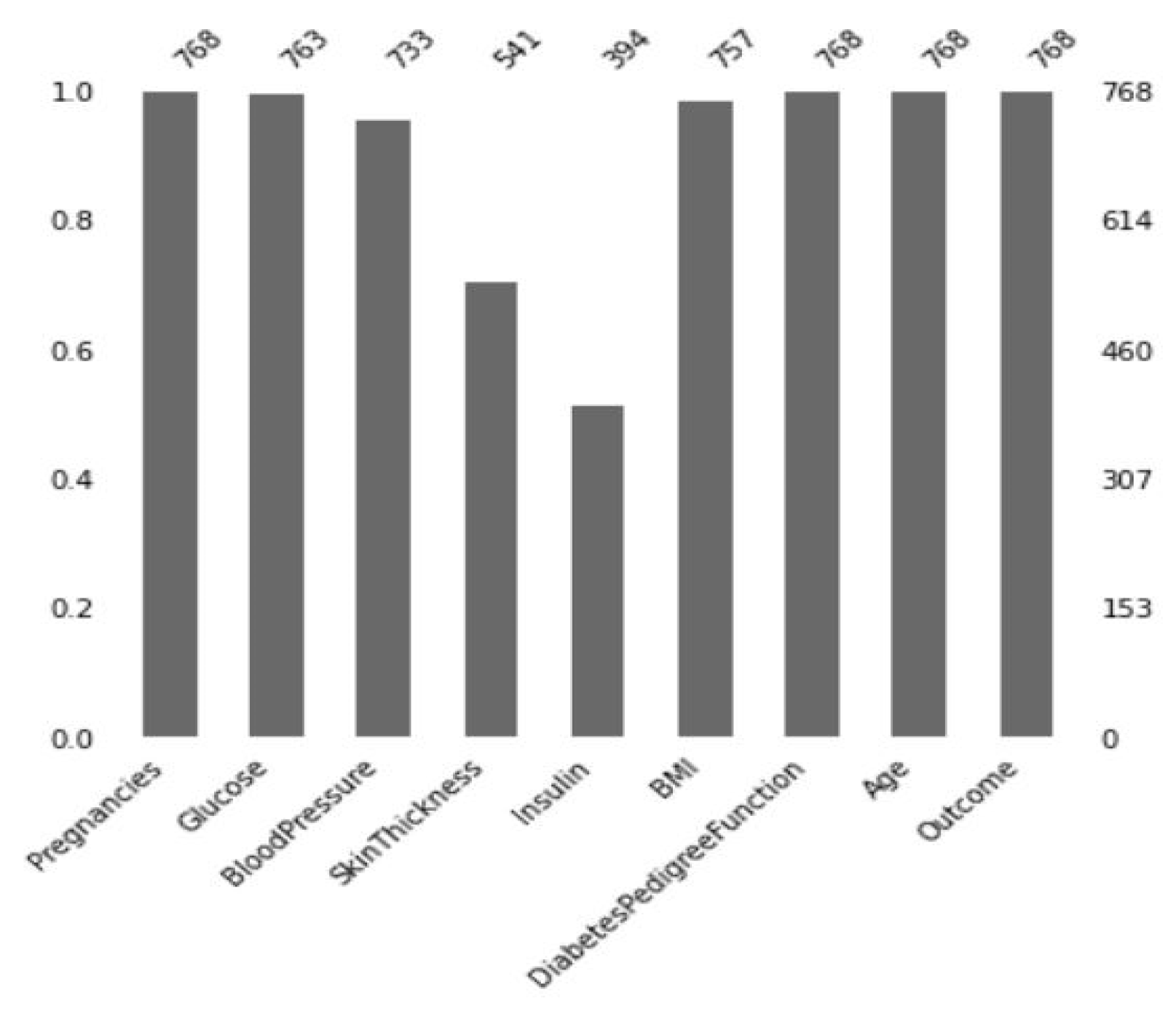
3.3.1. Missing Value Imputation
3.3.2. Data Partitioning
3.3.3. Handling Imbalanced Classes of a Dataset
3.3.4. Feature Scaling
3.3.5. Weighted Score Approach for the Ensemble Method
3.4. Models and Algorithms
3.4.1. Ensemble Learning
3.4.2. AdaBoost
3.4.3. Random Forest
3.4.4. XGBoost
3.4.5. Logistic Regression
3.4.6. Support Vector Machine
3.4.7. Artificial Neural Network
3.4.8. Reproducible Models
3.4.9. Shapley Additive Explanations (SHAP)
4. Performance Analysis and Experimental Results
4.1. Performance Parameter
4.2. Performance Results
4.3. Comparison with Previous Research
4.4. Model’s Explainability
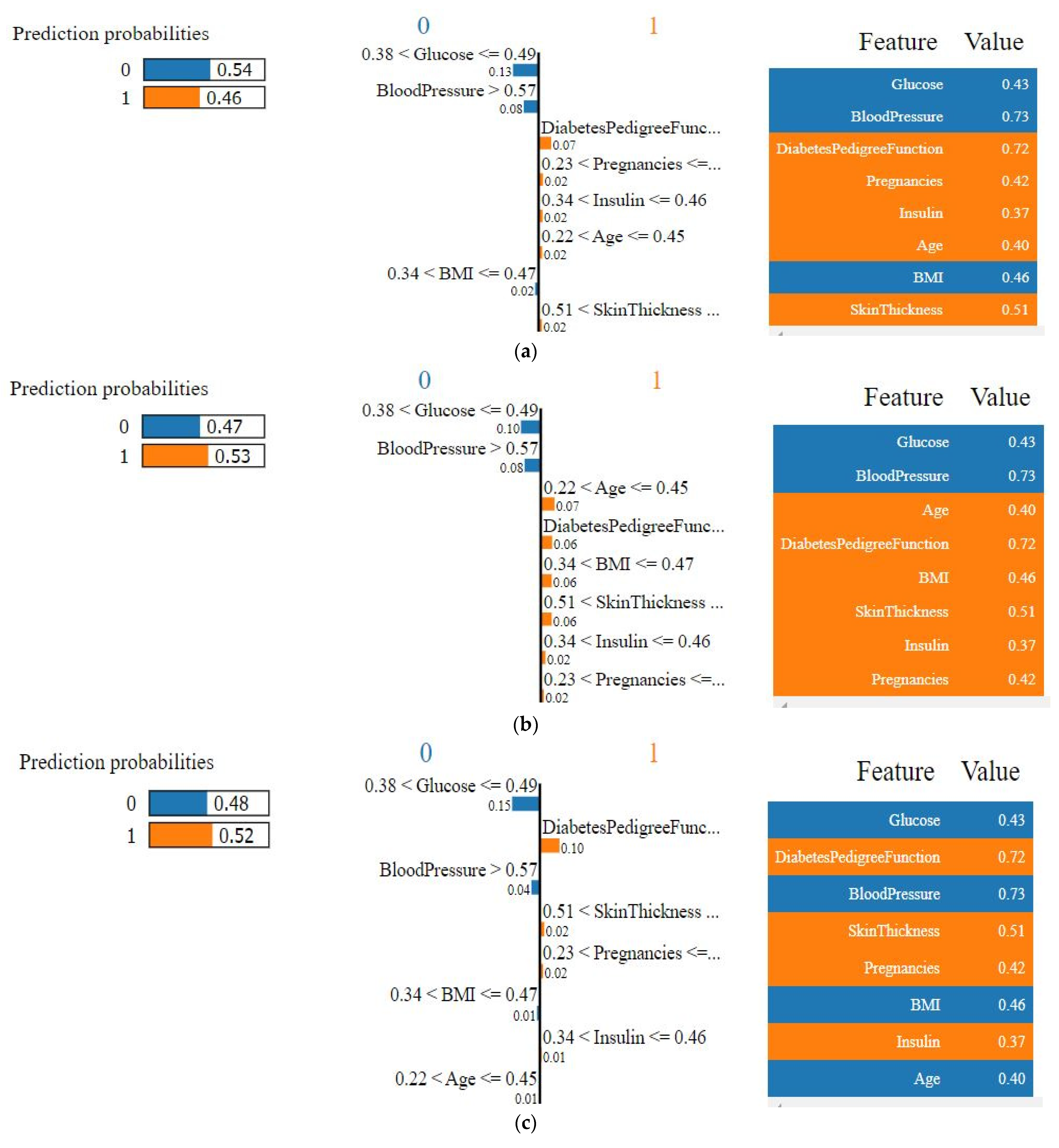
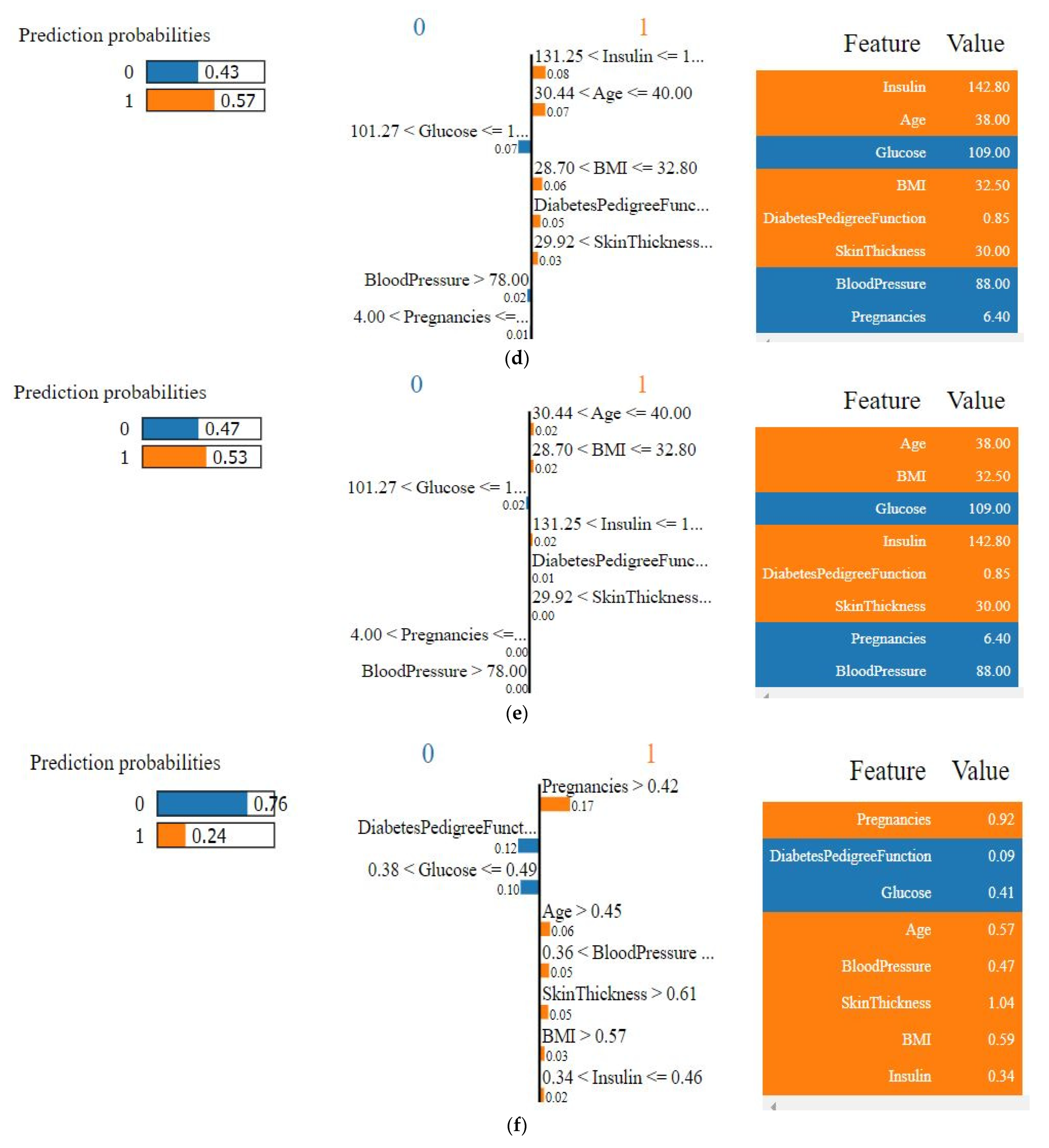
4.4.1. Explainability of the Outcome using LIME (Local)
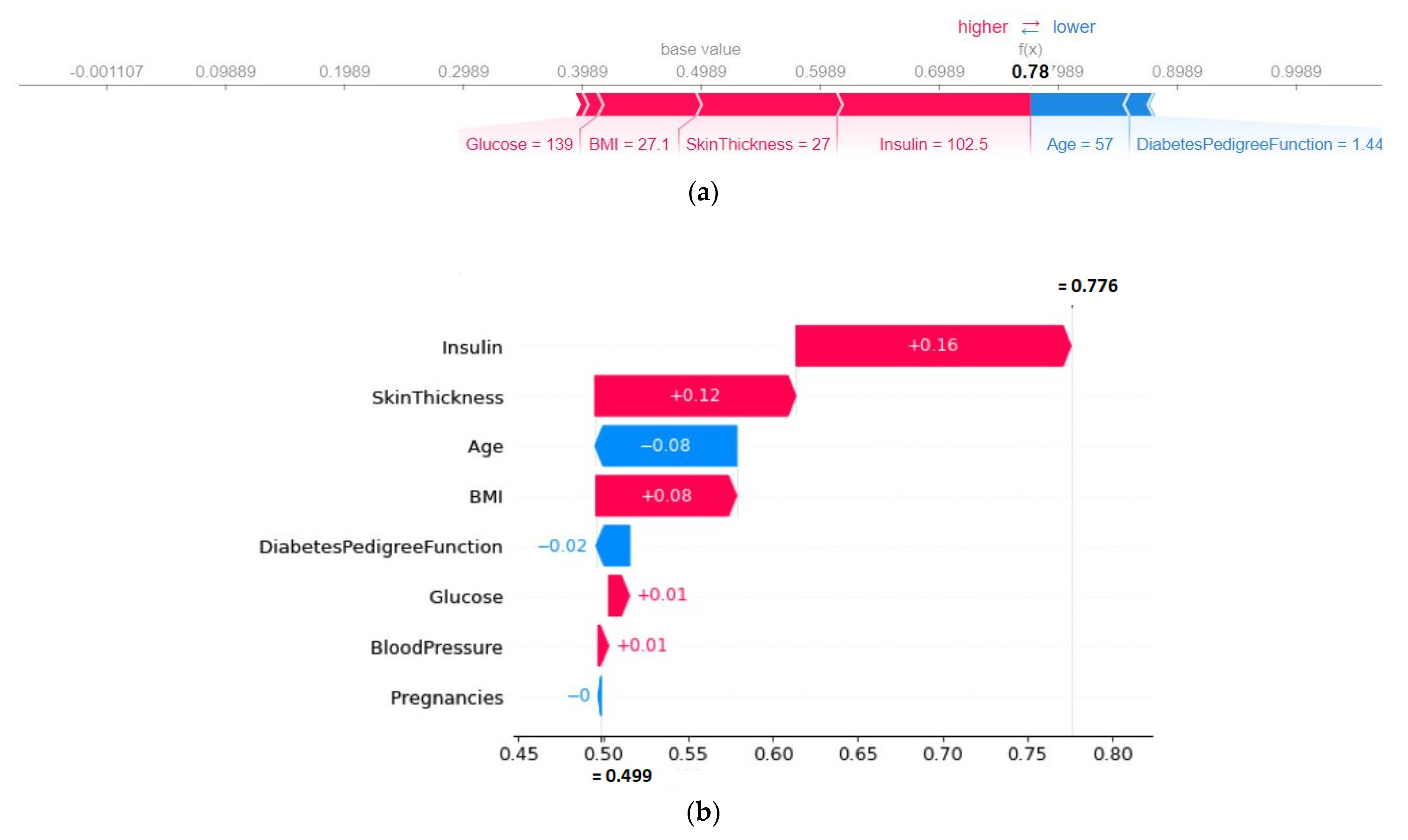
4.4.2. SHAP Force Plot of a Particular Test Set (Local)
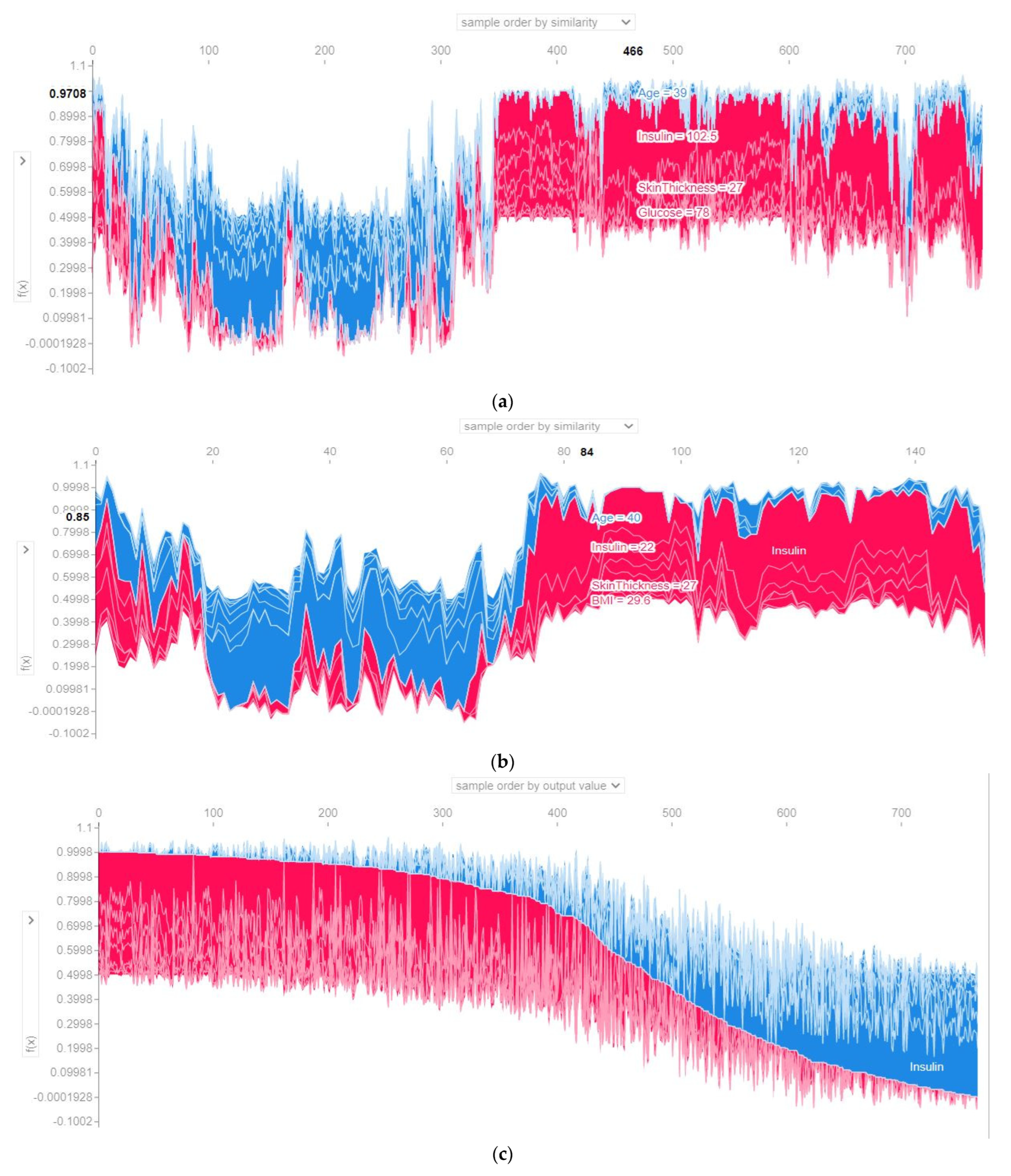
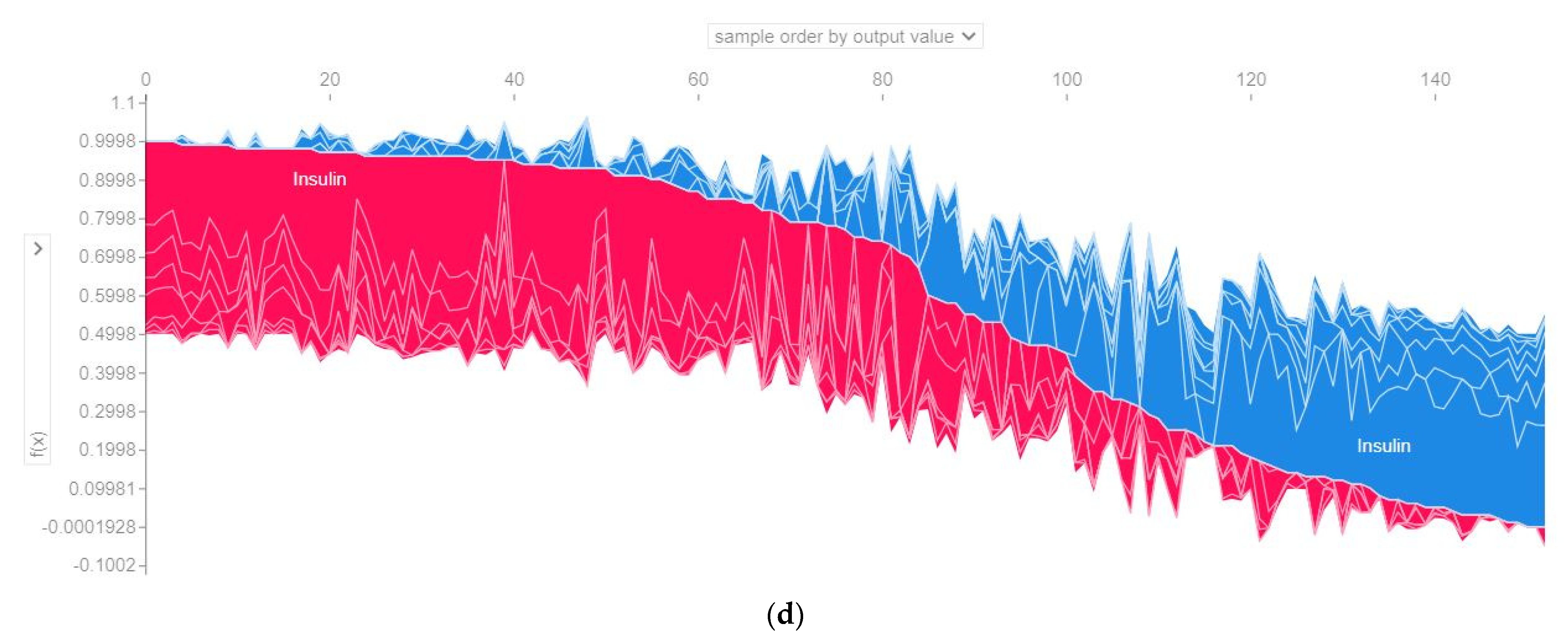
4.4.3. SHAP Force Plot of the Test Set (SHAP Supervised Clustering)
4.4.4. Permutation Importance of the Features (Global)
4.4.5. SHAP Summary Plot of the Violin Distribution
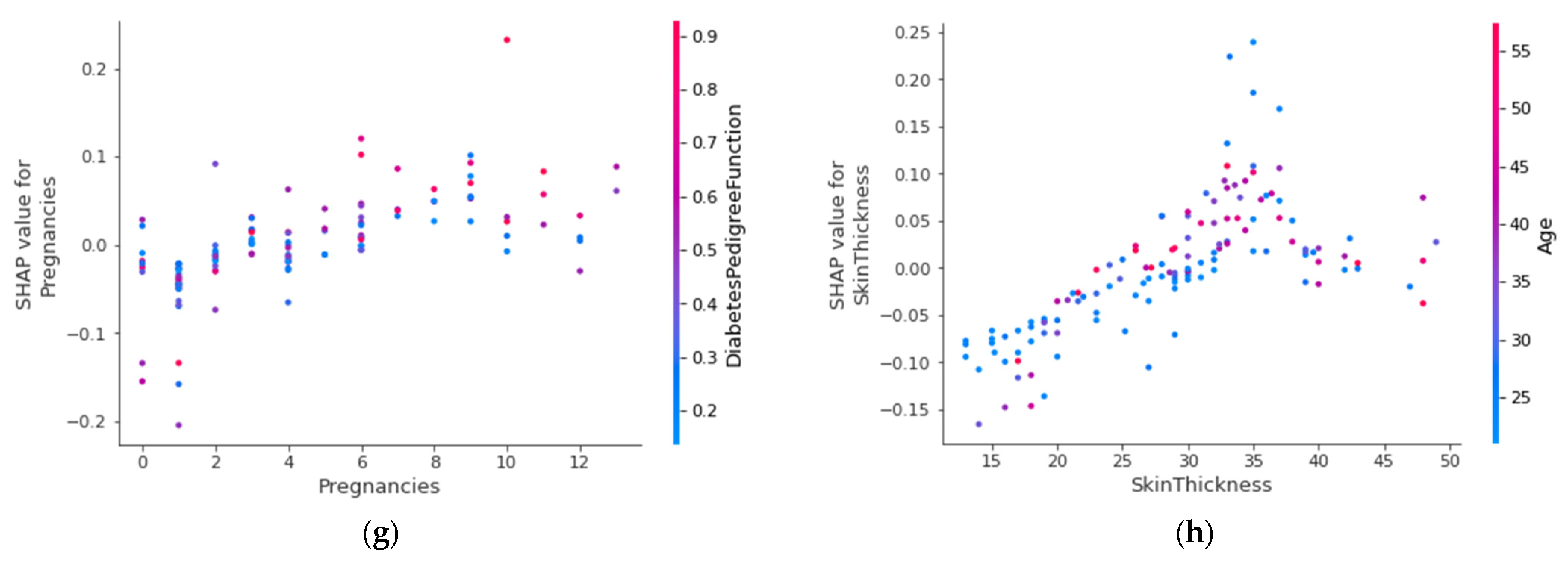
4.4.6. SHAP Dependence Plot (Global)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Target Class | Precision | Recall | F1-Score | AUC Score | Accuracy | Confusion Matrix | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | |||||||||
| Fold 1 | Class 0 | 0.83 | 0.79 | 0.81 | - | - | - | True label | Predicted label | |
| Class 1 | 0.64 | 0.70 | 0.67 | - | - | - | 79 | 21 | ||
| Average | 0.74 | 0.75 | 0.74 | 0.84 | 0.80 | 0.75 | 16 | 38 | ||
| Fold 2 | Class 0 | 0.84 | 0.74 | 0.79 | - | - | - | True label | Predicted label | |
| Class 1 | 0.61 | 0.74 | 0.67 | - | - | - | 74 | 26 | ||
| Average | 0.72 | 0.74 | 0.74 | 0.83 | 0.81 | 0.74 | 14 | 40 | ||
| Fold 3 | Class 0 | 0.85 | 0.85 | 0.85 | - | - | - | True label | Predicted label | |
| Class 1 | 0.72 | 0.72 | 0.72 | - | - | - | 85 | 15 | ||
| Average | 0.79 | 0.79 | 0.79 | 0.88 | 0.80 | 0.80 | 15 | 39 | ||
| Fold 4 | Class 0 | 0.88 | 0.87 | 0.87 | - | - | - | True label | Predicted label | |
| Class 1 | 0.76 | 0.77 | 0.77 | - | - | - | 87 | 13 | ||
| Average | 0.82 | 0.82 | 0.82 | 0.91 | 0.81 | 0.83 | 12 | 41 | ||
| Fold 5 | Class 0 | 0.91 | 0.81 | 0.86 | - | - | - | True label | Predicted label | |
| Class 1 | 0.70 | 0.85 | 0.77 | - | - | - | 81 | 19 | ||
| Average | 0.81 | 0.83 | 0.81 | 0.89 | 0.84 | 0.82 | 8 | 45 | ||
| All folds’ average | 0.77 | 0.78 | 0.78 | 0.87 | 0.81 | 0.79 | ||||
| Target Class | Precision | Recall | F1-Score | AUC Score | Accuracy | Confusion Matrix | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | |||||||||
| Fold 1 | Class 0 | 0.88 | 0.77 | 0.82 | - | - | - | True label | Predicted label | |
| Class 1 | 0.65 | 0.80 | 0.72 | - | - | - | 77 | 23 | ||
| Average | 0.76 | 0.78 | 0.77 | 0.85 | 0.88 | 0.78 | 11 | 43 | ||
| Fold 2 | Class 0 | 0.91 | 0.72 | 0.80 | - | - | - | True label | Predicted label | |
| Class 1 | 0.63 | 0.87 | 0.73 | - | - | - | 72 | 28 | ||
| Average | 0.77 | 0.80 | 0.77 | 0.87 | 0.87 | 0.72 | 7 | 47 | ||
| Fold 3 | Class 0 | 0.88 | 0.83 | 0.86 | - | - | - | True label | Predicted label | |
| Class 1 | 0.72 | 0.80 | 0.75 | - | - | - | 83 | 17 | ||
| Average | 0.80 | 0.81 | 0.81 | 0.90 | 0.87 | 0.81 | 11 | 43 | ||
| Fold 4 | Class 0 | 0.91 | 0.86 | 0.88 | - | - | - | True label | Predicted label | |
| Class 1 | 0.76 | 0.83 | 0.79 | - | - | - | 86 | 14 | ||
| Average | 0.83 | 0.85 | 0.85 | 0.89 | 0.86 | 0.85 | 9 | 44 | ||
| Fold 5 | Class 0 | 0.92 | 0.81 | 0.86 | - | - | - | True label | Predicted label | |
| Class 1 | 0.71 | 0.87 | 0.78 | - | - | - | 81 | 19 | ||
| Average | 0.81 | 0.84 | 0.82 | 0.90 | 0.86 | 0.83 | 7 | 46 | ||
| All folds’ average | 0.79 | 0.81 | 0.80 | 0.88 | 0.87 | 0.79 | ||||
| Target Class | Precision | Recall | F1-Score | AUC Score | Accuracy | Confusion Matrix | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | |||||||||
| Fold 1 | Class 0 | 0.86 | 0.77 | 0.81 | - | - | - | True label | Predicted label | |
| Class 1 | 0.64 | 0.76 | 0.69 | - | - | - | 77 | 23 | ||
| Average | 0.75 | 0.76 | 0.77 | 0.84 | 0.81 | 0.76 | 13 | 41 | ||
| Fold 2 | Class 0 | 0.88 | 0.76 | 0.82 | - | - | - | True label | Predicted label | |
| Class 1 | 0.65 | 0.81 | 0.72 | - | - | - | 76 | 24 | ||
| Average | 0.77 | 0.79 | 0.77 | 0.83 | 0.80 | 0.77 | 10 | 44 | ||
| Fold 3 | Class 0 | 0.86 | 0.83 | 0.85 | - | - | - | True label | Predicted label | |
| Class 1 | 0.71 | 0.76 | 0.73 | - | - | - | 83 | 17 | ||
| Average | 0.79 | 0.79 | 0.79 | 0.88 | 0.79 | 0.80 | 13 | 41 | ||
| Fold 4 | Class 0 | 0.91 | 0.84 | 0.87 | - | - | - | True label | Predicted label | |
| Class 1 | 0.74 | 0.85 | 0.79 | - | - | - | 84 | 16 | ||
| Average | 0.83 | 0.84 | 0.83 | 0.89 | 0.80 | 0.84 | 8 | 45 | ||
| Fold 5 | Class 0 | 0.87 | 0.77 | 0.81 | - | - | - | True label | Predicted label | |
| Class 1 | 0.64 | 0.77 | 0.70 | - | - | - | 77 | 23 | ||
| Average | 0.75 | 0.77 | 0.76 | 0.86 | 0.81 | 0.77 | 12 | 41 | ||
| All folds’ average | 0.78 | 0.79 | 0.78 | 0.86 | 0.80 | 0.78 | ||||
| Target Class | Precision | Recall | F1-Score | AUC Score | Accuracy | Confusion Matrix | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | |||||||||
| Fold 1 | Class 0 | 0.94 | 0.88 | 0.91 | - | - | - | True label | Predicted label | |
| Class 1 | 0.80 | 0.89 | 0.84 | - | - | - | 88 | 12 | ||
| Average | 0.87 | 0.88 | 0.87 | 0.94 | 1.00 | 0.88 | 6 | 48 | ||
| Fold 2 | Class 0 | 0.92 | 0.86 | 0.89 | - | - | - | True label | Predicted label | |
| Class 1 | 0.77 | 0.87 | 0.82 | - | - | - | 86 | 14 | ||
| Average | 0.85 | 0.87 | 0.85 | 0.93 | 1.00 | 0.86 | 7 | 47 | ||
| Fold 3 | Class 0 | 0.91 | 0.90 | 0.90 | - | - | - | True label | Predicted label | |
| Class 1 | 0.82 | 0.83 | 0.83 | - | - | - | 90 | 10 | ||
| Average | 0.86 | 0.87 | 0.87 | 0.94 | 1.00 | 0.87 | 9 | 45 | ||
| Fold 4 | Class 0 | 0.94 | 0.92 | 0.93 | - | - | - | True label | Predicted label | |
| Class 1 | 0.85 | 0.89 | 0.87 | - | - | - | 92 | 8 | ||
| Average | 0.90 | 0.90 | 0.90 | 0.95 | 1.00 | 0.91 | 6 | 47 | ||
| Fold 5 | Class 0 | 0.94 | 0.88 | 0.91 | - | - | - | True label | Predicted label | |
| Class 1 | 0.80 | 0.89 | 0.84 | - | - | - | 88 | 12 | ||
| Average | 0.87 | 0.88 | 0.87 | 0.95 | 1.00 | 0.88 | 6 | 47 | ||
| All folds’ average | 0.87 | 0.88 | 0.87 | 0.94 | 1.00 | 0.88 | ||||
| Target Class | Precision | Recall | F1-Score | AUC Score | Accuracy | Confusion Matrix | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | |||||||||
| Fold 1 | Class 0 | 0.92 | 0.90 | 0.91 | - | - | - | True label | Predicted label | |
| Class 1 | 0.82 | 0.85 | 0.84 | - | - | - | 90 | 10 | ||
| Average | 0.87 | 0.88 | 0.87 | 0.91 | 0.99 | 0.88 | 8 | 46 | ||
| Fold 2 | Class 0 | 0.93 | 0.86 | 0.90 | - | - | - | True label | Predicted label | |
| Class 1 | 0.77 | 0.89 | 0.83 | - | - | - | 86 | 14 | ||
| Average | 0.85 | 0.87 | 0.86 | 0.91 | 1.00 | 87 | 6 | 48 | ||
| Fold 3 | Class 0 | 0.94 | 0.90 | 0.92 | - | - | - | True label | Predicted label | |
| Class 1 | 0.83 | 0.89 | 0.86 | - | - | - | 90 | 10 | ||
| Average | 0.88 | 0.89 | 0.89 | 0.94 | 0.99 | 0.89 | 6 | 48 | ||
| Fold 4 | Class 0 | 0.94 | 0.91 | 0.92 | - | - | - | True label | Predicted label | |
| Class 1 | 0.84 | 0.89 | 0.86 | - | - | - | 91 | 9 | ||
| Average | 0.89 | 0.90 | 0.89 | 0.93 | 0.99 | 0.90 | 6 | 47 | ||
| Fold 5 | Class 0 | 0.93 | 0.93 | 0.93 | - | - | - | True label | Predicted label | |
| Class 1 | 0.87 | 0.87 | 0.87 | - | - | - | 93 | 7 | ||
| Average | 0.90 | 0.90 | 0.90 | 0.92 | 0.99 | 0.90 | 7 | 46 | ||
| All folds’ average | 0.88 | 0.89 | 0.88 | 0.92 | 0.99 | 0.88 | ||||
| Target Class | Precision | Recall | F1-Score | AUC Score | Accuracy | Confusion Matrix | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | |||||||||
| Fold 1 | Class 0 | 0.92 | 0.80 | 0.86 | - | - | - | True label | Predicted label | |
| Class 1 | 0.70 | 0.87 | 0.78 | - | - | - | 80 | 20 | ||
| Average | 0.81 | 0.84 | 0.82 | 0.95 | 0.86 | 0.82 | 7 | 47 | ||
| Fold 2 | Class 0 | 0.94 | 0.80 | 0.86 | - | - | - | True label | Predicted label | |
| Class 1 | 0.71 | 0.91 | 0.80 | - | - | - | 80 | 20 | ||
| Average | 0.83 | 0.85 | 0.83 | 0.95 | 0.87 | 0.83 | 5 | 49 | ||
| Fold 3 | Class 0 | 0.93 | 0.85 | 0.89 | - | - | - | True label | Predicted label | |
| Class 1 | 0.76 | 0.89 | 0.82 | - | - | - | 85 | 15 | ||
| Average | 0.85 | 0.87 | 0.86 | 0.95 | 0.87 | 0.86 | 6 | 48 | ||
| Fold 4 | Class 0 | 0.94 | 0.82 | 0.88 | - | - | - | True label | Predicted label | |
| Class 1 | 0.73 | 0.91 | 0.81 | - | - | - | 82 | 18 | ||
| Average | 0.83 | 0.86 | 0.84 | 0.95 | 0.88 | 0.85 | 5 | 48 | ||
| Fold 5 | Class 0 | 0.94 | 0.76 | 0.84 | - | - | - | True label | Predicted label | |
| Class 1 | 0.67 | 0.91 | 0.77 | - | - | - | 76 | 24 | ||
| Average | 0.80 | 0.83 | 0.80 | 0.96 | 0.86 | 0.81 | 5 | 48 | ||
| All folds’ average | 0.82 | 0.85 | 0.83 | 0.95 | 0.86 | 0.83 | ||||
| Target Class | Precision | Recall | F1-Score | AUC Score | Taken Weight | Accuracy | Confusion Matrix | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Train | Test | ||||||||||
| Fold 1 | Class 0 | 0.93 | 0.91 | 0.92 | - | 4 (Xgb) 3 (RF) | - | - | True label | Predicted label | |
| Class 1 | 0.84 | 0.87 | 0.85 | - | - | - | 91 | 9 | |||
| Average | 0.88 | 0.89 | 0.89 | 0.94 | .99 | 0.90 | 7 | 47 | |||
| Fold 2 | Class 0 | 0.93 | 0.87 | 0.90 | - | 1 2 | - | - | True label | Predicted label | |
| Class 1 | 0.78 | 0.87 | 0.82 | - | - | - | 87 | 13 | |||
| Average | 0.85 | 0.87 | 0.86 | 0.94 | 1.00 | 0.87 | 7 | 47 | |||
| Fold 3 | Class 0 | 0.91 | 0.91 | 0.91 | - | 1 1 | - | - | True label | Predicted label | |
| Class 1 | 0.83 | 0.83 | 0.83 | - | - | - | 91 | 9 | |||
| Average | 0.87 | 0.87 | 0.87 | 0.95 | 1.00 | 0.88 | 9 | 45 | |||
| Fold 4 | Class 0 | 0.96 | 0.91 | 0.93 | - | 1 4 | - | - | True label | Predicted label | |
| Class 1 | 0.84 | 0.92 | 0.88 | - | - | - | 91 | 9 | |||
| Average | 0.90 | 0.92 | 0.91 | 0.96 | 1.00 | 0.91 | 4 | 49 | |||
| Fold 5 | Class 0 | 0.95 | 0.94 | 0.94 | - | 2 2 | - | - | True label | Predicted label | |
| Class 1 | 0.89 | 0.91 | 0.90 | - | - | - | 94 | 6 | |||
| Average | 0.92 | 0.92 | 0.92 | 0.96 | 1.00 | 0.92 | 5 | 48 | |||
| All folds’ average | 0.88 | 0.89 | 0.89 | 0.95 | 0.99 | 0.90 | |||||
References
- Chatterjee, S.; Khunti, K.; Davies, M.J. Type 2 diabetes. Lancet 2017, 389, 2239–2251. [Google Scholar] [CrossRef]
- Alam, T.M.; Iqbal, M.A.; Ali, Y.; Wahab, A.; Ijaz, S.; Baig, T.I.; Hussain, A.; Malik, M.A.; Raza, M.M.; Ibrar, S.; et al. A model for early prediction of diabetes. Inform. Med. Unlocked 2019, 16, 100204. [Google Scholar] [CrossRef]
- Islam, M.M.F.; Ferdousi, R.; Rahman, S.; Bushra, H.Y. Likelihood Prediction of Diabetes at Early Stage Using Data Mining Techniques. In Computer Vision and Machine Intelligence in Medical Image Analysis; Springer: Singapore, 2019; pp. 113–125. [Google Scholar] [CrossRef]
- Wild, S.; Roglic, G.; Green, A.; Sicree, R.; King, H. Global Prevalence of Diabetes. Diabetes Care 2004, 27, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Rubino, F. Is Type 2 Diabetes an Operable Intestinal Disease? Diabetes Care 2008, 31, S290–S296. [Google Scholar] [CrossRef] [PubMed]
- Kibria, H.B.; Matin, A.; Jahan, N.; Islam, S. A Comparative Study with Different Machine Learning Algorithms for Diabetes Disease Prediction. In Proceedings of the 2021 18th International Conference on Electrical Engineering, Computing Science and Automatic Control (CCE), Mexico City, Mexico, 10–12 November 2021; pp. 1–8. [Google Scholar] [CrossRef]
- Kibria, H.B.; Matin, A. The severity prediction of the binary and multi-class cardiovascular disease − A machine learning-based fusion approach. Comput. Biol. Chem. 2022, 98, 107672. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthi, R.; Joshi, S.; Almarzouki, H.Z.; Shukla, P.K.; Rizwan, A.; Kalpana, C.; Tiwari, B. A Novel Diabetes Healthcare Disease Prediction Framework Using Machine Learning Techniques. J. Health Eng. 2022, 2022, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kibria, H.B.; Jyoti, O.; Matin, A. Forecasting the spread of the third wave of COVID-19 pandemic using time series analysis in Bangladesh. Inform. Med. Unlocked 2021, 28, 100815. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Kumar, D.; Mittal, M. An ensemble approach for classification and prediction of diabetes mellitus using soft voting classifier. Int. J. Cogn. Comput. Eng. 2021, 2, 40–46. [Google Scholar] [CrossRef]
- Sisodia, D.; Sisodia, D.S. Prediction of Diabetes using Classification Algorithms. Procedia Comput. Sci. 2018, 132, 1578–1585. [Google Scholar] [CrossRef]
- Tiwari, P.; Singh, V. Diabetes disease prediction using significant attribute selection and classification approach. J. Phys. Conf. Ser. 2021, 1714, 012013. [Google Scholar] [CrossRef]
- Chang, V.; Bailey, J.; Xu, Q.A.; Sun, Z. Pima Indians diabetes mellitus classification based on machine learning (ML) algorithms. Neural Comput. Appl. 2022, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Chen, S.; Zhang, H.; Wu, T. A hybrid prediction model for type 2 diabetes using K-means and decision tree. In Proceedings of the 2017 8th IEEE International Conference on Software Engineering and Service Science (ICSESS), Beijing, China, 24–26 November 2017; pp. 386–390. [Google Scholar] [CrossRef]
- Mir, A.; Dhage, S.N. Diabetes Disease Prediction Using Machine Learning on Big Data of Healthcare. In Proceedings of the 2018 Fourth International Conference on Computing Communication Control and Automation (ICCUBEA), Pune, India, 16–18 August 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Sangien, T.; Bhat, T.; Khan, M.S. Diabetes Disease Prediction Using Classification Algorithms. In Internet of Things and Its Applications; Springer: Singapore, 2022; pp. 185–197. [Google Scholar] [CrossRef]
- Ramesh, J.; Aburukba, R.; Sagahyroon, A. A remote healthcare monitoring framework for diabetes prediction using machine learning. Health Technol. Lett. 2021, 8, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, U.; Issa, G.F.; Khan, M.A.; Aftab, S.; Said, R.A.T.; Ghazal, T.M.; Ahmad, M. Prediction of Diabetes Empowered With Fused Machine Learning. IEEE Access 2022, 10, 8529–8538. [Google Scholar] [CrossRef]
- Abdollahi, J.; Nouri-Moghaddam, B. Hybrid stacked ensemble combined with genetic algorithms for diabetes prediction. Iran J. Comput. Sci. 2022, 1–16. [Google Scholar] [CrossRef]
- Fitriyani, N.L.; Syafrudin, M.; Alfian, G.; Rhee, J. Development of Disease Prediction Model Based on Ensemble Learning Approach for Diabetes and Hypertension. IEEE Access 2019, 7, 144777–144789. [Google Scholar] [CrossRef]
- Kibria, H.B.; Matin, A. An Efficient Machine Learning-Based Decision-Level Fusion Model to Predict Cardiovascular Disease. In International Conference on Intelligent Computing & Optimization; Springer: Berlin/Heidelberg, Germany, 2021; pp. 1097–1110. [Google Scholar] [CrossRef]
- Santos, M.S.; Soares, J.P.; Abreu, P.H.; Araujo, H.; Santos, J. Cross-Validation for Imbalanced Datasets: Avoiding Overoptimistic and Overfitting Approaches [Research Frontier]. IEEE Comput. Intell. Mag. 2018, 13, 59–76. [Google Scholar] [CrossRef]
- Bucholc, M.; Ding, X.; Wang, H.; Glass, D.H.; Wang, H.; Prasad, G.; Maguire, L.P.; Bjourson, A.J.; McClean, P.L.; Todd, S.; et al. A practical computerized decision support system for predicting the severity of Alzheimer’s disease of an individual. Expert Syst. Appl. 2019, 130, 157–171. [Google Scholar] [CrossRef]
- Das, D.; Ito, J.; Kadowaki, T.; Tsuda, K. An interpretable machine learning model for diagnosis of Alzheimer’s disease. PeerJ 2019, 7, e6543. [Google Scholar] [CrossRef]
- Burrell, J. How the machine ‘thinks’: Understanding opacity in machine learning algorithms. Big Data Soc. 2016, 3. [Google Scholar] [CrossRef]
- Adadi, A.; Berrada, M. Peeking Inside the Black-Box: A Survey on Explainable Artificial Intelligence (XAI). IEEE Access 2018, 6, 52138–52160. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Nair, B.; Vavilala, M.S.; Horibe, M.; Eisses, M.J.; Adams, T.; Liston, D.E.; Low, D.K.-W.; Newman, S.-F.; Kim, J.; et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat. Biomed. Eng. 2018, 2, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, R.; Monreale, A.; Ruggieri, S.; Turini, F.; Giannotti, F.; Pedreschi, D. A Survey of Methods for Explaining Black Box Models. ACM Comput. Surv. 2019, 51, 1–42. [Google Scholar] [CrossRef]
- Tephen, S.; Ranks, F. Polycystic Ovary Syndrome. N. Engl. J. Med. 1995, 333, 853–861. [Google Scholar] [CrossRef]
- Saxena, P.; Prakash, A.; Nigam, A. Efficacy of 2-hour post glucose insulin levels in predicting insulin resistance in polycystic ovarian syndrome with infertility. J. Hum. Reprod. Sci. 2011, 4, 20. [Google Scholar] [CrossRef]
- García, S.; Ramírez-Gallego, S.; Luengo, J.; Benítez, J.M.; Herrera, F. Big data preprocessing: Methods and prospects. Big Data Anal. 2016, 1, 9. [Google Scholar] [CrossRef]
- Batista, G.E.A.P.A.; Prati, R.C.; Monard, M.C. A study of the behavior of several methods for balancing machine learning training data. ACM SIGKDD Explor. Newsl. 2004, 6, 20–29. [Google Scholar] [CrossRef]
- Li, L.; Hu, Q.; Wu, X.; Yu, D. Exploration of classification confidence in ensemble learning. Pattern Recognit. 2014, 47, 3120–3131. [Google Scholar] [CrossRef]
- Kibria, H.B.; Matin, A.; Islam, S. Comparative Analysis of Two Artificial Intelligence Based Decision Level Fusion Models for Heart Disease Prediction. 2020. Available online: http://ceur-ws.org (accessed on 1 July 2022).
- Agatonovic-Kustrin, S.; Beresford, R. Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J. Pharm. Biomed. Anal. 2000, 22, 717–727. [Google Scholar] [CrossRef]
- Hart, S. Shapley Value. In Game Theory; Palgrave Macmillan: London, UK, 1989; pp. 210–216. [Google Scholar] [CrossRef]
- Deegan, J.; Packel, E.W. A new index of power for simplen-person games. Int. J. Game Theory 1978, 7, 113–123. [Google Scholar] [CrossRef]
- El-Sappagh, S.; Alonso, J.M.; Islam, S.M.R.; Sultan, A.M.; Kwak, K.S. A multilayer multimodal detection and prediction model based on explainable artificial intelligence for Alzheimer’s disease. Sci. Rep. 2021, 11, 2660. [Google Scholar] [CrossRef]
- Fryer, D.; Strumke, I.; Nguyen, H. Shapley values for feature selection: The good, the bad, and the axioms. IEEE Access 2021, 9, 144352–144360. [Google Scholar] [CrossRef]
- Sundararajan, M.; Najmi, A. The Many Shapley Values for Model Explanation. PMLR; pp. 9269–9278, 21 November 2020. Available online: https://proceedings.mlr.press/v119/sundararajan20b.html (accessed on 28 August 2022).
- An Introduction to Explainable AI with Shapley Values—SHAP Latest Documentation. Available online: https://shap.readthedocs.io/en/latest/example_notebooks/overviews/An%20introduction%20to%20explainable%20AI%20with%20Shapley%20values.html (accessed on 20 September 2022).
- Gupta, S.; Sandhu, S.V.; Bansal, H.; Sharma, D. Comparison of salivary and serum glucose levels in diabetic patients. J. Diabetes Sci. Technol. 2015, 9, 91–96. [Google Scholar] [CrossRef] [PubMed]




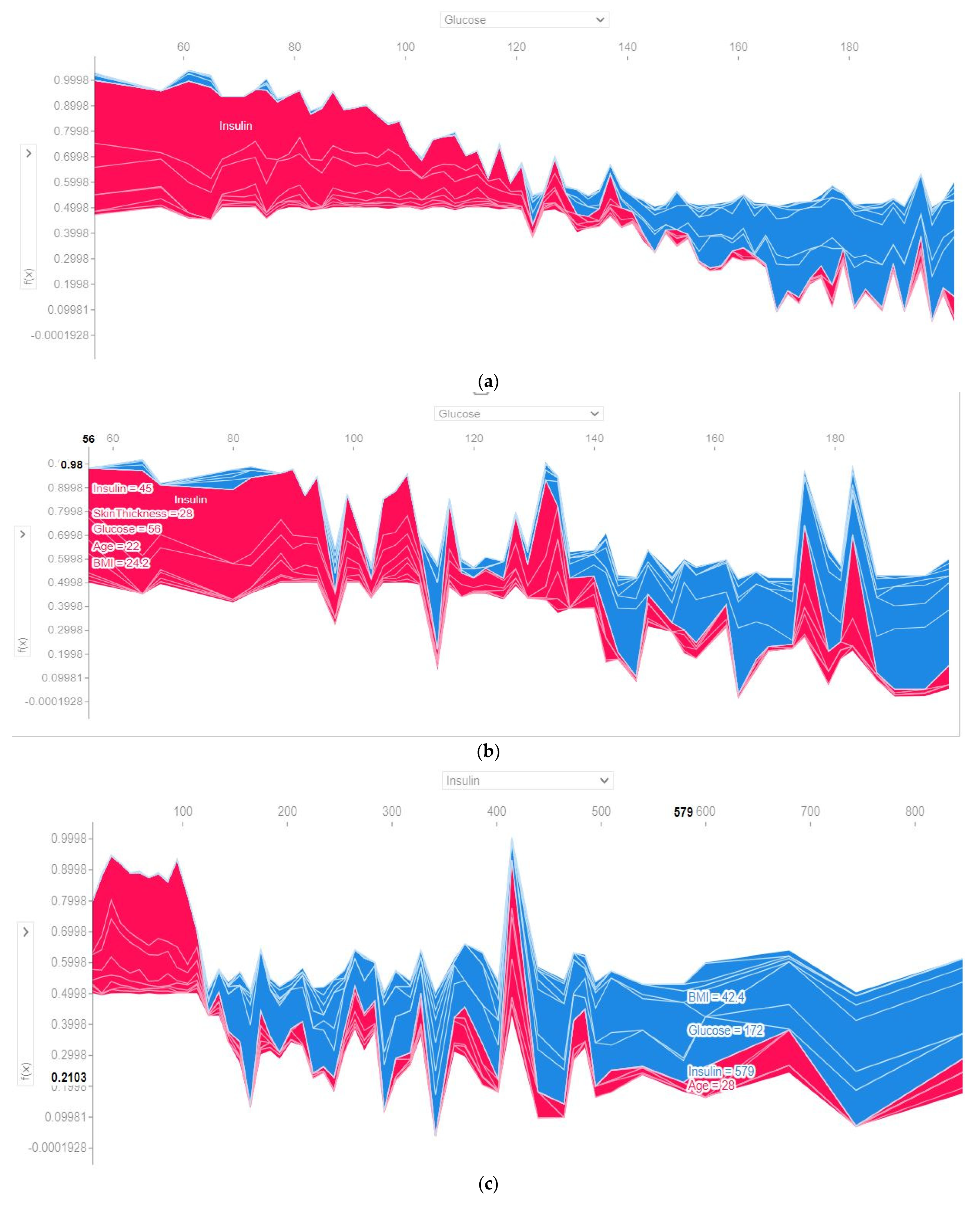
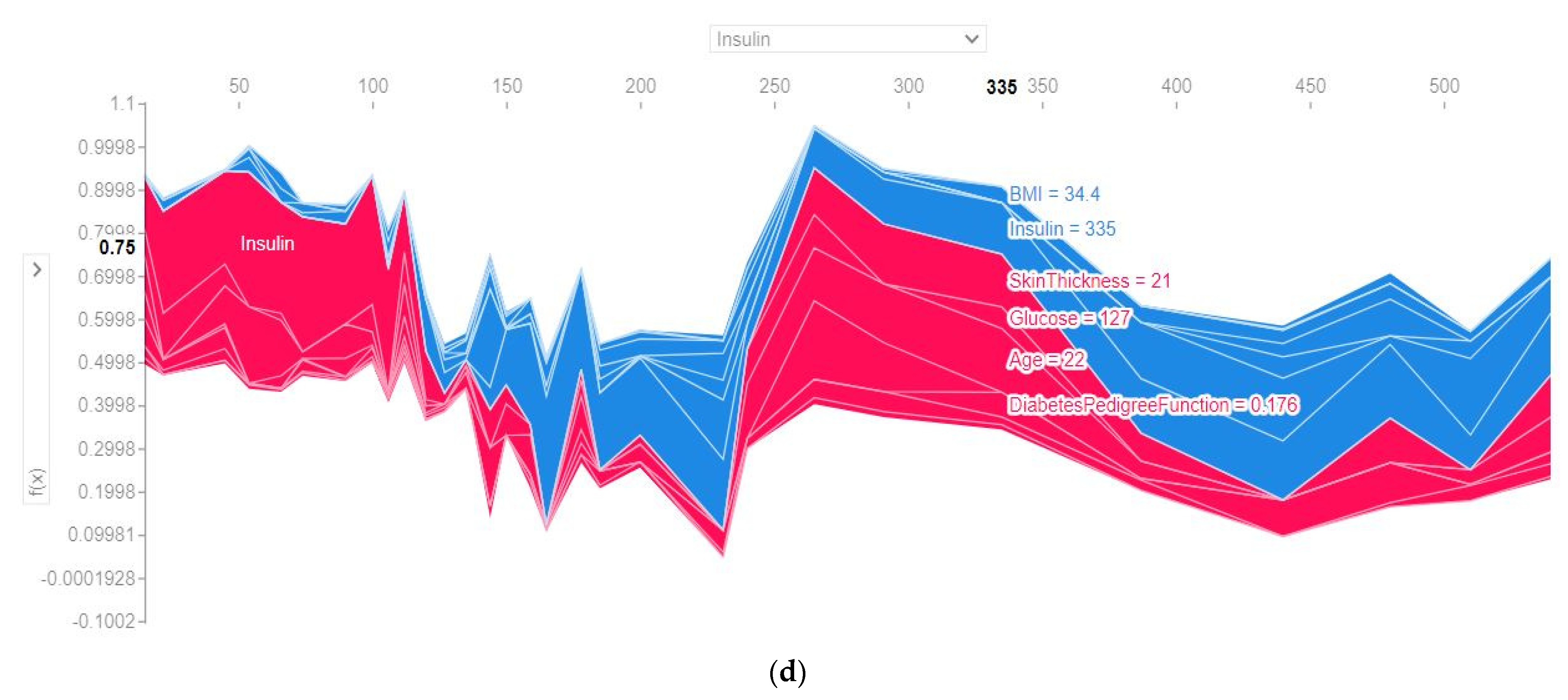

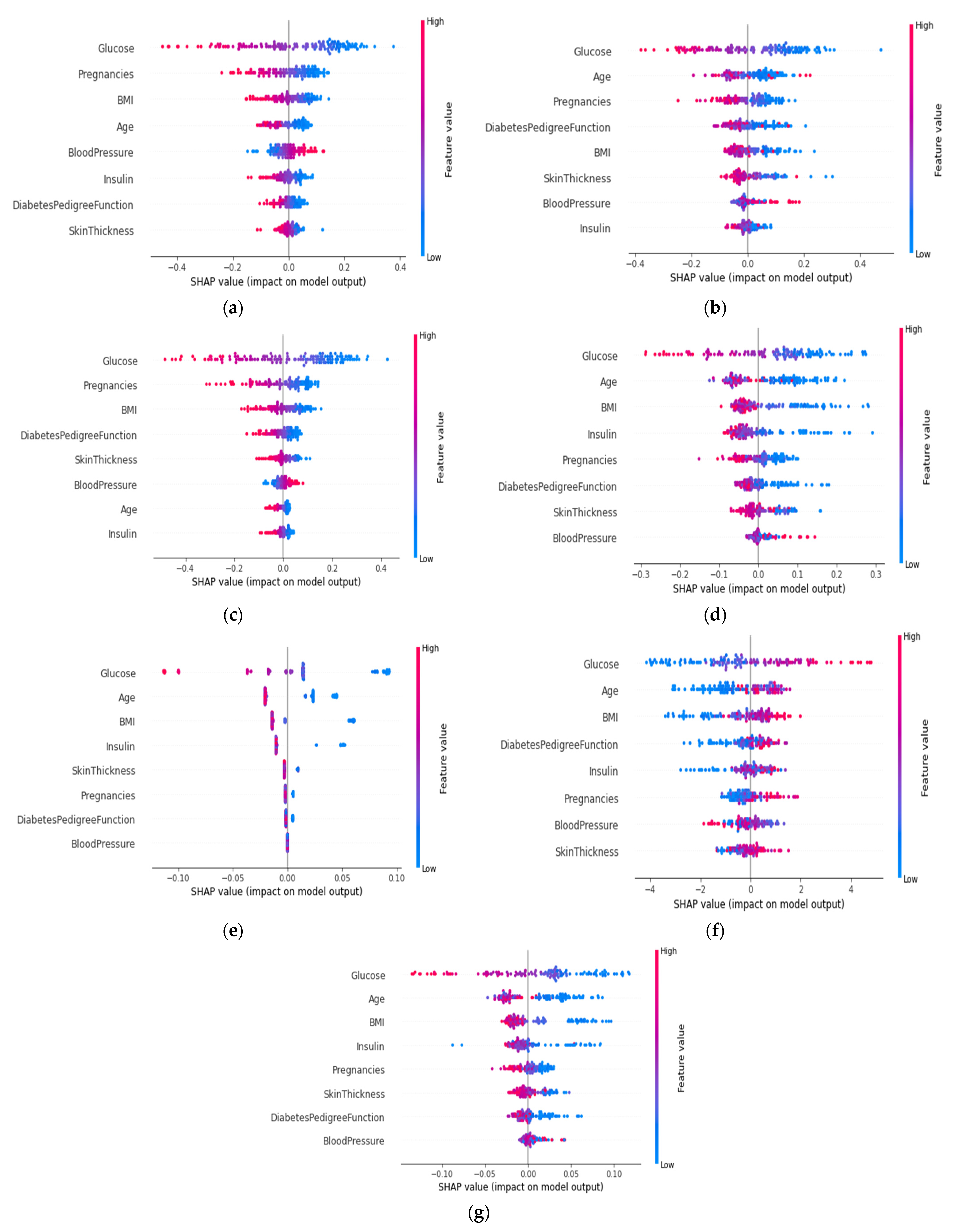

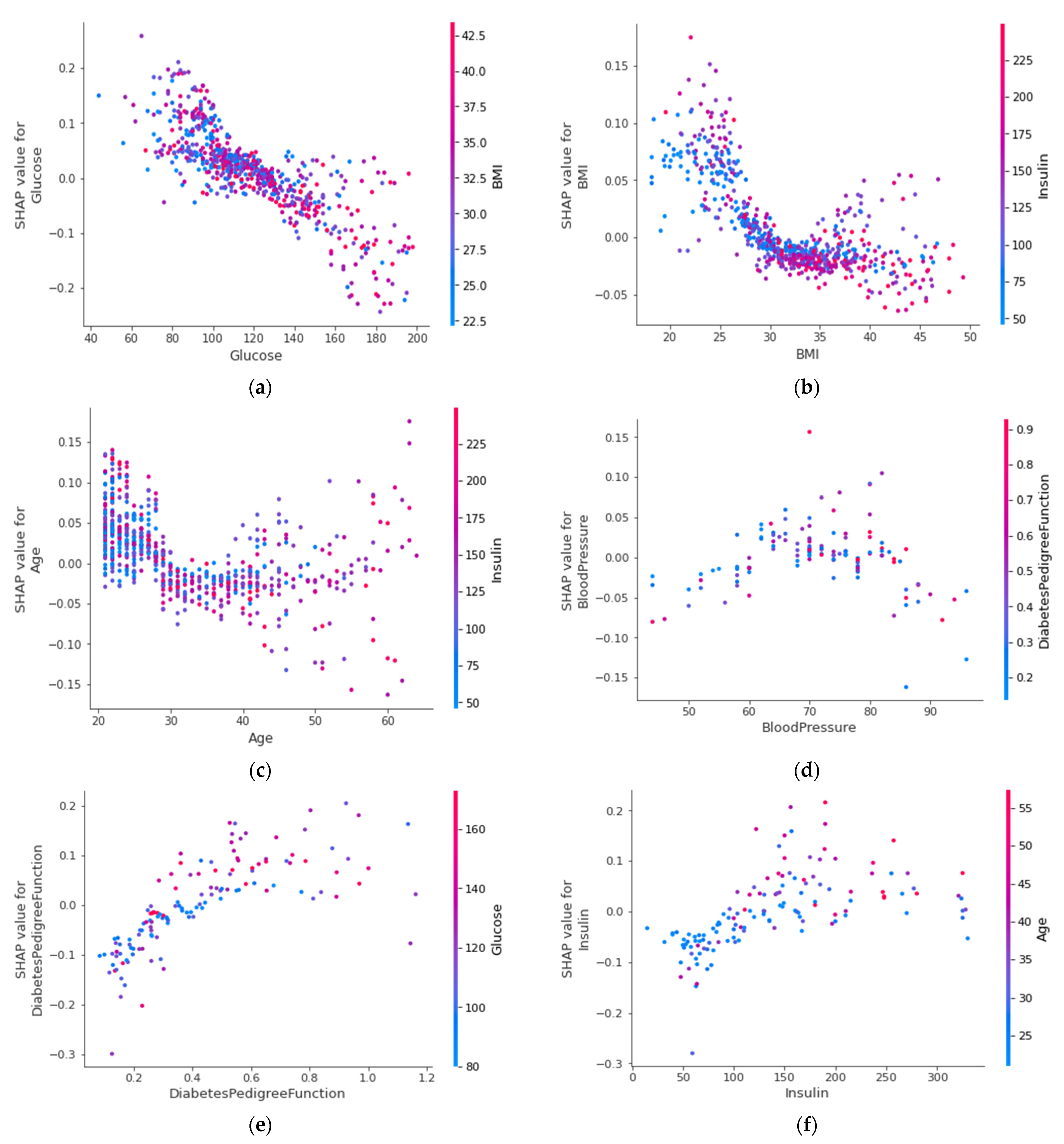
| Attribute | Attribute Type | Attribute Description |
|---|---|---|
| Pregnancies | Numeric | Number of times pregnant |
| Glucose | Numeric | Plasma glucose concentration (mmol/L) a 2 h in an oral glucose tolerance test |
| Blood Pressure | Numeric | Diastolic blood pressure (mm Hg) |
| Skin Thickness | Numeric | Triceps skin fold thickness (mm) |
| Insulin | Numeric | 2 h serum insulin (mu U/mL): Insulin-resistant (IR) cells lead to prediabetes and type-2 diabetes. “2 h post glucose insulin level” is a cost-effective, convenient, and efficient indicator to diagnose IR [29,30] |
| BMI | Numeric | Body mass index weight in kg/(height in m) |
| Diabetes PF | Numeric | Diabetes pedigree function: indicates the function which measures the chance of diabetes based on family history. |
| Age | Numeric | Age (years) |
| Pregnancies | Glucose | Blood Pressure | Skin Thickness | Insulin | BMI | Diabetes- Pedigree Function | Age | Outcome | |
|---|---|---|---|---|---|---|---|---|---|
| count | 768 | 768 | 768 | 768 | 768 | 768 | 768 | 768 | 768 |
| mean | 3.84 | 121.59 | 72.37 | 29.11 | 153.18 | 32.42 | 0.47 | 33.24 | 0.34 |
| std | 3.36 | 30.49 | 12.2 | 9.42 | 98.38 | 6.88 | 0.33 | 11.76 | 0.47 |
| min | 0 | 44 | 24 | 7 | 14 | 18.2 | 0.07 | 21 | 0 |
| 25% | 1 | 99 | 64 | 23 | 87.9 | 27.5 | 0.24 | 24 | 0 |
| 50% | 3 | 117 | 72 | 29 | 133.7 | 32.09 | 0.37 | 29 | 0 |
| 75% | 6 | 140.25 | 80 | 35 | 190.15 | 36.6 | 0.62 | 41 | 1 |
| max | 17 | 199 | 122 | 99 | 846 | 67.1 | 2.42 | 81 | 1 |
| Before the SMOTETomek | After the SMOTETomek | |
|---|---|---|
| Numbers in class 0 (non-diabetic) | 400 | 393 |
| Numbers in class 1 (diabetic) | 214 | 393 |
| Algorithms | Optimal Parameters |
|---|---|
| Artificial neural network | Batch size = 5, epochs = 20 |
| Support vector machine | default |
| Logistic regression | C = 10 |
| Random forest | default |
| XGBoost | number of estimators = 20 |
| AdaBoost | number of estimators = 300, learning rate = 0.01 |
| Algorithms | Precision | Recall | F1-Score | AUC Score | Accuracy | |
|---|---|---|---|---|---|---|
| Train | Test | |||||
| ANN | 0.77 | 0.78 | 0.78 | 0.87 | 0.81 | 0.79 |
| SVM | 0.79 | 0.81 | 0.80 | 0.88 | 0.87 | 0.79 |
| LR | 0.78 | 0.79 | 0.78 | 0.86 | 0.80 | 0.78 |
| RF | 0.87 | 0.88 | 0.87 | 0.94 | 1.00 | 0.88 |
| XGB | 0.88 | 0.89 | 0.88 | 0.92 | 0.99 | 0.88 |
| Ada | 0.82 | 0.85 | 0.83 | 0.95 | 0.86 | 0.83 |
| Voting Classifier (XGB + RF) | 0.88 | 0.89 | 0.89 | 0.95 | 0.99 | 0.90 |
| Approach | Train Test Split | Result (%) | Ref. | |||
|---|---|---|---|---|---|---|
| Decision tree Random forest Naive Bayes | 70:30 train test ratio | DT | RF | NB | [13] | |
| Accuracy Precision Sensitivity Specificity F1 score AUC | 74.78 70.86 88.43 59.63 78.68 78.55 | 79.57 89.40 81.33 75.00 85.17 86.24 | 78.67 81.88 86.75 63.29 84.24 84.63 | |||
| RF AdaBoost Soft voting classifier | 70:30 train test ratio | RF | Ada | Voting classifier | [10] | |
| Accuracy Precision F1 score Recall AUC | 77.48 71.21 64.38 58.75 78.10 | 75.32 68.25 60.13 53.75 74.98 | 79.08 73.13 71.56 70.00 80.98 | |||
| RF | Not mentioned | RF | ANN | K mean clustering | [2] | |
| Accuracy AUC | 74.70 80.60 | 75.70 81.60 | 73.60 - | |||
| ANN XGB | Not mentioned | ANN | XGB | [12] | ||
| Accuracy Sensitivity Specificity AUC | 71.35 45.22 85.20 65.00 | 78.91 59.33 89.40 88.00 | ||||
| Naive Bayes SVM DT | 10-fold Cross-validation | NB | SVM | DT | [11] | |
| Precision Recall F1 score Accuracy | 75.9 76.3 76 76.3 | 42.4 65.1 51.3 65.1 | 73.50 73.80 73.60 73.82 | |||
| Proposed soft voting classifier (XgBoost + RF) | 5 fold Cross-validation | Accuracy Precision Recall F1 score AUC | 90 88 89 95 95 | - | ||
| Features | Values (Natural Unit) |
|---|---|
| Glucose | 109.00 |
| Blood pressure | 88.00 |
| Insulin | 142.80 |
| Skin thickness | 30.00 |
| Pregnancies | 6.40 |
| BMI | 32.50 |
| Diabetes pedigree function | 0.85 |
| Age | 38.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kibria, H.B.; Nahiduzzaman, M.; Goni, M.O.F.; Ahsan, M.; Haider, J. An Ensemble Approach for the Prediction of Diabetes Mellitus Using a Soft Voting Classifier with an Explainable AI. Sensors 2022, 22, 7268. https://doi.org/10.3390/s22197268
Kibria HB, Nahiduzzaman M, Goni MOF, Ahsan M, Haider J. An Ensemble Approach for the Prediction of Diabetes Mellitus Using a Soft Voting Classifier with an Explainable AI. Sensors. 2022; 22(19):7268. https://doi.org/10.3390/s22197268
Chicago/Turabian StyleKibria, Hafsa Binte, Md Nahiduzzaman, Md. Omaer Faruq Goni, Mominul Ahsan, and Julfikar Haider. 2022. "An Ensemble Approach for the Prediction of Diabetes Mellitus Using a Soft Voting Classifier with an Explainable AI" Sensors 22, no. 19: 7268. https://doi.org/10.3390/s22197268
APA StyleKibria, H. B., Nahiduzzaman, M., Goni, M. O. F., Ahsan, M., & Haider, J. (2022). An Ensemble Approach for the Prediction of Diabetes Mellitus Using a Soft Voting Classifier with an Explainable AI. Sensors, 22(19), 7268. https://doi.org/10.3390/s22197268












