A Rapid Antigen Detection Test to Diagnose SARS-CoV-2 Infection Using Exhaled Breath Condensate by A Modified Inflammacheck® Device
Abstract
:1. Introduction
2. Materials and Methods
2.1. RT-PCR Swabs
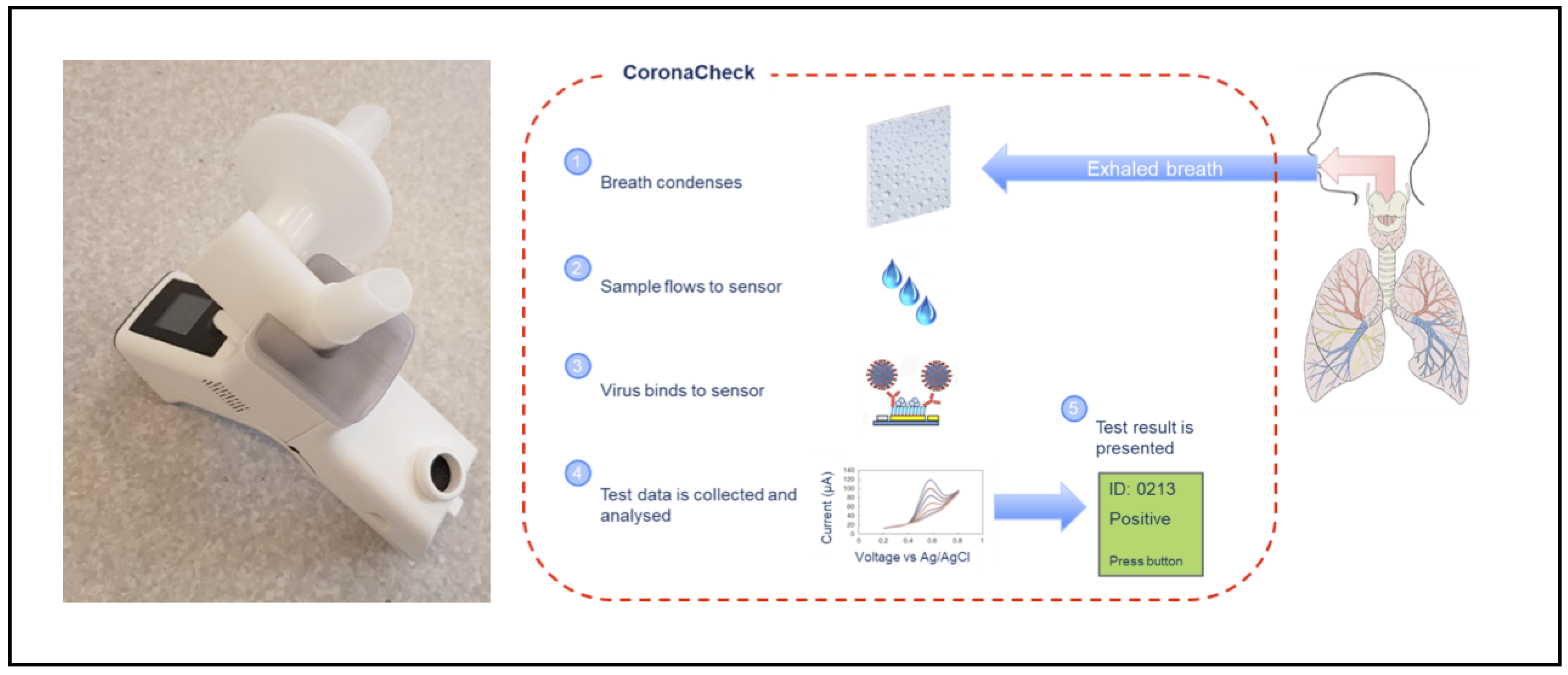
2.2. Inflammacheck®
2.3. Statistics
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, H.; Wei, L.; Niu, P. The novel coronavirus outbreak in Wuhan, China. Glob. Health Res. Policy 2020, 5, 6. [Google Scholar] [CrossRef] [Green Version]
- Lovato, E.C.W.; Barboza, L.N.; Wietzikoski, S.; de Souza, A.N.V.; Auth, P.A.; Junior, A.G.; Dos Reis Livero, F.A. Repurposing Drugs for the Management of Patients with Confirmed Coronavirus Disease 2019 (COVID-19). Curr. Pharm. Des. 2021, 27, 115–126. [Google Scholar] [CrossRef]
- Lauer, S.A.; Grantz, K.H.; Bi, Q.; Jones, F.K.; Zheng, Q.; Meredith, H.R.; Azman, A.S.; Reich, N.G.; Lessler, J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann. Intern. Med. 2020, 172, 577–582. [Google Scholar] [CrossRef] [Green Version]
- CDC. Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons for Coronavirus Disease 2019—COVID-19. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html (accessed on August 23, 2021).
- Vijayan, T.; Klausner, J.D. Hepatitis C: Challenges and opportunities in the laboratory diagnosis of infection. MLO Med. Lab. Obs. 2016, 48, 18. [Google Scholar]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58, e00512-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhimraj, A.; Morgan, R.L.; Shumaker, A.H.; Lavergne, V.; Baden, L.; Cheng, V.C.; Edwards, K.M.; Gandhi, R.; Muller, W.J.; O’Horo, J.C.; et al. Infectious Diseases Society of America Guidelines on the Treatment and Management of Patients with COVID-19. Clin. Infect. Dis. 2020, ciaa478. [Google Scholar] [CrossRef] [PubMed]
- Sawano, M.; Takeshita, K.; Ohno, H.; Oka, H. RT-PCR diagnosis of COVID-19 from exhaled breath condensate: A clinical study. J. Breath Res. 2021, 15, 037103. [Google Scholar] [CrossRef] [PubMed]
- Khoubnasabjafari, M.; Jouyban-Gharamaleki, V.; Ghanbari, R.; Jouyban, A. Exhaled breath condensate as a potential specimen for diagnosing COVID-19. Bioanalysis 2020, 12, 1195–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef] [Green Version]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Med. 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Arevalo-Rodriguez, I.; Buitrago-Garcia, D.; Simancas-Racines, D.; Zambrano-Achig, P.; Del Campo, R.; Ciapponi, A.; Sued, O.; Martinez-Garcia, L.; Rutjes, A.W.; Low, N.; et al. False-negative results of initial RT-PCR assays for COVID-19: A systematic review. PLoS ONE 2020, 15, e0242958. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, M.; Iacovelli, A.; Olmati, F.; Menichini, I.; Oliva, A.; Carnevalini, M.; Graziani, E.; Mastroianni, C.M.; Palange, P. False-negative RT-PCR in SARS-CoV-2 disease: Experience from an Italian COVID-19 unit. ERJ Open Res. 2020, 6, 00324-2020. [Google Scholar] [CrossRef] [PubMed]
- Artesi, M.; Bontems, S.; Gobbels, P.; Franckh, M.; Maes, P.; Boreux, R.; Meex, C.; Melin, P.; Hayette, M.P.; Bours, V.; et al. A Recurrent Mutation at Position 26340 of SARS-CoV-2 Is Associated with Failure of the E Gene Quantitative Reverse Transcription-PCR Utilized in a Commercial Dual-Target Diagnostic Assay. J. Clin. Microbiol. 2020, 58, e01598-20. [Google Scholar] [CrossRef]
- He, X.; Lau, E.H.Y.; Wu, P.; Deng, X.; Wang, J.; Hao, X.; Lau, Y.C.; Wong, J.Y.; Guan, Y.; Tan, X.; et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020, 26, 672–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Muller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, D.J.; Toomey, S.; Madden, S.F.; Casey, M.; Breathnach, O.S.; Morris, P.G.; Grogan, L.; Branagan, P.; Costello, R.W.; De Barra, E.; et al. Use of exhaled breath condensate (EBC) in the diagnosis of SARS-COV-2 (COVID-19). Thorax 2021, 76, 86–88. [Google Scholar] [CrossRef]
- Sethuraman, N.; Jeremiah, S.S.; Ryo, A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA 2020, 323, 2249–2251. [Google Scholar] [CrossRef]
- Sohn, Y.; Jeong, S.J.; Chung, W.S.; Hyun, J.H.; Baek, Y.J.; Cho, Y.; Kim, J.H.; Ahn, J.Y.; Choi, J.Y.; Yeom, J.S. Assessing Viral Shedding and Infectivity of Asymptomatic or Mildly Symptomatic Patients with COVID-19 in a Later Phase. J. Clin. Med. 2020, 9, 2924. [Google Scholar] [CrossRef]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambrosino, P.; Papa, A.; Maniscalco, M.; Di Minno, M.N.D. COVID-19 and functional disability: Current insights and rehabilitation strategies. Postgrad. Med. J. 2021, 97, 469–470. [Google Scholar] [CrossRef] [PubMed]
- Struble-Fitzsimmons, D.; Feld-Glazman, R.; Dominick, E.; Alexandrou, S.; Rider, E.; Bogosian, C.; Norton, J.; Pacheco, L.; Andreassi, E. A Retrospective Quality Improvement Study to Describe Operational Management Strategies in an Inpatient Rehabilitation Facility during the COVID-19 Pandemic. Arch. Phys. Med. Rehabil. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, Z.; Zhou, Y.; Onoda, K.; Maruyama, H.; Hu, C.; Liu, Z. Summary of respiratory rehabilitation and physical therapy guidelines for patients with COVID-19 based on recommendations of World Confederation for Physical Therapy and National Association of Physical Therapy. J. Phys. Ther. Sci. 2020, 32, 545–549. [Google Scholar] [CrossRef] [PubMed]

| Total Number of Valid RT-PCR Tests | 105 |
|---|---|
| Positive RT-PCR, n (%) | 13 (12.4) |
| Age (years) | 58.4 ± 14.4 |
| Female gender (%) | 41 (39.0) |
| Clinically suspected COVID-19, n (%) | 21 (20.0) |
| Convalescent COVID-19, n (%) | 20 (19.0) |
| Asymptomatic with high clinical probability, n (%) | 14 (13.3) |
| Asymptomatic with low clinical probability, n (%) | 50 (47.6) |
| RT-PCR | Inflammacheck® Device | ||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 12 | 1 | 13 |
| Negative | 1 | 91 | 92 |
| Total | 13 | 92 | 105 |
| Positive Subject | Age | Gender | Study Group | Ct Value |
|---|---|---|---|---|
| 1 | 76 | Female | 1 | 33 |
| 2 | 70 | Male | 1 | 28 |
| 3 | 49 | Male | 1 | 33 |
| 4 | 41 | Male | 1 | 32 |
| 5 | 78 | Male | 1 | 31 |
| 6 | 54 | Male | 1 | 38 |
| 7 | 65 | Male | 1 | 26 |
| 8 | 68 | Male | 3 | 28 |
| 9 | 59 | Female | 1 | 34 |
| 10 | 74 | Male | 3 | 38 |
| 11 | 44 | Male | 1 | 37 |
| 12 | 57 | Male | 1 | 33 |
| 13 | 36 | Male | 1 | 31 |
| Value | 95% CI | |
|---|---|---|
| Total cohort (n = 105) | ||
| Sensitivity | 92.3% | 64.0% to 99.8% |
| Specificity | 98.9% | 94.1% to 100% |
| Positive Likelihood Ratio | 84.9 | 12.0 to 600.3 |
| Negative Likelihood Ratio | 0.08 | 0.01 to 0.51 |
| Positive Predictive Value | 92.3% | 62.9% to 98.8% |
| Negative Predictive Value | 98.9% | 93.3% to 99.8% |
| Accuracy | 98.1% | 93.3% to 99.8% |
| Cohen’s κ score | 0.91 | 0.79 to 1.00 |
| Females (n = 41) | ||
| Sensitivity | 100% | 15.8% to 100% |
| Specificity | 97.4% | 85.5% to 99.9% |
| Positive Likelihood Ratio | 39.0 | 5.6 to 270.0 |
| Negative Likelihood Ratio | 0 | - |
| Positive Predictive Value | 66.7% | 22.4% to 93.3% |
| Negative Predictive Value | 100% | - |
| Accuracy | 97.6% | 87.1% to 99.9% |
| Cohen’s κ score | 0.79 | 0.39 to 1.00 |
| Males (n = 64) | ||
| Sensitivity | 90.9% | 58.7% to 100% |
| Specificity | 100% | 93.3% to 100% |
| Positive Likelihood Ratio | - | - |
| Negative Likelihood Ratio | 0.09 | 0.01 to 0.59 |
| Positive Predictive Value | 100% | 62.9% to 98.8% |
| Negative Predictive Value | 98.1% | 89.1% to 99.7% |
| Accuracy | 98.4% | 91.6% to 100% |
| Cohen’s κ score | 0.94 | 0.83 to 1.00 |
| ≥65 years (n = 42) | ||
| Sensitivity | 100% | 47.8% to 100% |
| Specificity | 100% | 90.5% to 100% |
| Positive Likelihood Ratio | - | - |
| Negative Likelihood Ratio | 0 | - |
| Positive Predictive Value | 100% | - |
| Negative Predictive Value | 100% | - |
| Accuracy | 100% | 91.6% to 100% |
| Cohen’s κ score | 1.00 | - |
| <65 years (n = 63) | ||
| Sensitivity | 87.5% | 47.3% to 99.7% |
| Specificity | 98.2% | 90.3% to 99.9% |
| Positive Likelihood Ratio | 48.1 | 6.8 to 341.5 |
| Negative Likelihood Ratio | 0.13 | 0.02 to 0.81 |
| Positive Predictive Value | 87.5% | 49.7% to 98.0% |
| Negative Predictive Value | 98.2% | 89.6% to 99.7% |
| Accuracy | 96.8% | 89.6% to 99.7% |
| Cohen’s κ score | 0.86 | 0.67 to 1.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maniscalco, M.; Ambrosino, P.; Ciullo, A.; Fuschillo, S.; Valente, V.; Gaudiosi, C.; Paris, D.; Cobuccio, R.; Stefanelli, F.; Motta, A. A Rapid Antigen Detection Test to Diagnose SARS-CoV-2 Infection Using Exhaled Breath Condensate by A Modified Inflammacheck® Device. Sensors 2021, 21, 5710. https://doi.org/10.3390/s21175710
Maniscalco M, Ambrosino P, Ciullo A, Fuschillo S, Valente V, Gaudiosi C, Paris D, Cobuccio R, Stefanelli F, Motta A. A Rapid Antigen Detection Test to Diagnose SARS-CoV-2 Infection Using Exhaled Breath Condensate by A Modified Inflammacheck® Device. Sensors. 2021; 21(17):5710. https://doi.org/10.3390/s21175710
Chicago/Turabian StyleManiscalco, Mauro, Pasquale Ambrosino, Anna Ciullo, Salvatore Fuschillo, Valerio Valente, Carlo Gaudiosi, Debora Paris, Raffaele Cobuccio, Francesco Stefanelli, and Andrea Motta. 2021. "A Rapid Antigen Detection Test to Diagnose SARS-CoV-2 Infection Using Exhaled Breath Condensate by A Modified Inflammacheck® Device" Sensors 21, no. 17: 5710. https://doi.org/10.3390/s21175710
APA StyleManiscalco, M., Ambrosino, P., Ciullo, A., Fuschillo, S., Valente, V., Gaudiosi, C., Paris, D., Cobuccio, R., Stefanelli, F., & Motta, A. (2021). A Rapid Antigen Detection Test to Diagnose SARS-CoV-2 Infection Using Exhaled Breath Condensate by A Modified Inflammacheck® Device. Sensors, 21(17), 5710. https://doi.org/10.3390/s21175710









