Sandwich-Type DNA Micro-Optode Based on Gold–Latex Spheres Label for Reflectance Dengue Virus Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Instrumentation
2.3. Synthesis of Poly(n-butyl acrylate-co-N-acryloxysuccinimide) Microspheres
2.4. Preparation of Gold–Latex Particles
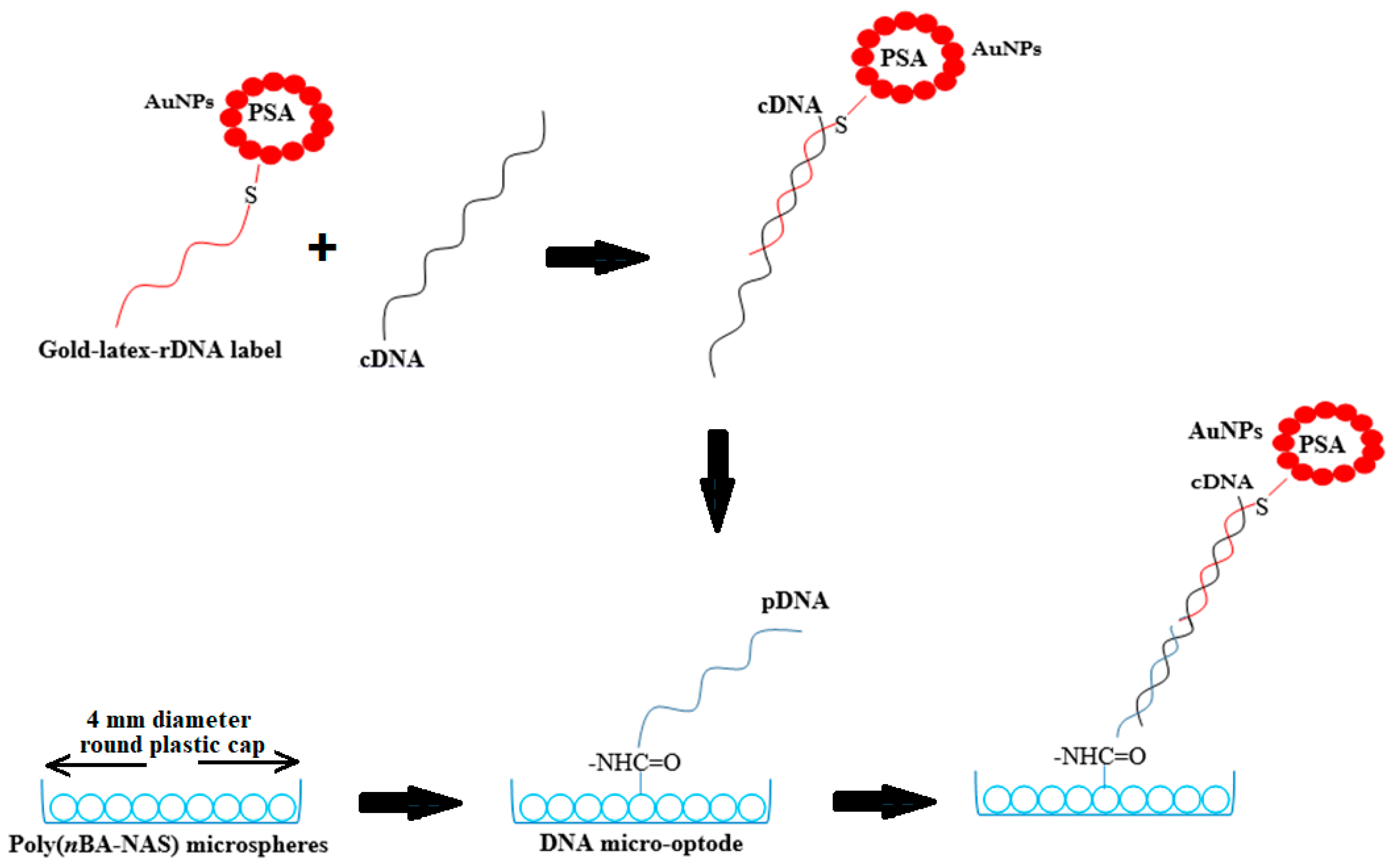
2.5. Fabrication of Sandwich-Type DNA Micro-Optode
2.6. Optimizing the DNA Micro-Optode for Dengue Virus Detection
2.7. Validation Study of DNA Micro-Optode with RT-PCR for Dengue Virus Detection in Blood, Urine, and Saliva Samples
3. Results and Discussion
3.1. Characterization of Gold Nanoparticles, Polyacrylate, and Latex Microspheres
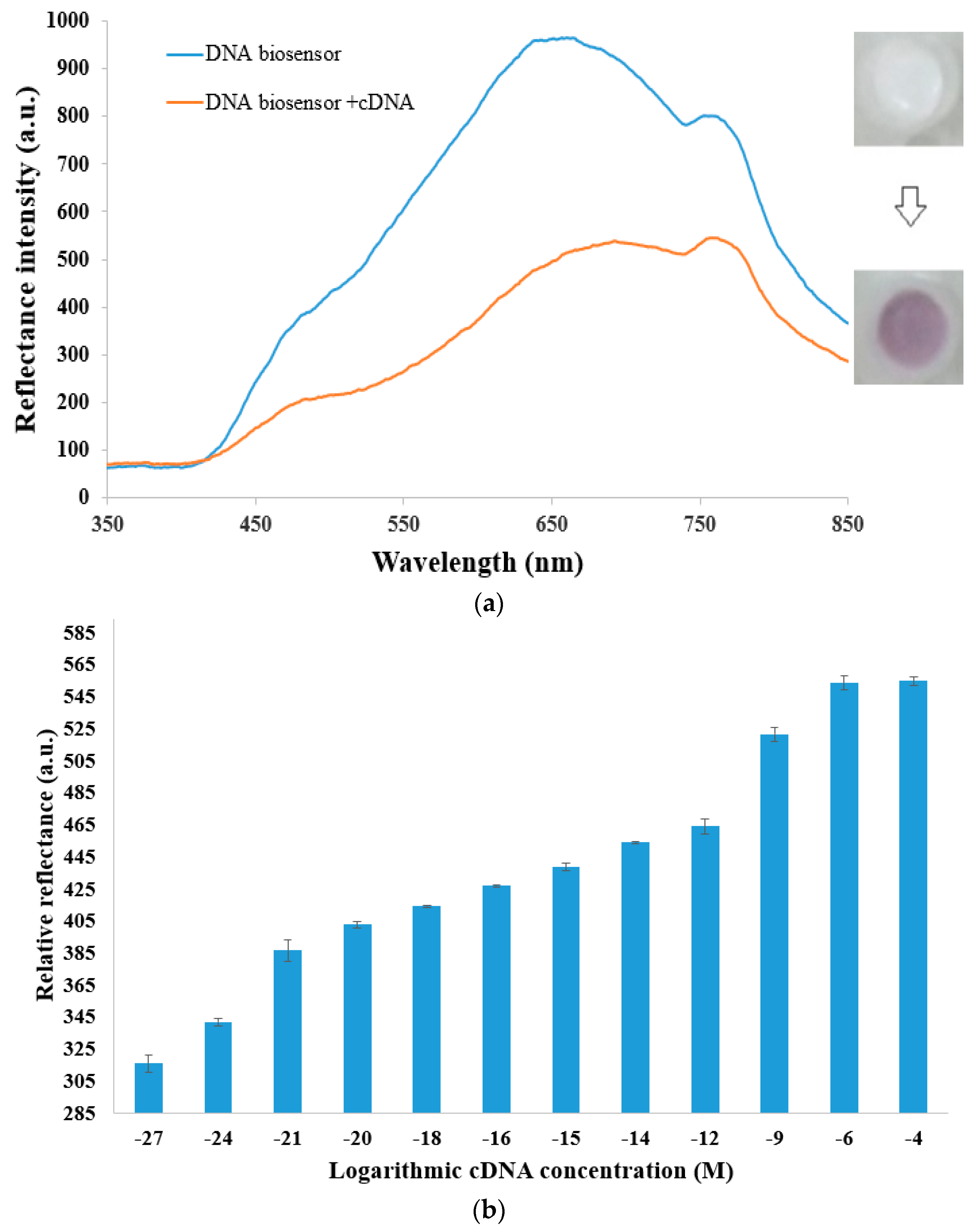
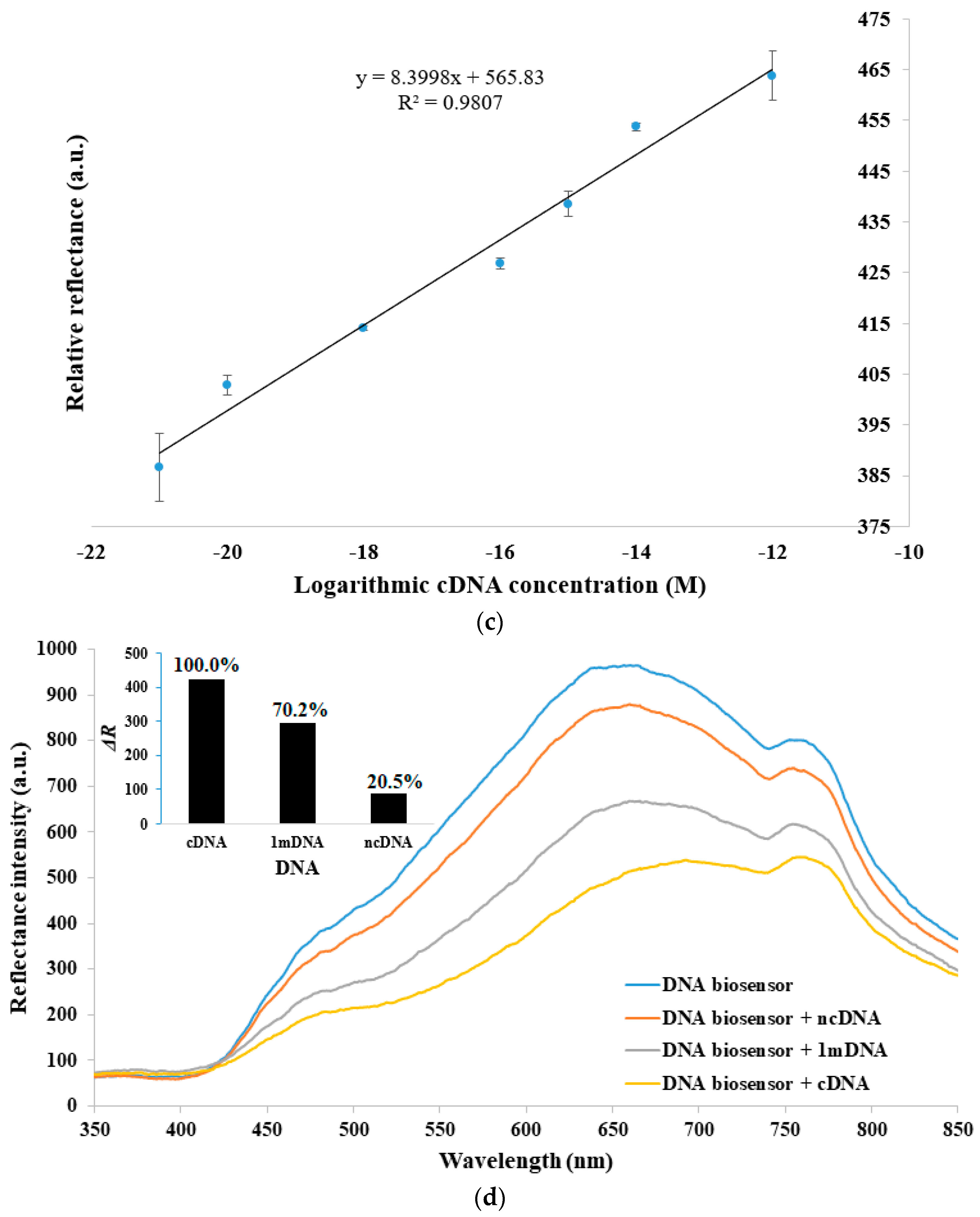
3.2. Characterization of DNA Micro-Optode with Fiber-Optic Reflectance Spectrophotometer
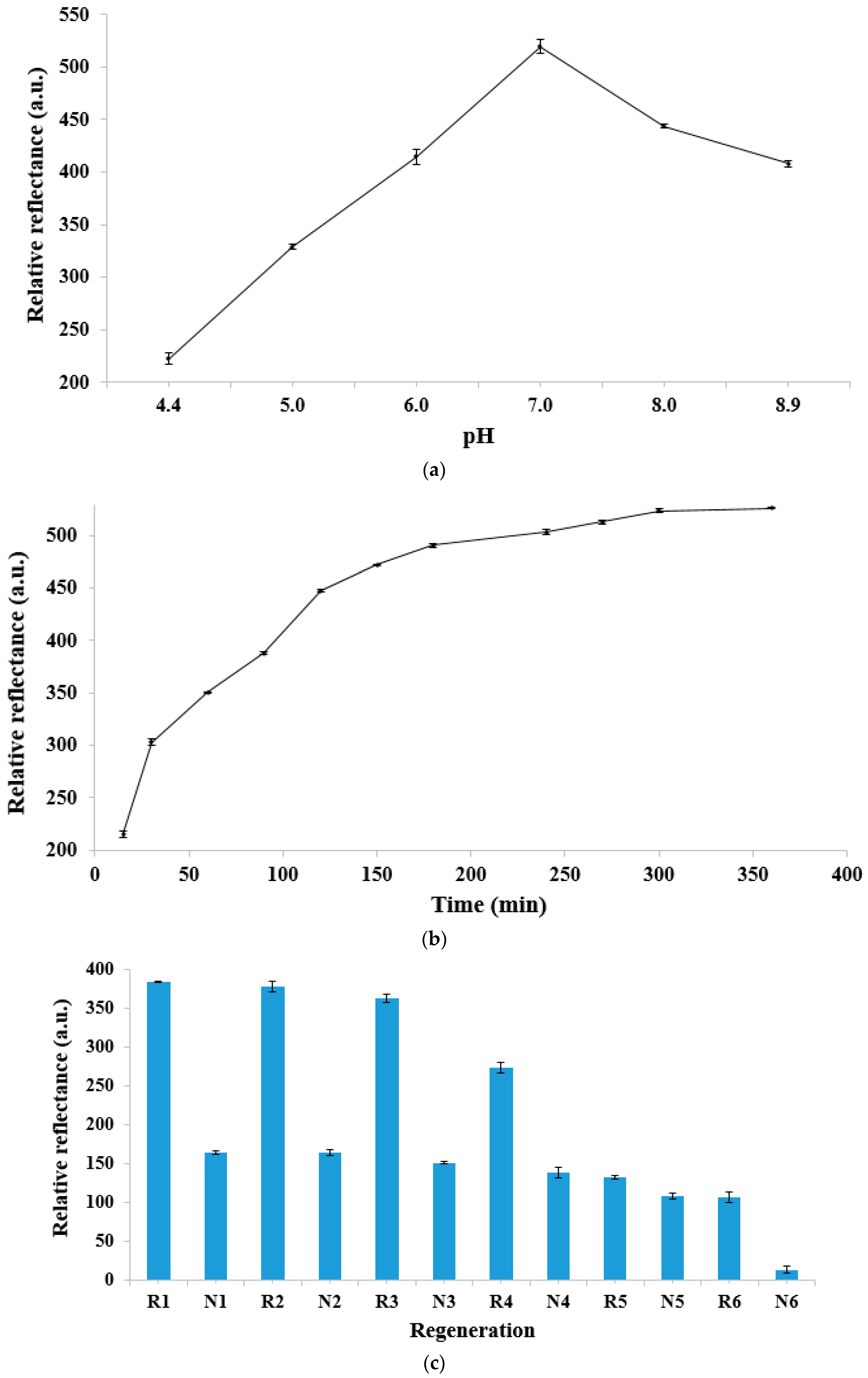
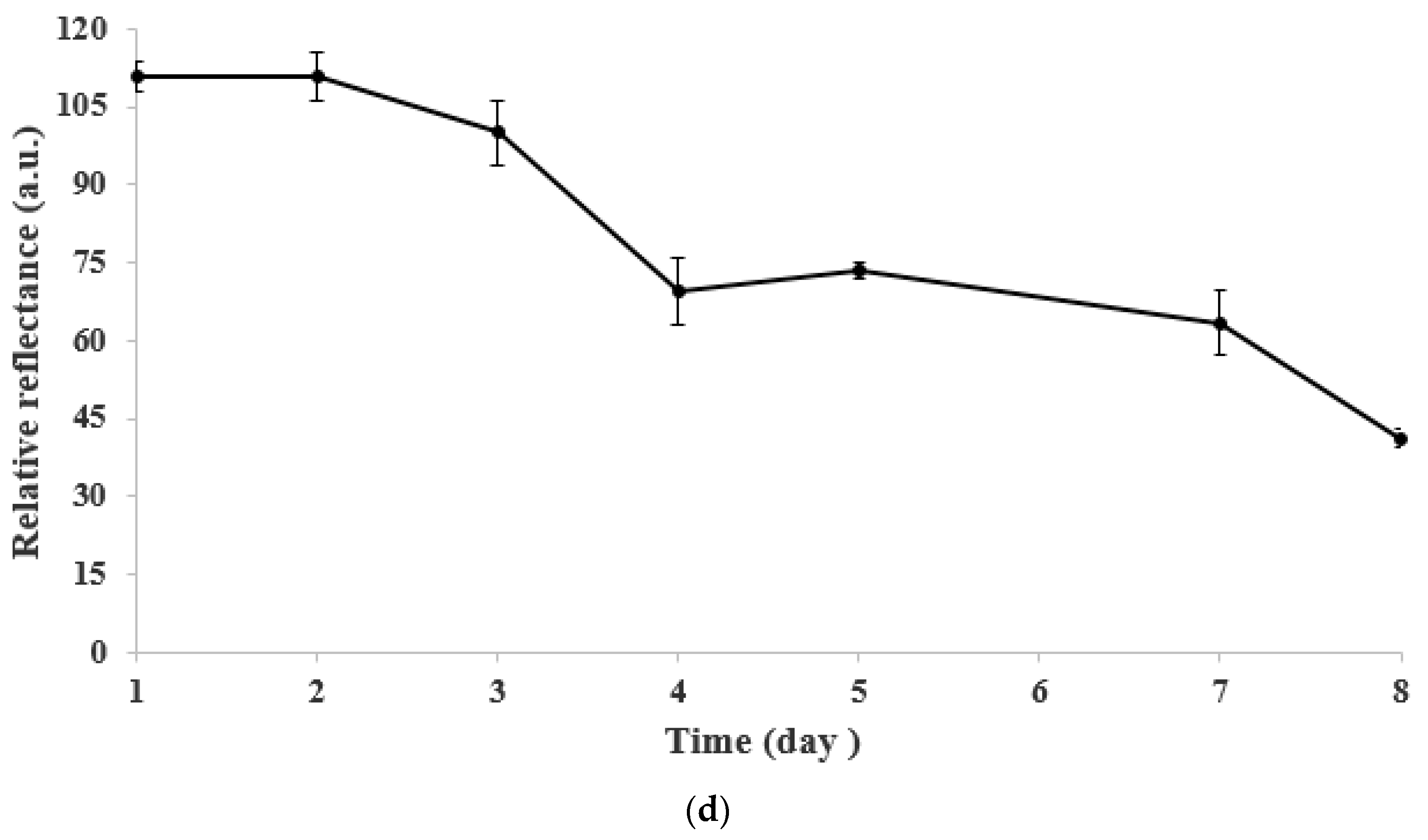
3.3. pH Effect, Response Time, Regeneration, and Long-Term Stability of the DNA Micro-Optode
3.4. Validation of Sandwich-Type DNA Biosensor with RT-PCR for Dengue Virus Detection
3.5. Comparison with Other Reported Dengue Virus DNA Biosensors
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Darwish, T.N.; Alias, Y.; Khor, S.M. An introduction to dengue-disease diagnostics. Trends Anal. Chem. 2015, 67, 45–55. [Google Scholar] [CrossRef]
- World Health Organization, Global Health Estimates 2014 Summary Tables: Deaths by Cause, Age and sex, by World Bank Income Group Category. 2000–2012. 2014. Available online: http://www.who.int/healthinfo/global_burden_disease/en/ (accessed on 4 May 2016).
- Parkash, Q.; Shueb, R.H. Diagnosis of dengue infection using conventional and biosensor based techniques. Viruses 2015, 7, 5410–5427. [Google Scholar] [CrossRef]
- Cheng, M.S.; Ho, J.S.; Tan, C.H.; Wong, J.P.; Ng, L.C.; Toh, C.S. Development of an electrochemical membrane-based nanobiosenor for ultrasensitive detection of dengue virus. Anal. Chim. Acta 2012, 725, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, L.; Huang, M.J.; Luo, Z.H.H.; Tay, G.K.I.; Lim, E.A.; Kang, T.G.; Chen, Y. Silicon nanowire biosensor for highly sensitive and rapid detection of dengue virus. Sens. Actuators B Chem. 2010, 146, 138–144. [Google Scholar] [CrossRef]
- Nascimento, H.P.O.; Oliveira, M.D.L.; Melo, C.P.; Silva, G.J.L.; Cordeiro, M.T.; Andrade, C.A.S. An impedimetric biosensor for detection of dengue serotype at picomolar concentration based on gold nanoparticles-polyaniline hybrid composites. Colloids Surf. B 2011, 86, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, S.J.; Philips, L.W.; Milligan, A.S.; Rodda, S.J. Toward specific detection of dengue virus serotypes using a novel modular biosensor. Biosens. Bioelectron. 2010, 26, 1696–1700. [Google Scholar] [CrossRef]
- Baeumner, A.J.; Schlesinger, N.A.; Slutzki, N.S.; Romano, J.; Lee, E.M.; Montagna, R.A. Biosensor for dengue virus detection: Sensitive, rapid, and serotype specific. Anal. Chem. 2002, 74, 1442–1448. [Google Scholar] [CrossRef]
- Ulianas, A.; Lee, Y.H.; Sharina, A.H.; Tan, L.L. An electrochemical DNA microbiosensor based on succinimide-modified acrylic microspheres. Sensors 2012, 12, 5445–5460. [Google Scholar] [CrossRef]
- Liang, Z.; Zhang, L.; Wang, L.; Song, S.; Fan, C.; Li, G. A centrifugation-based method for preparation of gold nanoparticles and its application in biodetection. Int. J. Mol. Sci. 2007, 8, 526–532. [Google Scholar] [CrossRef]
- Kuan, G.C.; Sheng, L.P.; Rijiravanich, P.; Marimuthu, K.; Ravichandran, M.; Yin, L.S.; Lertanantawong, B.; Surareungchai, W. Gold-nanoparticle based electrochemical DNA sensor for the detection of fish pathogen Aphanomyces invadans. Talanta 2013, 117, 312–317. [Google Scholar] [CrossRef]
- Mizuno, Y.; Kotaki, A.; Harada, F.; Tajima, S.; Kurane, I.; Takasaki, T. Confirmation of dengue virus infection by detection of dengue virus type 1 genome in urine and saliva but not in plasma. Trans. R. Soc. Trop. Med. Hyg. 2007, 101, 738–739. [Google Scholar] [CrossRef] [PubMed]
- Ulianas, A.; Lee, Y.H.; Musa, A. A biosensor for urea from succinimide-modified acrylic microspheres based on reflectance transduction. Sensors 2011, 11, 8323–8338. [Google Scholar] [CrossRef] [PubMed]
- Bastús, N.G.; Comenge, J.; Puntes, V. Kinetically controlled seeded growth synthesis of citrate-stabilized gold nanoparticles of up to 200 nm: Size focusing versus ostwald ripening. Langmuir 2011, 27, 11098–11105. [Google Scholar] [CrossRef] [PubMed]
- Hames, B.B.; Higgins, S.J. Nucleic Acid Hybridisation—A Practical Approach; Oxford University Press: Oxford, UK, 1985. [Google Scholar]
- Metzenberg, S. Working with DNA: The Basics; Taylor & Francis Group: Florence, Italy, 2007. [Google Scholar]
- Chan, C.P.; Choi, J.W.; Cao, K.Y.; Wang, M.; Gao, Y.; Zhuo, D.H.; Di, B.; Xu, H.F.; Leung, M.F.; Bergmann, A.; et al. Detection of serum neopterin for early assessment of dengue virus infection. J. Infect. 2006, 53, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Na, H.; Min, D.; Kim, D. Desorption of single-stranded nucleic acids from graphene oxide by disruption of hydrogen bonding. Analyst 2013, 138, 1745–1749. [Google Scholar] [CrossRef] [PubMed]
- Simonian, M.H.; Smith, J.A. Spectrophotometric and colorimetric determination of protein concentration. Curr. Protoc. Mol. Biol. 2006, 76, 10.1.1–10.1A.9. [Google Scholar] [CrossRef] [PubMed]
- Stollar, V.; Schlesinger, R.W.; Stevens, T.M. Studies on the nature of dengue viruses: III. RNA synthesis in cells infected with type 2 dengue virus. Virology 1967, 33, 650–658. [Google Scholar] [CrossRef]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Current Protocols in Molecular Biology; John Wiley and Sons Inc.: Hoboken, NJ, USA, 1999. [Google Scholar]
- Wilfinger, W.; Mackey, K.; Chomczynski, P. Effect of pH and ionic strength on the spectrophotometric assessment of nucleic acid purity. Biotechniques 1997, 22, 474–481. [Google Scholar] [CrossRef]
- Li, Z.; Reimers, S.; Pandit, S.; Deutscher, M.P. RNA quality control: Degradation of defective transfer RNA. EMBO J. 2002, 21, 1132–1138. [Google Scholar] [CrossRef]
- Radonic, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef]
- Zaytseva, N.V.; Montagna, R.A.; Baeumner, A.J. Microfluidic biosensor for the serotype-specific detection of dengue virus RNA. Anal. Chem. 2005, 77, 7520–7527. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Toh, C.S. Impedimetric DNA biosensor based on a nanoporous alumina membrane for the detection of the specific oligonucleotide sequence of dengue virus. Sensors 2013, 13, 7774–7785. [Google Scholar] [CrossRef] [PubMed]
- Rai, V.C.; Hapuarachchi, H.C.; Ng, L.C.; Soh, S.H.; Leo, Y.S.; Toh, C.S. Ultrasensitive cDNA detection of dengue virus rna using electrochemical nanoporous membrane-based biosensor. PLoS ONE 2012, 7, e42346. [Google Scholar] [CrossRef] [PubMed]
- Priye, A.; Ball, C.S.; Meagher, R.J. Colorimetric-luminance readout for quantitative analysis of fluorescence signals with a smartphone CMOS sensor. Anal. Chem. 2018, 90, 12385–12389. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Hagen, J.A.; Papautsky, I. Point-of-care colorimetric detection with a smartphone. Lab Chip 2012, 21, 4240–4243. [Google Scholar] [CrossRef] [PubMed]
- Priye, A.; Bird, S.W.; Light, Y.K.; Ball, C.S.; Negrete, O.A.; Meagher, R.J. A smartphone-based diagnostic platform for rapid detection of Zika, chikungunya, and dengue viruses. Sci. Rep. 2017, 7, 44778. [Google Scholar] [CrossRef]
| DNA | Base Sequences (from 5′ to 3′) |
|---|---|
| pDNA | NH2–(CH2)6–TTT TGT CCT GCT CTT |
| rDNA | CAT TTA GGC TGG GTT–(CH2)3–SH |
| cDNA (dengue serotype 2) | AAC CCA GCC TAA ATG AAG AGC AGG ACA AAA |
| 1mDNA | AAC CCA TCC TAA ATG AAG AGC AGG ACA AAA |
| ncDNA (Escherichia coli) | AAC GCC GAT ACC ATT ACT TAT ACC GCG ACG |
| Sample Type | Sample Code | cDNA Concentration Determined by Sandwich-Type DNA Micro-Optode (µM) | cDNA Concentration Determined by RT-PCR (µM) | t-Test |
|---|---|---|---|---|
| P1B | 7.5 ± 0.424 | 7.5 ± 0.109 | ||
| Blood | P2B | 3.8 ± 0.937 | 3.4 ± 0.174 | 2.664 |
| P3B | 5.3 ± 1.381 | 5.3 ± 0.226 | ||
| P1U | 4.9 ± 1.024 | 4.9 ± 0.339 | ||
| Urine | P2U | 5.4 ± 1.446 | 5.3 ± 0.283 | 0.582 |
| P3U | 2.3 ± 1.232 | 2.4 ± 0.073 | ||
| P1S | 4.8 ± 1.206 | 4.8 ± 1.659 | ||
| Saliva | P2S | 3.4 ± 0.975 | 3.4 ± 0.248 | 0.407 |
| P3S | 1.9 ± 2.101 | 1.9 ± 0.319 |
| Biosensor | Transducer | Linear Range (M) | LOD (M) | DNA Hybridization Time (min) | Reference |
|---|---|---|---|---|---|
| Succinimide-functionalized acrylic microsphere-based sandwich-type reflectance DNA biosensor with gold–latex sphere optical label | Fiber-optic reflectance spectrophotometer | 1.0 × 10−21−1.0 × 10−12 | 1.00 × 10−21 | 90 | Present work |
| Streptavidin-modified polyethersulfone membrane-based optical DNA biosensor with dye-entrapping liposomal nanovesicle rDNA label | Reflectometer | 1.0 × 10−9−7.5 × 10−7 | 1.00 × 10−9 | 20 | [8] |
| Microfluidic RNA biosensor based on PDMS microfluidic channel–glass interface | Fluorescence microscopy | 1.0 × 10−11−2.5 × 10−8 | 1.25 × 10−10 | 20 | [25] |
| Label-free electrochemical DNA biosensor based on platinum-coated ananoporous alumina membrane | EIS | 1.0 × 10−12−1.0 × 10−6 | 2.70 × 10−12 | 60 | [26] |
| Electrochemical DNA biosensor based on nanoporous alumina membrane and Fe(CN)64− redox label | DPV | 1.0 × 10−12−1.0 × 10−6 | 9.55 × 10−12 | 45 | [27] |
| Silicon nanowire-based DNA biosensor | FET | - | 10.00 × 10−15 | 30 | [5] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeningsih; Tan, L.L.; Ulianas, A.; Heng, L.Y.; Mazlan, N.-F.; Jamaluddin, N.D.; Mohd. Yusof, N.Y.; Khalid, B.; Ta, G.C. Sandwich-Type DNA Micro-Optode Based on Gold–Latex Spheres Label for Reflectance Dengue Virus Detection. Sensors 2020, 20, 1820. https://doi.org/10.3390/s20071820
Jeningsih, Tan LL, Ulianas A, Heng LY, Mazlan N-F, Jamaluddin ND, Mohd. Yusof NY, Khalid B, Ta GC. Sandwich-Type DNA Micro-Optode Based on Gold–Latex Spheres Label for Reflectance Dengue Virus Detection. Sensors. 2020; 20(7):1820. https://doi.org/10.3390/s20071820
Chicago/Turabian StyleJeningsih, Ling Ling Tan, Alizar Ulianas, Lee Yook Heng, Nur-Fadhilah Mazlan, Nur Diyana Jamaluddin, Nurul Yuziana Mohd. Yusof, Bahariah Khalid, and Goh Choo Ta. 2020. "Sandwich-Type DNA Micro-Optode Based on Gold–Latex Spheres Label for Reflectance Dengue Virus Detection" Sensors 20, no. 7: 1820. https://doi.org/10.3390/s20071820
APA StyleJeningsih, Tan, L. L., Ulianas, A., Heng, L. Y., Mazlan, N.-F., Jamaluddin, N. D., Mohd. Yusof, N. Y., Khalid, B., & Ta, G. C. (2020). Sandwich-Type DNA Micro-Optode Based on Gold–Latex Spheres Label for Reflectance Dengue Virus Detection. Sensors, 20(7), 1820. https://doi.org/10.3390/s20071820






