1. Introduction
Skin cancers are the most common types of cancer in the Caucasian population [
1]. Melanoma is the most lethal skin cancer due to its possible evolution into metastasis [
1]. Among pigmented lesions, it is particularly difficult to differentiate in melanoma between nevi and seborrheic keratosis [
2,
3,
4]. Typical pigmented melanoma, nevi, and seborrheic keratosis can be distinguished easily.
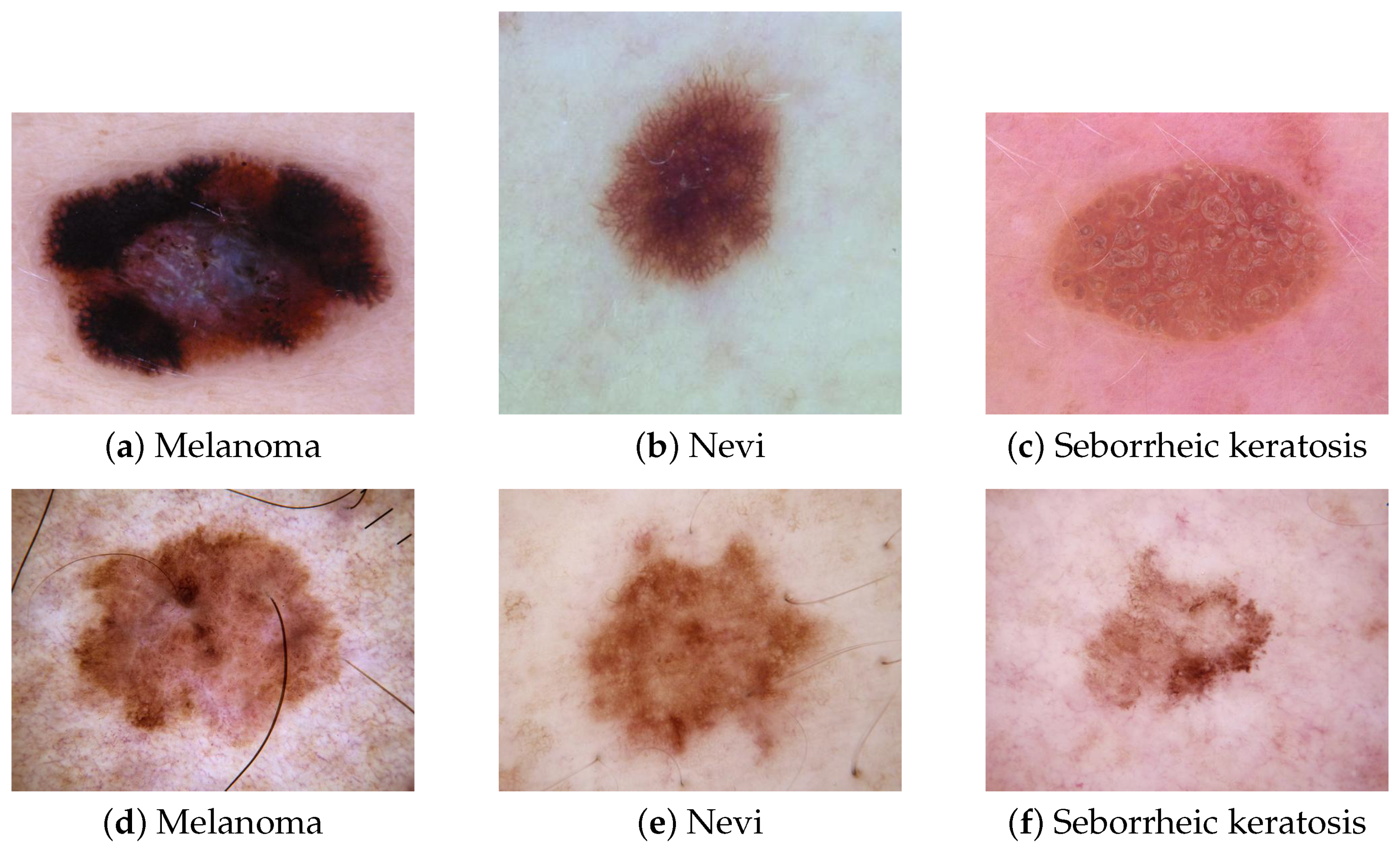
Figure 1 depicts these lesions and a typical melanoma (
Figure 1a), a typical nevi, (
Figure 1b), and typical seborrheic keratosis (
Figure 1c), which do not raise any diagnosis issues for dermatologists. However, atypical nevi or seborrheic keratosis can be confused with melanoma.
Figure 1d–f show some atypical melanoma, nevi, and seborrheic keratosis and highlight how it can be challenging for a dermatologist to rule out a melanoma among these types of pigmented lesions. Faced with atypical pigmented lesions, dermatologists require excision with histological analysis to confirm or reject a diagnosis of melanoma.
These types of atypical pigmented lesions explain the high ratio of the number of lesions excised to the number of melanomas diagnosed [
6]. A total of 9.6 suspicious benign lesions are excised before reaching a confirmed diagnosis of melanoma [
7]. Each excision can lead to scarring and post-surgery complications. The principal objective for dermatologists is to decrease this number and excise only true melanomas. Thus, differentiating early melanoma from nevi and seborrheic keratosis not only constitutes a daily problem for dermatologists, but also has the potential to decrease cancer deaths since melanoma can be cured with a simple excision at an early stage [
8].
Most dermatologists are currently using a dermoscopic sensor during dermatological examination for skin cancer screening. It is a non-invasive dermatological tool allowing the visualization of the lesions’ patterns and structures with a high resolution. It involves a magnification lens and different lighting schemes, such as non-polarized and polarized light. Polarized light helps to minimize the light reflection of the skin’s surface and highlights the detailed patterns and vascularization of the lesion. A dermoscopic sensor helps dermatologists to recognize specific features for the early diagnosis of skin cancer that are sometimes not visible to the naked eye [
9].
Figure 2 illustrates the use of a dermoscopic sensor.
The contribution of dermatoscopy has been investigated by many authors and dermatologists [
10,
11], demonstrating its efficiency in increasing melanoma diagnostic accuracy by 5% to 30% over clinical visual inspection alone. Frequent skin cancer screening of the general population with a dermoscopic examination of pigmented lesions is necessary to detect early melanoma; unfortunately, the lack of dermatologists prevents the development of large screening programs. Therefore, CAD systems have been developed to assist dermatologists to achieve the early diagnosis of melanoma.
To help dermatologists diagnose melanoma early and to reduce the number of unnecessary excisions of benign lesions, the computer vision community has developed several CAD systems. A CAD system is an automatic tool used to support dermatologists in their diagnosis. Before 2015, CAD development was mainly based on handcrafted features. It consisted of extracting features such as shape, color, and texture. These approaches were inspired by the ABCDE criteria (A, asymmetry; B, irregular borders; C, inhomogeneous color; D, diameter > 6 mm; and E, evolution) [
12]. The extracted characteristics were then used as input vectors for a machine learning algorithm (multilayer perceptron (MLP), SVM, KNN, logistic regression, etc.). Celebi et al. proposed an approach to classify dermoscopic images involving border detection and handcrafted extraction of features (texture, color, and shape). These features were then used to train an SVM for classification with feature selection [
13]. However, the ABCDE criteria are not the best features to use for melanoma detection [
14]. Moreover, these features are assimilated to low-level features in CAD systems, which can limit the overall accuracy of the CADs. CNNs attempt to model high-level abstractions in data using multiple processing layers. Due to the availability of public datasets and the advances in computing capacity, there is a growing trend in their use in skin lesion classification. Esteva et al. [
15] were the first to compare CNNs’ diagnostic accuracy with that of dermatologists. They found that most dermatologists, and especially the less experienced ones, were outperformed by CNNs.
The computer vision community relies on the ensemble method to achieve highly accurate performance in the multiclass classification of skin lesions. The ensemble method is based on fusing a finite set of classifiers [
16]. Harangi et al., for example, combined the output of the classification layer from four CNNs using a weighted majority voting strategy for a three-class classification task [
17]. Pacheco et al. tested different approaches including simple majority voting, maximum probability, and the average of probabilities to merge the output of 13 CNNs in an eight-class classification task [
18]. The average probability achieved the best results. Mahbod et al. proposed a framework based on three CNN backbones, where each model was trained on images of skin lesions of six different sizes, ranging from 224 × 224 to 450 × 450 pixels. All the models constructed were then assembled on a three-level ensemble strategy based on the average of predicted probabilities [
19]. In [
20,
21,
22,
23,
24], the average of probabilities has also been used as an aggregated method to improve the performance of CAD.
Broadly, current studies applying ensemble methods follow a similar workflow. First, several multiclass CNNs are trained for a specific task and then their outputs are merged using an aggregation approach. An overview of related works applying ensemble methods is provided in
Table 1.
The most used aggregation methods are:
Max-Win strategy: The class selected by the Max-Win strategy is the class that receives the maximum number of votes.
Product of probabilities strategy: The product of the individual outputs of the CNNs is calculated and the selected class is determined by the maximum of the normalized products.
Average probability strategy: The arithmetic mean of the confidence values of each CNN is calculated, and the selected class is determined by the maximum of the normalized means.
Max confidence strategy: The class selected by the max confidence strategy is the class that received the maximum confidence score.
Geometric mean strategy: The geometric mean of the confidence values of each CNN is calculated, and the selected class is determined by the maximum of normalized means.
The multiclass classification of pigmented lesions remains a challenging task because skin lesions have a high degree of similarity, making their classification a complex task that requires an extensive amount of labeled data and careful definition of the network’s free parameters to train an accurate CNN. Additionally, CNNs behave as black boxes, making it difficult for dermatologists to interpret their prediction.
Rather than simply merging several multiclass CNNs, as has often been the case in most work using the ensemble method, an innovative approach involves decomposing the initial multiclass problem into several less complex classification tasks. Galar et al. stated that multiclass classification is typically more difficult than binary classification [
26]. They explained that the decision boundary of a multiclass classification problem tends to be more complex than a binary classification problem. Therefore, researchers have investigated decomposition and ensemble methods as an alternative to resolve these problems. The idea behind the decomposition and ensemble method is to split the multiclass problem into a set of binary problems and then aggregate the results. The two well-known approaches to developing a decomposition and ensemble strategy are one-versus-rest and one-versus-one [
26]. For an N-class classification, each approach is described as follows:
The one-versus-rest approach consists of constructing a set of N binary classifiers. Each classifier is trained with one class as the positive and all the others as the negatives. The final decision corresponds to the class associated with the classifier with the highest output value.
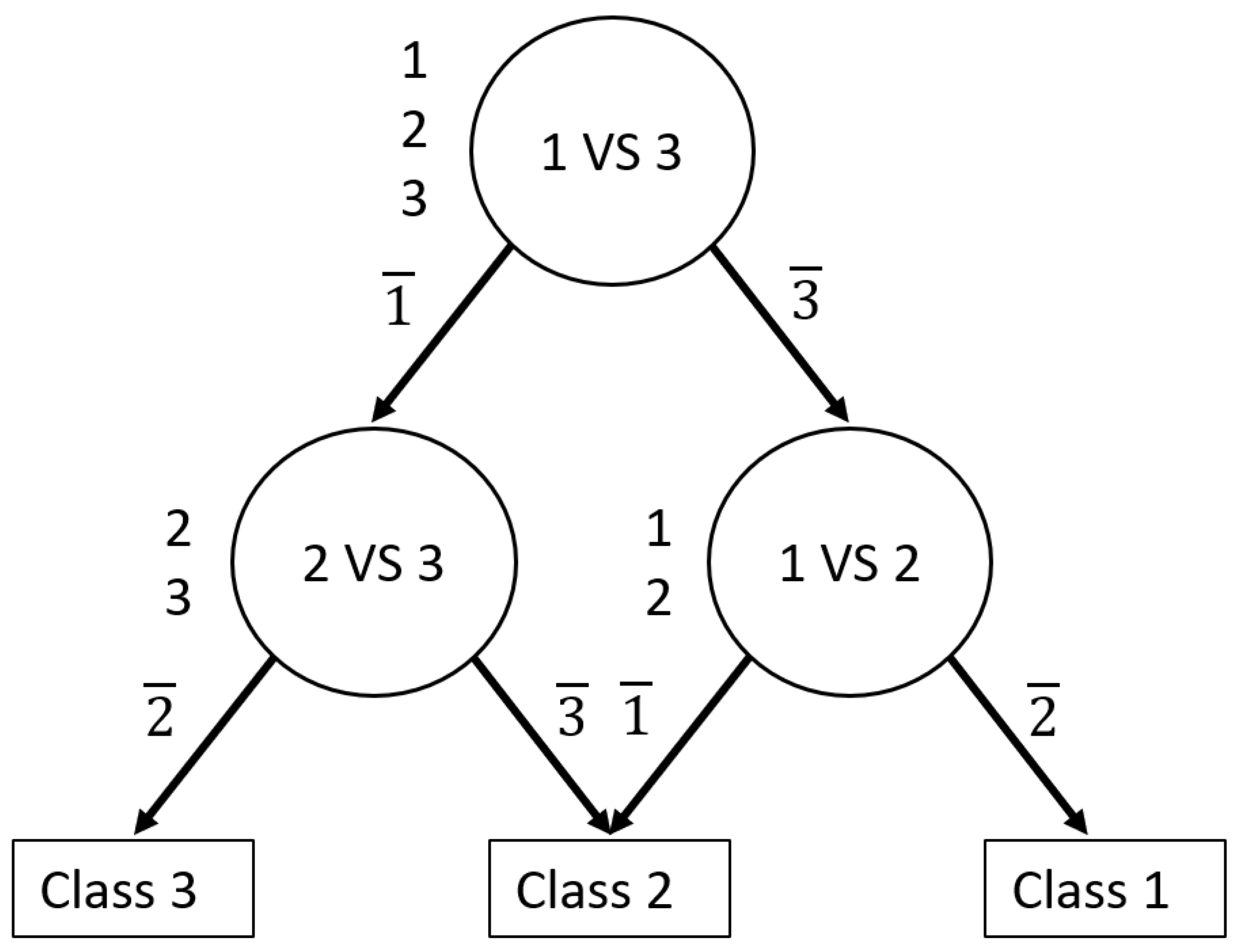
The one-versus-one approach consists of constructing all possible binary classifiers from the N classes. Each classifier is trained on only two classes out of the N initial classes. Thus, there will be N(N – 1)/2 classifiers. The outputs of these base classifiers are combined to predict the final decision.
The main limitations of these approaches are that the one-versus-one approach tends to overfit the overall N-class classifier, and the Max-Win algorithm used does not have bounds on the generalization error [
27]. Therefore, to remedy these disadvantages, Platt [
27] proposed a decision-tree-based pairwise classification called the decision directed acyclic graph (DDAG). Platt demonstrated that DDAGs provide good generalization performance and their structure is efficient to train and evaluate.
In this study, we relied on the decomposition and ensemble method to develop an accurate automated diagnosis of melanoma, nevi, and seborrheic keratosis. For this purpose, we constructed a novel ensemble of CNNs based on DDAGs. We hypothesized that decomposing a multiclass problem into a binary problem would reduce the complexity of the initial multiclass problem faced by CNNs and simultaneously increase the overall performance. The DDAG follows a hierarchical workflow mimicking the multi-step reasoning used by dermatologists faced with pigmented lesions to make a diagnosis [
28]. Thus, following a hierarchical structure can ensure that CAD decision-making is understandable for dermatologists and increase their use in a clinical setting. To the best of our knowledge, this is the first attempt to use a DDAG as a decomposition and ensemble strategy with CNNs. The main contributions of this work are:
Decomposing the initial multiclass classification of pigmented lesions into a binary problem to reduce the complexity of the task and increase the overall classification performance;
Using a directed acyclic graph as an ensemble method to perform multiclass classification with CNNs;
Following a hierarchical workflow provides more transparent decision-making of the computer-aided diagnosis system, thus making it more understandable for dermatologists.
The remainder of this paper is organized as follows:
Section 2 describes the methods applied. In
Section 3, we present the results of the experiments conducted on the 2018 International Skin Imaging Collaboration (ISIC) public dataset, and we discuss in detail the results of our proposed method. Finally, we conclude the work and discuss its future scope in
Section 4.
3. Results and Discussion
We evaluated our novel approach based on the combination of DDAG and binary CNNs. First, we tested the performance of the individual binary classifiers. Second, we analyzed the effect of varying the root node after aggregating the outputs with the DDAG approach. Third, we compared the result of our method with three well-known CNN architectures on a three-class classification task. Then, we analyzed the performance of our best DDAG structure. Finally, we evaluated our approach against other conventional aggregation strategies. We used a three-fold cross-validation on the training set and present the average and standard deviation for the BACC, the sensitivity (S), and the AUROC. In the following, we refer to melanoma, nevi, and seborrheic keratosis as MEL, NEV, and SEK, respectively.
3.1. Performance of Binary CNNs
Table 4 shows the results obtained with a three-fold cross-validation on the training set with resnet50_2, VGG16_2, and VGG19_2 for each individual task: MEL versus NEV, MEL versus SEK, and NEV versus SEK. The BACC and the sensitivity for each class are presented. Mostly, we observed that the classifiers performed very well in binary classification. For our task, we observed that the backbone model VGG19 performed better than Resnet50 and VGG16. These results can be explained by the deeper VGG19 architecture compared with VGG16 and Resnet50. Therefore, VGG19 can learn more discriminating features. Interestingly, among these three tasks, seborrheic keratosis and nevi were easiest to distinguish, with the best performance obtained by the binary CNNs NEV vs. SEK. Seborrheic keratosis is very dark and composed of completely different patterns to melanocytic lesions, such as keratin structures, horn cysts, or a cerebriform pattern. However, melanoma can be confused with seborrheic keratosis; melanoma can be very dark, similar to seborrheic keratosis, and can sometimes mimic seborrheic keratosis by having atypical structures. The most challenging tasks for our framework are distinguishing benign melanocytic lesions (nevi) from malignant melanocytic lesions. When the melanoma is excised at an early stage with a thin Breslow (thickness of the melanoma), the difficulty of differentiating melanoma from nevi is high even for dermatologist experts. Moreover, some melanoma are raised on nevi, so they may share the same patterns and structures (reticular pattern or dotted pattern); however, for melanomas, the pattern is more irregular than that of nevi.
3.2. Impact of Root Node
The second aspect that we investigated was the effect of variations in the root node on the overall performance of our approach. This was conducted based on the BACC. The results of this analysis are presented in
Table 5. Regardless of the type of DDAG, we noticed that the overall performance of the framework depends on the performance of each individual classifier, explaining why DDAGs based on VGG19 performed better. The DDAG structure based on VGG19 reached BACCs between 73.7% and 76.6%, compared with the 72.55–73.25% for the VGG16 and 70.1–71.1% for ResNet50 backbone models. Moreover, the choice of the DDAG structure may slightly affect the final accuracy of the framework, which is similar to the observation of [
36] with support vector machine. Thus, inspired by [
37], the optimal structure of the DDAG was obtained by placing the classifier with the greatest generalization ability in the root node. This explains why DDAG structures with the SEK vs. NEV classifier as the root node performed better on VGG19_2 and VGG16_2, and the best performance was achieved with root MEL vs. NEV for resnet50_2 (
Section 3.1). The best structure with the most accurate performance (BACC = 76.6
%) was obtained with the DDAG structure based on VGG19 and having a binary CNN on the task with NEV vs. SEK as the root node.
3.3. Multiclass CNNs Versus DDAG Model
Table 6 shows the evaluation of our main hypothesis. We compared our approach with ResNet50, VGG16, and VGG19 trained on a three-class classification task based on the results of the 3-fold cross-validation. For a faithful comparison, only the classification layer was modified to adapt it to a three-class classification (
Section 2.2.3). We refer to these adapted models as ResNet50_3, VGG16_3, and VGG19_3. The DDAG-based approach achieved the best BACCs compared to multiclass CNNs.
The best models obtained for each configuration were then selected and evaluated on the test set for an in-depth analysis. We observed that the DDAG structure with the BEK vs. NEV classifier as the root node and the VGG19 architecture as the backbone model obtained the best performance, reaching a balanced accuracy of 76.6% on the test set. However, we highlight that the performance of the DDAG structure is closely linked to the choice of the backbone model, as illustrated by our results obtained with VGG16 and ResNet50. Models with potentially better performance, such as EfficienNet [
38] and SeNet [
39], may improve the accuracy of the DDAG structure.
On the other hand, binary CNNs aggregated with a DDAG structure achieved better performance than 3-class CNNs. These results matched with the previous analysis (see
Table 6). We performed statistical analyses using a paired
t test on the predicted probabilities of each model and, interestingly, we found that scores from the DDAG models were significantly different from those of multiclass CNNs (
Table 7). We thus concluded that decomposing a multiclass problem into a binary problem reduces the complexity of the initial problem and increases the overall performance.
3.4. Performance Analysis of Our Best DDAG Structure
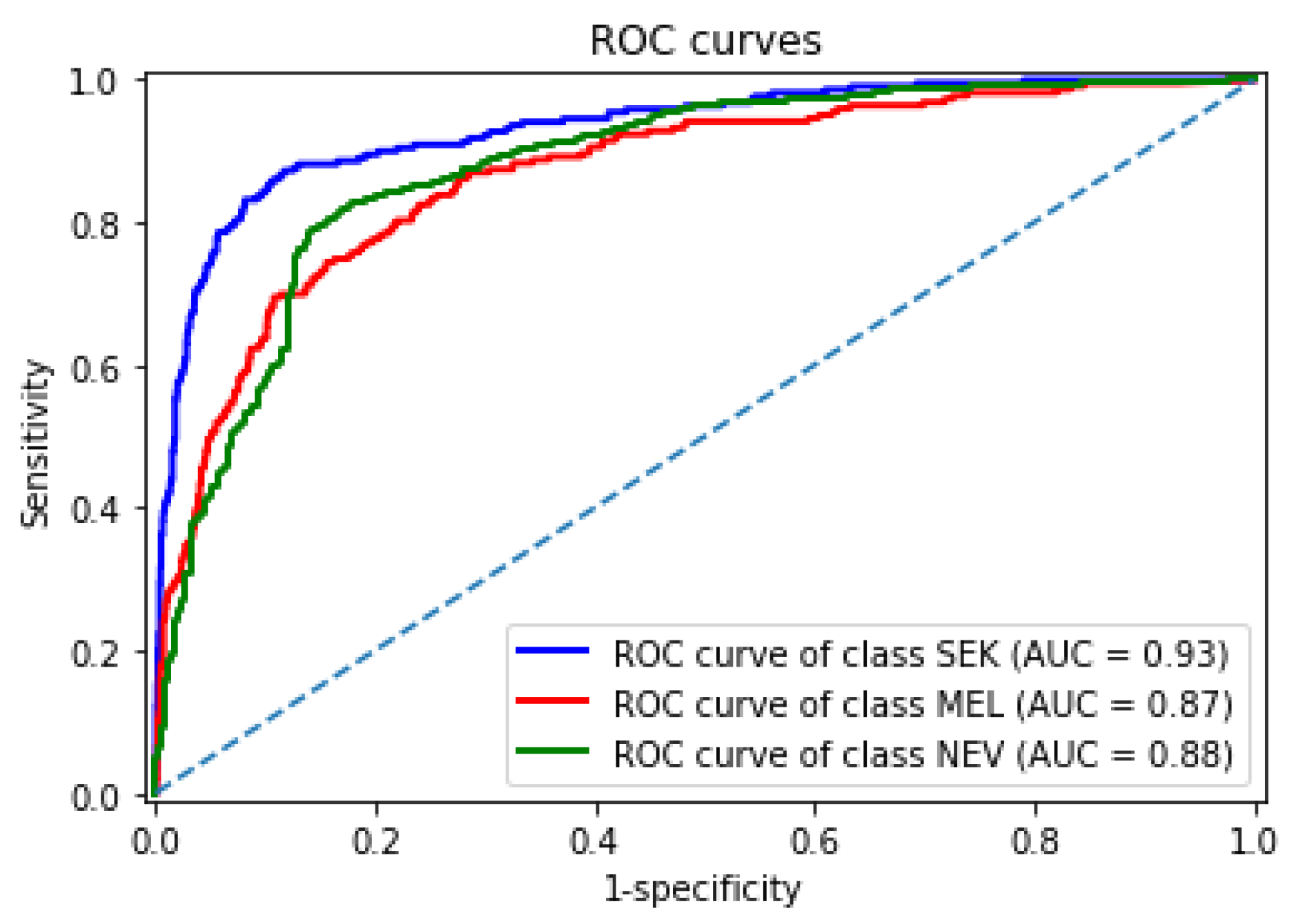
Figure 8 shows the receiver operating characteristic curves obtained by our best DDAG structure for each lesion in the test set. Our framework achieved an AUROC of 0.93, 0.87, and 0.88 for seborrheic keratosis, melanoma, and nevi, respectively. We observed that melanoma remained the most challenging class.
We presented the structure of our framework to a dermatologist for an in-depth analysis. To facilitate the dermatologist’s analysis, we associated each prediction provided by a classifier and its corresponding heatmap, allowing visualization of the regions contributing to the prediction; heatmap generation was implemented with the Grad-CAM method [
40].
Figure 9 illustrates the decision strategy of our best DDAG structure. As an example, we present a challenging pigmented lesion that was classified as a melanoma at the end of this framework. The arrows in green represent the evaluation path in this case. The dermoscopic image (input image in
Figure 9) shows a pigmented lesion that is slightly suspicious. The reticular network is irregular and enlarged on the left part. On this part and in the middle, we can also observe a blue white veil color with some dots corresponding to a regression area, which is associated with melanoma diagnosis. Interestingly, the heatmap shows the decision-making area of the CNN, focusing its prediction on the atypical left part of the lesion, the most suspicious for melanoma diagnosis.
3.5. Comparison with Other Methods
We further compared our approach based on DDAG with commonly used aggregation methods (avg, mconf, prod, gmean, and max-win). For this, the best models obtained with ResNet50_3, VGG16_3, and VGG19_3 during cross-validation were merged following these aggregation methods. The results presented in
Table 8 summarize for each method the performance obtained on the test set and highlight the outcome of Kruskal–Wallis’s test and post hoc multiple-comparison on the predicted probabilities. Here, “g.r” denotes the group rank of methods with stastistically similar predicted scores, and “s.o.g” is the set of other groups that are statistically worse. An empty set indicates that a particular method was not statistically better than any other group.
The DDAG structure achieved the best BACC (76.6%) amongst the ensemble of multiclass CNNs with classical aggregation methods. Moreover, the probability scores generated by our approach were statistically different (p < 0.05) from those of other classical aggregation methods, which confirms the robustness of our DDAG structure and its ability to improve the performance of a computer-aided diagnosis system. We also found that, among the classical aggregation methods, avg, max_conf, and gmean achieved the best performance, with no statistically difference amongst their predicted score. These results suggest that these are the best of the classical aggregation approaches to use for building a CAD on dermoscopic images. The product of the probabilities strategy was the worst performer. Thus, the product significantly enhances the propagation of the worst prediction probabilities. To reduce this effect in the application of ensemble methods, we recommend merging only classifiers with similar performance.
We also compared our method with existing methods on the same three-class classification task [
41,
42]. Based on the BACC, our approach outperformed these related methods.
Our approach is much simpler to interpret by dermatologists because it follows a hierarchical workflow similar to two-step reasoning [
28], whereas conventional approaches simply aggregate several CNNs without providing transparency in the decision-making process.