Sensor-Integrated Microfluidic Approaches for Liquid Biopsies Applications in Early Detection of Cancer
Abstract
:1. Introduction
2. Lab-on-a-Chip Platforms for Liquid Biopsies Applications
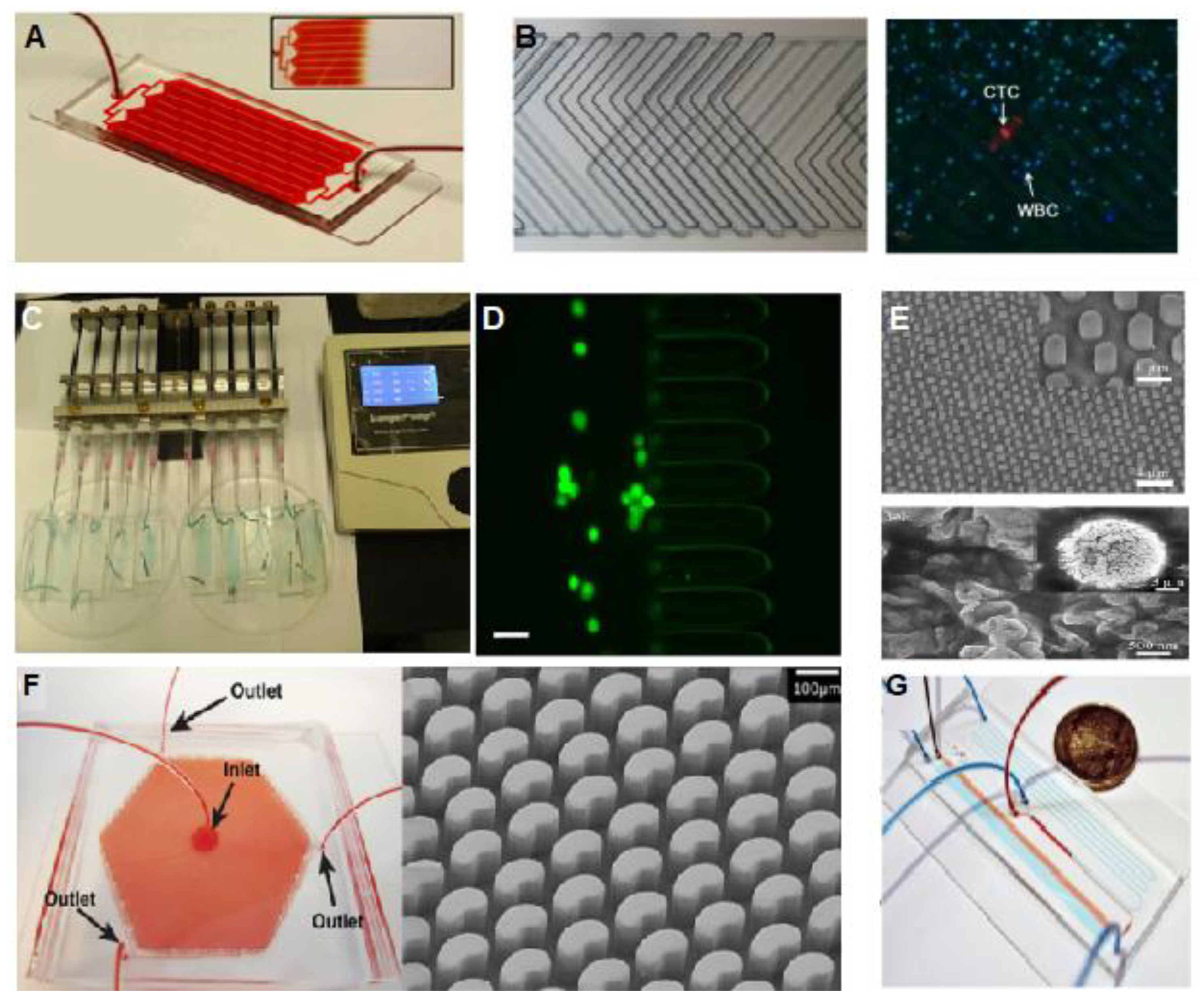
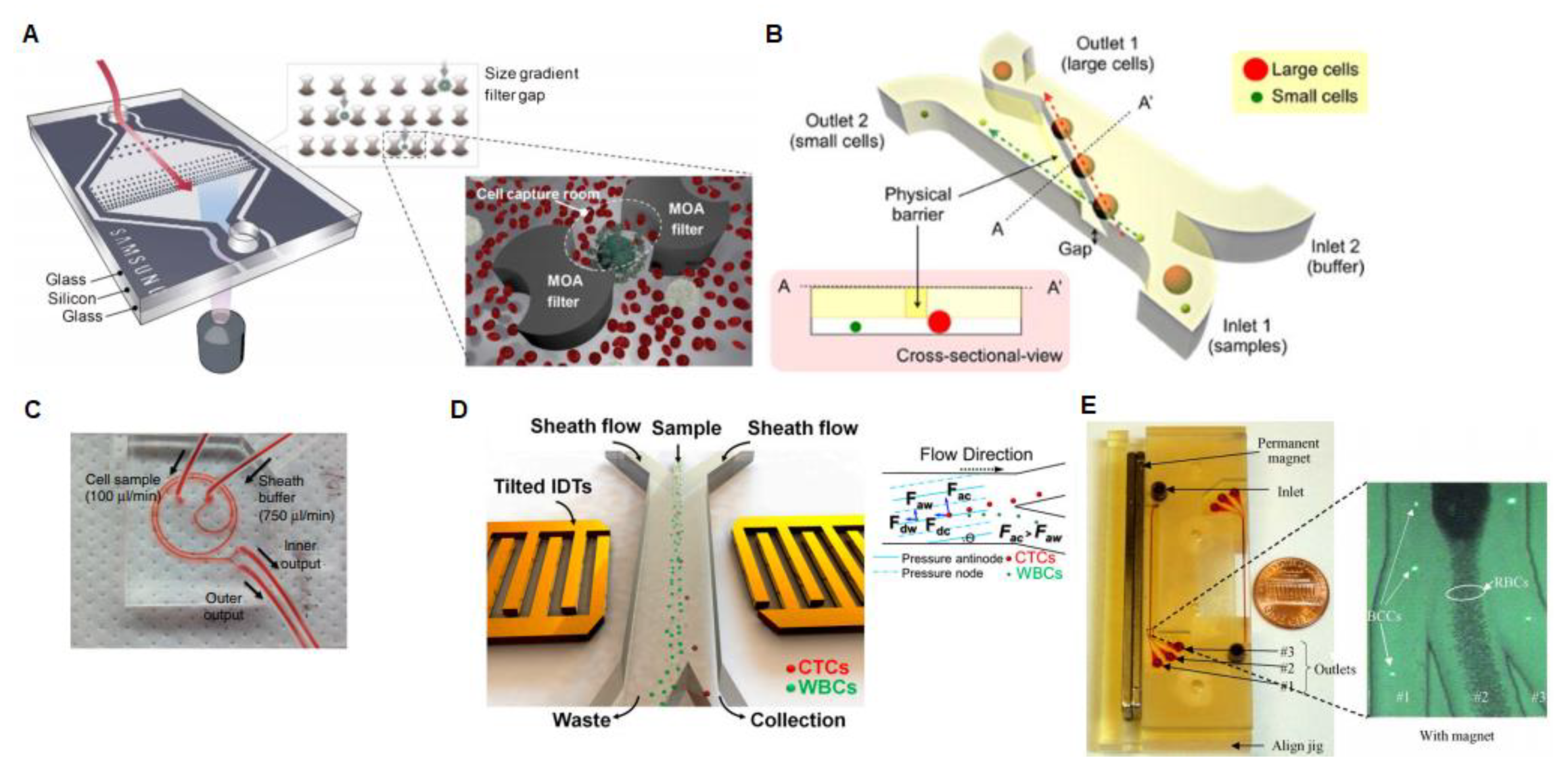
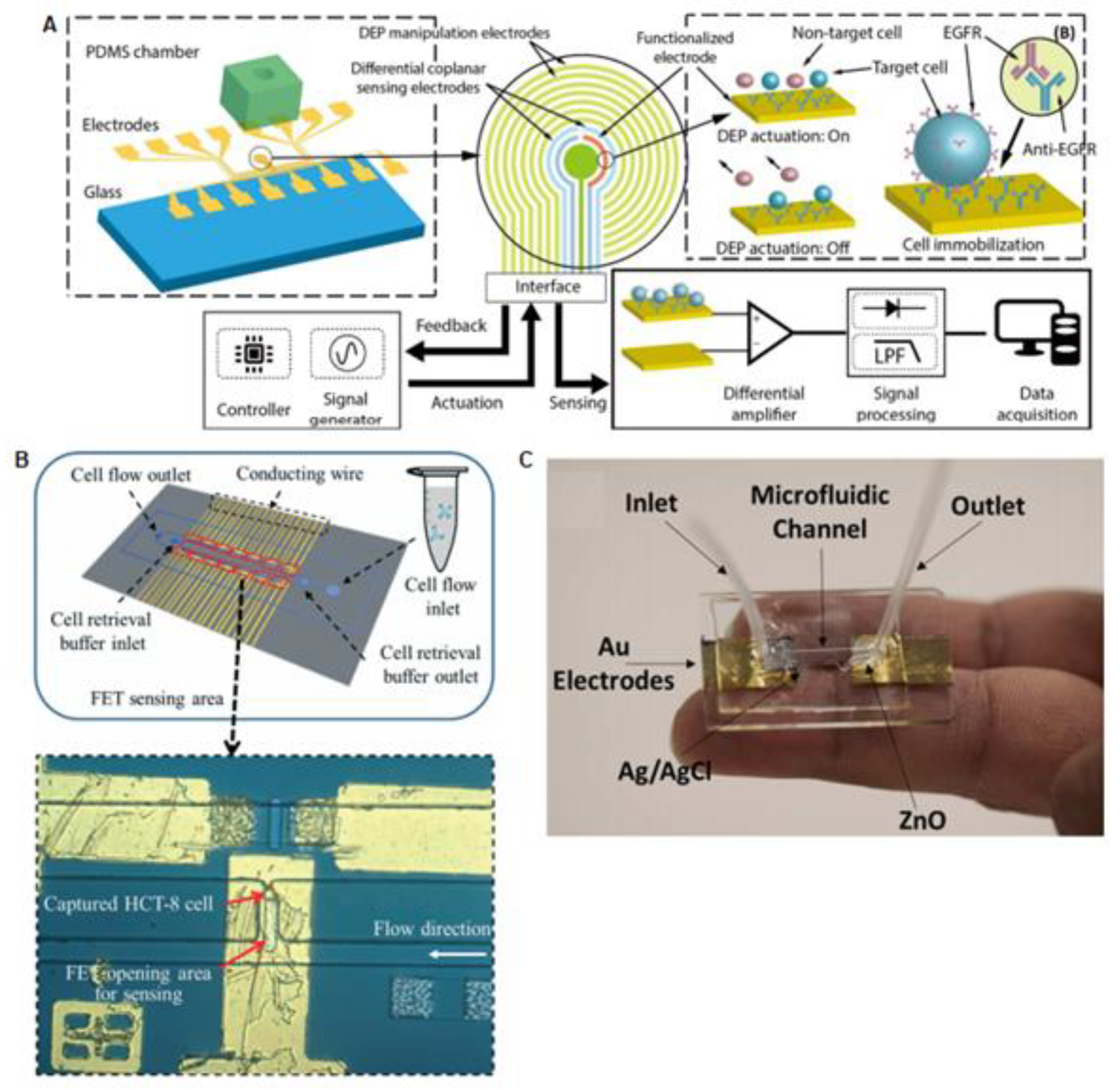
2.1. Platforms Based on Circulating Tumor Cells Analysis
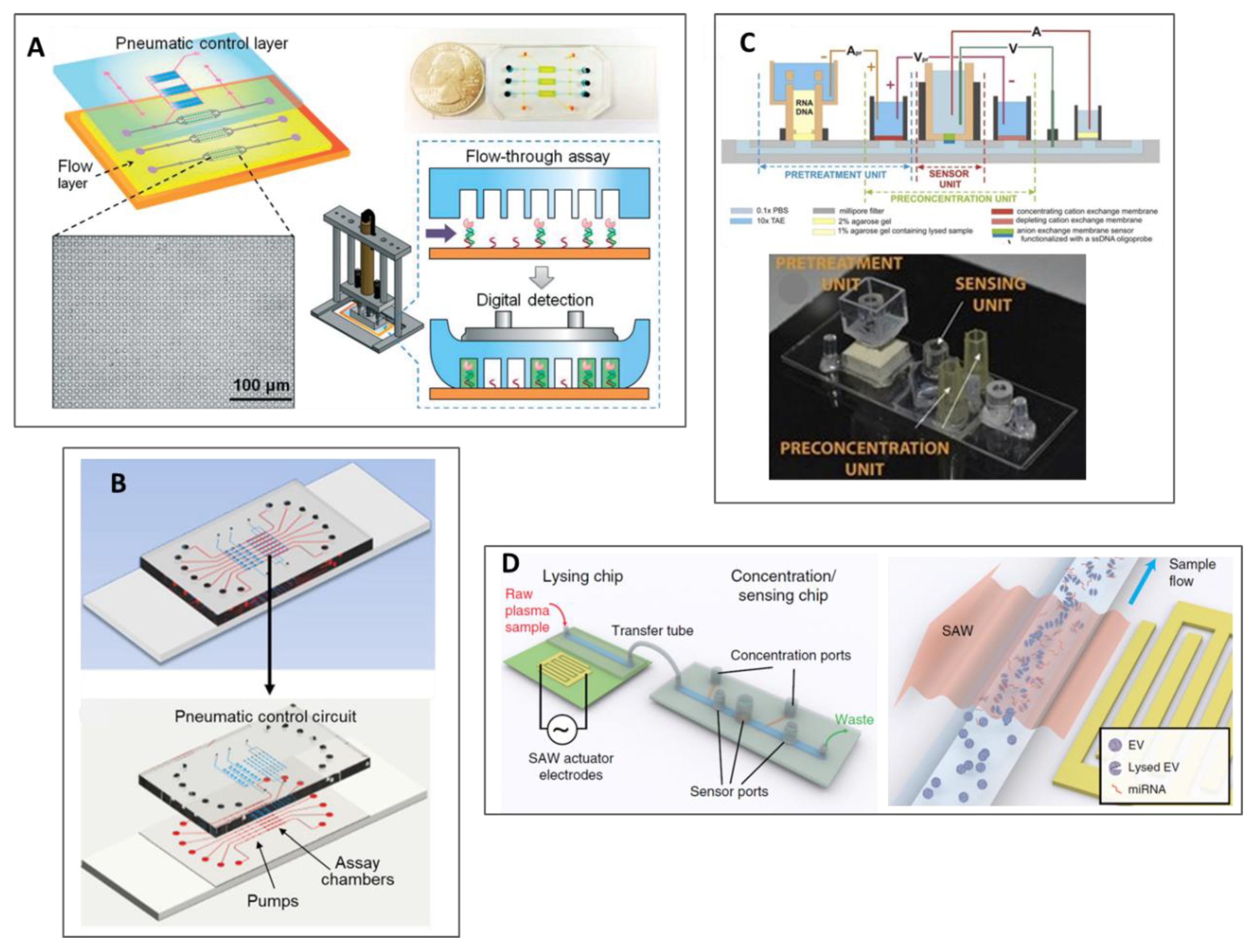
2.2. Platforms Based on Exosomes Analysis
2.3. Platforms Based on Circulant Tumor Nucleic Acids Analysis
3. Conclusions and Future Trends
Funding
Conflicts of Interest
References
- World Health Organization Global Cancer Observatory. Available online: https://gco.iarc.fr/today/home (accessed on 20 February 2020).
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Marrugo-Ramírez, J.; Mir, M.; Samitier, J. Blood-based cancer biomarkers in liquid biopsy: A promising non-invasive alternative to tissue biopsy. Int. J. Mol. Sci. 2018, 19, 2877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalli, M.; Stylianopoulos, T. Defining the Role of Solid Stress and Matrix Stiffness in Cancer Cell Proliferation and Metastasis. Front. Oncol. 2018, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, E.G.; Baxter, G. Tumour seeding following percutaneous needle biopsy: The real story! Clin. Radiol. 2011, 66, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Campos, C.D.M.; Jackson, J.M.; Witek, M.A.; Soper, S.A. Molecular Profiling of Liquid Biopsy Samples for Precision Medicine. Cancer J. 2018, 24, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Rofi, E.; Vivaldi, C.; Del Re, M.; Arrigoni, E.; Crucitta, S.; Funel, N.; Fogli, S.; Vasile, E.; Musettini, G.; Fornaro, L.; et al. The emerging role of liquid biopsy in diagnosis, prognosis and treatment monitoring of pancreatic cancer. Pharmacogenomics 2019, 20, 49–68. [Google Scholar] [CrossRef] [PubMed]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Thiery, J.P. Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef]
- Atelli, A.; Mitra, A.; Brownlee, Z.; Xia, X.; Bellister, S.; Overman, M.J.; Kopetz, S.; Ellis, L.M.; Meng, Q.H.; Li, S. Epithelial-mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin. Cancer Res. 2015, 21, 899–906. [Google Scholar] [CrossRef] [Green Version]
- Bardelli, A.; Pantel, K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017, 31, 172–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alix-Panabières, C.; Pantel, K. Circulating Tumor Cells: Liquid Biopsy of Cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef] [Green Version]
- Haber, D.A.; Velculescu, V.E. Blood-based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014, 4, 650–661. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.T.; Delijani, K.; Goldkorn, A. Current status of liquid biopsies for the detection and management of prostate cancer. Cancer Manag Res. 2019, 11, 5271–5291. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373–382. [Google Scholar] [CrossRef]
- Pantel, K.; Speicher, M.R. The biology of circulating tumor cells. Oncogene 2016, 35, 1216–1224. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating Tumor DNA as a Liquid Biopsy for Cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Ulz, P.; Geigl, J.B.; Speicher, M.R. Non-invasive detection of genome-wide somatic copy number alterations by liquid biopsies. Mol. Oncol. 2016, 10, 494–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Perakis, S.; Speicher, M.R. Emerging concepts in liquid biopsies. Bmc Med. 2017, 15, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulz, P.; Heitzer, E.; Geigl, J.B.; Speicher, M.R. Patient monitoring through liquid biopsies using circulating tumor DNA. Int. J. Cancer 2017, 141, 887–896. [Google Scholar] [CrossRef] [Green Version]
- Bach, D.-H.; Hong, J.-Y.; Park, H.J.; Lee, S.K. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int. J. Cancer 2017, 141, 220–230. [Google Scholar] [CrossRef] [Green Version]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef] [Green Version]
- Tian, W.; Liu, S.; Li, B. Potential Role of Exosomes in Cancer Metastasis. Biomed. Res. Int. 2019, 4649705, 1–15. [Google Scholar] [CrossRef]
- Zhu, Q.; Heon, M.; Zhao, Z.; He, M. Microfluidic engineering of exosomes: Editing cellular messages for precision therapeutics. Lab A Chip 2018, 18, 1690–1703. [Google Scholar] [CrossRef] [Green Version]
- Verlingue, L.; Alt, M.; Kamal, M.; Sablin, M.-P.; Zoubir, M.; Bousetta, N.; Pierga, J.-Y.; Servant, N.; Paoletti, X.; Tourneau, C.L. Challenges for the implementation of high-throughput testing and liquid biopsies in personalized medicine cancer trials. Pers. Med. 2014, 11, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Kulasinghe, A.; Wu, H.; Punyadeera, C.; Warkiani, M.E. The Use of Microfluidic Technology for Cancer Applications and Liquid Biopsy. Micromachines 2018, 9, 397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sequist, L.V.; Nagrath, S.; Toner, M.; Haber, D.A.; Lynch, T.J. The CTC-chip: An exciting new tool to detect circulating tumor cells in lung cancer patients. J. Thorac. Oncol. 2009, 4, 281–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stott, S.L.; Hsu, C.-H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Wang, Z.; Zhang, M.; Guo, W.; Li, N.; Deng, Y.; Shi, Q. A microfluidic chip with double-sided herringbone microstructures for enhanced capture of rare tumor cells. J. Mater. Chem. B 2017, 5, 9114–9120. [Google Scholar] [CrossRef]
- Xue, P.; Wu, Y.; Guo, J.; Kang, Y. Highly efficient capture and harvest of circulating tumor cells on a microfluidic chip integrated with herringbone and micropost arrays. Biomed. Microdevices 2015, 17, 39. [Google Scholar] [CrossRef]
- Chen, H.; Cao, B.; Sun, B.; Cao, Y.; Yang, K.; Lin, Y.-S. Author Correction: Highly-sensitive capture of circulating tumor cells using micro-ellipse filters. Sci. Rep. 2018, 8, 5269. [Google Scholar] [CrossRef] [Green Version]
- Yoon, H.J.; Shanker, A.; Wang, Y.; Kozminsky, M.; Jin, Q.; Palanisamy, N.; Burness, M.L.; Azizi, E.; Simeone, D.M.; Wicha, M.S.; et al. Tunable Thermal-Sensitive Polymer–Graphene Oxide Composite for Efficient Capture and Release of Viable Circulating Tumor Cells. Adv. Mater. 2016, 28, 4891–4897. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Wan, Y.; Liu, Y. Effects of nanopillar array diameter and spacing on cancer cell capture and cell behaviors. Nanoscale 2014, 6, 12482–12489. [Google Scholar] [CrossRef] [Green Version]
- Murlidhar, V.; Zeinali, M.; Grabauskiene, S.; Ghannad-Rezaie, M.; Wicha, M.S.; Simeone, D.M.; Ramnath, N.; Reddy, R.M.; Nagrath, S. A radial flow microfluidic device for ultra-high-throughput affinity-based isolation of circulating tumor cells. Small 2014, 10, 4895–4904. [Google Scholar] [CrossRef] [Green Version]
- Au, S.H.; Edd, J.; Stoddard, A.E.; Wong, K.H.K.; Fachin, F.; Maheswaran, S.; Haber, D.A.; Stott, S.L.; Kapur, R.; Toner, M. Microfluidic Isolation of Circulating Tumor Cell Clusters by Size and Asymmetry. Sci. Rep. 2017, 7, 2433. [Google Scholar] [CrossRef] [PubMed]
- Saucedo-Zeni, N.; Mewes, S.; Niestroj, R.; Gasiorowski, L.; Murawa, D.; Nowaczyk, P.; Tomasi, T.; Weber, E.; Dworacki, G.; Morgenthaler, N.G.; et al. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int. J. Oncol. 2012, 41, 1241–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor Cells Circulate in the Peripheral Blood of All Major Carcinomas but not in Healthy Subjects or Patients With Nonmalignant Diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raimondi, C.; Gradilone, A.; Naso, G.; Cortesi, E.; Gazzaniga, P. Clinical utility of circulating tumor cell counting through CellSearch(®): The dilemma of a concept suspended in Limbo. Onco. Targets Ther. 2014, 7, 619–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karabacak, N.M.; Spuhler, P.S.; Fachin, F.; Lim, E.J.; Pai, V.; Ozkumur, E.; Martel, J.M.; Kojic, N.; Smith, K.; Chen, P.-i.; et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat. Protoc. 2014, 9, 694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.S.; Sim, T.S.; Kim, Y.J.; Kim, S.S.; Jeong, H.; Park, J.-M.; Moon, H.-S.; Kim, S.I.; Gurel, O.; Lee, S.S.; et al. SSA-MOA: A novel CTC isolation platform using selective size amplification (SSA) and a multi-obstacle architecture (MOA) filter. Lab Chip 2012, 12, 2874–2880. [Google Scholar] [CrossRef]
- Chung, J.; Shao, H.; Reiner, T.; Issadore, D.; Weissleder, R.; Lee, H. Microfluidic cell sorter (μFCS) for on-chip capture and analysis of single cells. Adv. Healthc. Mater. 2012, 1, 432–436. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/adhm.201200046 (accessed on 26 February 2020). [CrossRef] [Green Version]
- Coumans, F.A.W.; van Dalum, G.; Beck, M.; Terstappen, L.W.M.M. Filtration parameters influencing circulating tumor cell enrichment from whole blood. PLoS ONE 2013, 8, e61774. [Google Scholar] [CrossRef] [Green Version]
- Andersson, E.; Steven, K.; Guldberg, P. Size-Based Enrichment of Exfoliated Tumor Cells in Urine Increases the Sensitivity for DNA-Based Detection of Bladder Cancer. PLoS ONE 2014, 9, e94023. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.; Wu, A.; Chen, X. Current detection technologies for circulating tumor cells. Chem. Soc. Rev. 2017, 46, 2038–2056. [Google Scholar] [CrossRef]
- Rosenberg, R.; Gertler, R.; Friederichs, J.; Fuehrer, K.; Dahm, M.; Phelps, R.; Thorban, S.; Nekarda, H.; Siewert, J.R. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry 2002, 49, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Warkiani, M.E.; Khoo, B.L.; Wu, L.; Tay, A.K.P.; Bhagat, A.A.S.; Han, J.; Lim, C.T. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat. Protoc. 2015, 11, 134. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Mao, Z.; Peng, Z.; Zhou, L.; Chen, Y.; Huang, P.-H.; Truica, C.I.; Drabick, J.J.; El-Deiry, W.S.; Dao, M.; et al. Acoustic separation of circulating tumor cells. Proc. Natl. Acad. Sci. USA 2015, 112, 4970–4975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cushing, K.; Undvall, E.; Ceder, Y.; Lilja, H.; Laurell, T. Reducing WBC background in cancer cell separation products by negative acoustic contrast particle immuno-acoustophoresis. Anal. Chim. Acta 2018, 1000, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Han, K.H.; Han, A.; Frazier, A.B. Microsystems for isolation and electrophysiological analysis of breast cancer cells from blood. Biosens. Bioelectron. 2006, 21, 1907–1914. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Pulikkathodi, A.K.; Ma, Y.D.; Wang, Y.-L.; Lee, G.-B.A. Microfluidic platform integrated with field-effect transistors for enumeration of circulating tumor cells. Lab Chip 2019, 19, 618–625. [Google Scholar] [CrossRef]
- Pulikkathodi, A.K.; Sarangadharan, I.; Hsu, C.-P.; Chen, Y.-H.; Hung, L.-Y.; Lee, G.-Y.; Chyi, J.-I.; Lee, G.-B.; Wang, Y.-L. Enumeration of circulating tumor cells and investigation of cellular responses using aptamer-immobilized AlGaN/GaN high electron mobility transistor sensor array. Sens. Actuators B Chem. 2018, 257, 96–104. [Google Scholar] [CrossRef]
- Gu, W.; ZhaoYi, Y. Cellular electrical impedance spectroscopy: An emerging technology of microscale biosensors. Expert Review of Medical Devices 2010, 7, 767–779. [Google Scholar] [CrossRef]
- Nguyen, N.-V.; Jen, C.-P. Impedance detection integrated with dielectrophoresis enrichment platform for lung circulating tumor cells in a microfluidic channel. Biosens. Bioelectron. 2018, 121, 10–18. [Google Scholar] [CrossRef]
- Kumeria, T.; Kurkuri, M.D.; Diener, K.R.; Parkinson, L.; Losic, D. Label-free reflectometric interference microchip biosensor based on nanoporous alumina for detection of circulating tumour cells. Biosens. Bioelectron. 2012, 35, 167–173. [Google Scholar] [CrossRef]
- Chiu, T.-K.; Chao, A.C.; Chou, W.-P.; Liao, C.-J.; Wang, H.-M.; Chang, J.-H.; Chen, P.-H.; Wu, M.-H. Optically-induced-dielectrophoresis (ODEP)-based cell manipulation in a microfluidic system for high-purity isolation of integral circulating tumor cell (CTC) clusters based on their size characteristics. Sens. Actuators B Chem. 2018, 258, 1161–1173. [Google Scholar] [CrossRef]
- Chiu, T.-K.; Lei, K.-F.; Hsieh, C.-H.; Hsiao, H.-B.; Wang, H.-M.; Wu, M.-H. Development of a microfluidic-based optical sensing device for label-free detection of circulating tumor cells (CTCs) through their lactic acid metabolism. Sensors 2015, 15, 6789–6806. [Google Scholar] [CrossRef] [PubMed]
- Mani, G.K.; Morohoshi, M.; Yasoda, Y.; Yokoyama, S.; Kimura, H.; Tsuchiya, K. ZnO-Based Microfluidic pH Sensor: A Versatile Approach for Quick Recognition of Circulating Tumor Cells in Blood. ACS Applied Mater. Interfaces 2017, 9, 5193–5203. [Google Scholar] [CrossRef] [PubMed]
- Do, L.; Thuy, H.; Bui, T.; Dau, V.; Nguyen, N.; Duc, T.; Jen, C. Dielectrophoresis Microfluidic Enrichment Platform with Built-In Capacitive Sensor for Rare Tumor Cell Detection. Biochip J. 2018, 12, 114–122. [Google Scholar] [CrossRef]
- Hendrix, A.; Hume, A.N. f development and cancer. Int. J. Dev. Biol. 2011, 55, 879–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Whiteside, T.L. The potential of tumor-derived exosomes for noninvasive cancer monitoring: An update. Expert Rev. Mol. Diagn. 2018, 18, 1029–1040. [Google Scholar] [CrossRef]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–359. [Google Scholar] [CrossRef]
- Vader, P.; Breakefield, X.O.; Wood, M.J.A. Extracellular vesicles: Emerging targets for cancer therapy. Trends Mol. Med. 2014, 20, 385–393. [Google Scholar] [CrossRef] [Green Version]
- Liga, A.; Vliegenthart, A.D.B.; Oosthuyzen, W.; Dear, J.W.; Kersaudy-Kerhoas, M. Exosome isolation: A microfluidic road-map. Lab Chip 2015, 15, 2388–2394. [Google Scholar] [CrossRef] [Green Version]
- Boukouris, S.; Mathivanan, S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteom. Clin. Appl. 2015, 9, 358–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caby, M.-P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like Vesicles are present in Human Blood Plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- André, F.; Schartz, N.E.C.; Chaput, N.; Flament, C.; Raposo, G.; Amigorena, S.; Angevin, E.; Zitvogel, L. Tumor-derived exosomes: A new source of tumor rejection antigens. Proc. Vaccine 2002, 20, A28–A31. [Google Scholar] [CrossRef]
- Antfolk, M.; Kim, S.H.; Koizumi, S.; Fujii, T.; Laurell, T. Label-free single-cell separation and imaging of cancer cells using an integrated microfluidic system. Sci. Rep. 2017, 7, 46507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, V.; Jafferji, I.; Garza, M.; Melnikova, V.O.; Hasegawa, D.K.; Pethig, R.; Davis, D.W. ApoStreamTM, a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics 2012, 6, 024133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Runz, S.; Keller, S.; Rupp, C.; Stoeck, A.; Issa, Y.; Koensgen, D.; Mustea, A.; Sehouli, J.; Kristiansen, G.; Altevogt, P. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol. Oncol. 2007, 107, 563–571. [Google Scholar] [CrossRef]
- Clayton, A.; Al-Taei, S.; Webber, J.; Mason, M.D.; Tabi, Z. Cancer Exosomes Express CD39 and CD73, Which Suppress T Cells through Adenosine Production. J. Immunol. 2011, 187, 676–683. [Google Scholar] [CrossRef]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab A Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef] [Green Version]
- Im, H.; Lee, K.; Weissleder, R.; Lee, H.; Castro, C.M. Novel nanosensing technologies for exosome detection and profiling. Lab A Chip 2017, 17, 2892–2898. [Google Scholar] [CrossRef]
- Zhao, Z.; Yang, Y.; Zeng, Y.; He, M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab A Chip 2016, 16, 489–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, S.; Park, J.; Pathania, D.; Castro, C.M.; Weissleder, R.; Lee, H. Integrated Magneto-Electrochemical Sensor for Exosome Analysis. Acs Nano 2016, 10, 1802–1809. [Google Scholar] [CrossRef] [Green Version]
- Ko, J.; Hemphill, M.A.; Gabrieli, D.; Wu, L.; Yelleswarapu, V.; Lawrence, G.; Pennycooke, W.; Singh, A.; Meaney, D.F.; Issadore, D. Smartphone-enabled optofluidic exosome diagnostic for concussion recovery. Sci. Rep. 2016, 6, 31215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebrahimi Warkiani, M.; Lou, C.-P.; Gong, H.-Q. Fabrication of multi-layer polymeric micro-sieve having narrow slot pores with conventional ultraviolet-lithography and micro-fabrication techniques. Biomicrofluidics 2011, 5, 36504–365049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warkiani, M.E.; Lou, C.-P.; Liu, H.-B.; Gong, H.-Q. A high-flux isopore micro-fabricated membrane for effective concentration and recovering of waterborne pathogens. Biomed. Microdevices 2012, 14, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Davies, R.T.; Kim, J.; Jang, S.C.; Choi, E.-J.; Gho, Y.S.; Park, J. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab A Chip 2012, 12, 5202–5210. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.; Jo, W.; Heo, Y.; Kang, J.Y.; Kwak, R.; Park, J. Isolation of extracellular vesicle from blood plasma using electrophoretic migration through porous membrane. Sens. Actuators B Chem. 2016, 233, 289–297. [Google Scholar] [CrossRef]
- Contreras-Naranjo, J.C.; Wu, H.-J.; Ugaz, V.M. Microfluidics for exosome isolation and analysis: Enabling liquid biopsy for personalized medicine. Lab A Chip 2017, 17, 3558–3577. [Google Scholar] [CrossRef]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A., Jr.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef] [Green Version]
- Egatz-Gomez, A.; Wang, C.; Klacsmann, F.; Pan, Z.; Marczak, S.; Wang, Y.; Sun, G.; Senapati, S.; Chang, H.C. Future microfluidic and nanofluidic modular platforms for nucleic acid liquid biopsy in precision medicine. Biomicrofluidics 2016, 10, 032902. [Google Scholar] [CrossRef]
- Palmieri, V.; Lucchetti, D.; Gatto, I.; Maiorana, A.; Marcantoni, M.; Maulucci, G.; Papi, M.; Pola, R.; De Spirito, M.; Sgambato, A. Dynamic light scattering for the characterization and counting of extracellular vesicles: A powerful noninvasive tool. J. Nanoparticle Res. 2014, 9, 2583. [Google Scholar] [CrossRef]
- Wiklander, O.P.B.; Bostancioglu, R.B.; Welsh, J.A.; Zickler, A.M.; Murke, F.; Corso, G.; Felldin, U.; Hagey, D.W.; Evertsson, B.; Liang, X.M.; et al. Systematic methodological evaluation of a multiplex bead-based flow cytometry assay for detection of extracellular vesicle surface signatures. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sina, A.A.I.; Vaidyanathan, R.; Dey, S.; Carrascosa, L.G.; Shiddiky, M.J.A.; Trau, M. Real time and label free profiling of clinically relevant exosomes. Sci. Rep. 2016, 6, 30460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pugholm, L.-H.; Revenfeld, A.-L.-S.; Jørgensen, M.-M. Antibody-Based Assays for Phenotyping of Extracellular Vesicles. Biomed Res Int. 2015, 524817, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Cavallaro, S.; Horak, J.; HÅÅg, P.; Gupta, D.; Stiller, C.; Sahu, S.S.; Görgens, A.; Gatty, H.K.; Viktorsson, K.; El Andaloussi, S.; et al. Label-Free Surface Protein Profiling of Extracellular Vesicles by an Electrokinetic Sensor. Acs Sens. 2019, 4, 1399–1408. [Google Scholar] [CrossRef]
- Guo, S.C.; Tao, S.C.; Dawn, H. Microfluidics-based on-a-chip systems for isolating and analysing extracellular vesicles. J. Extracell. Vesicles 2018, 7, 1508271. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.; Deng, C.-X. Current Progresses of Exosomes as Cancer Diagnostic and Prognostic Biomarkers. Int J Biol Sci. 2019, 15, 1–11. [Google Scholar] [CrossRef]
- Vaidyanathan, R.; Naghibosadat, M.; Rauf, S.; Korbie, D.; Carrascosa, L.G.; Shiddiky, M.J.A.; Trau, M. Detecting exosomes specifically: A multiplexed device based on alternating current electrohydrodynamic induced nanoshearing. Anal. Chem. 2014, 86, 11125–11132. [Google Scholar] [CrossRef] [Green Version]
- Akagi, T.; Kato, K.; Kobayashi, M.; Kosaka, N.; Ochiya, T.; Ichiki, T. On-chip immunoelectrophoresis of extracellular vesicles released from human breast cancer cells. PLoS ONE 2015, 10, e0123603. [Google Scholar] [CrossRef]
- Shao, H.; Chung, J.; Lee, K.; Balaj, L.; Min, C.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Lee, H.; Weissleder, R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 2015, 6, 6999. [Google Scholar] [CrossRef] [Green Version]
- Oliveira-Rodríguez, M.; López-Cobo, S.; Reyburn, H.T.; Costa-García, A.; López-Martín, S.; Yáñez-Mó, M.; Cernuda-Morollón, E.; Paschen, A.; Valés-Gómez, M.; Blanco-López, M.C. Development of a rapid lateral flow immunoassay test for detection of exosomes previously enriched from cell culture medium and body fluids. J. Extracell. Vesicles 2016, 5, 31803. [Google Scholar] [CrossRef] [PubMed]
- Gagni, P.; Cretich, M.; Benussi, L.; Tonoli, E.; Ciani, M.; Ghidoni, R.; Santini, B.; Galbiati, E.; Prosperi, D.; Chiari, M. Combined mass quantitation and phenotyping of intact extracellular vesicles by a microarray platform. Anal. Chim. Acta 2016, 902, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liao, C.; Zuo, P.; Liu, Z.; Ye, B.C. Magnetic-Based Microfluidic Device for On-Chip Isolation and Detection of Tumor-Derived Exosomes. Anal. Chem. 2018, 90, 13451–13458. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cheng, S.B.; Cao, P.; Qiu, Q.F.; Chen, Y.; Xie, M.; Xu, Y.; Huang, W.H. Detection of exosomes by ZnO nanowires coated three-dimensional scaffold chip device. Biosens. Bioelectron. 2018, 122, 211–216. [Google Scholar] [CrossRef]
- Ortega, F.G.; Piguillem, S.V.; Messina, G.A.; Tortella, G.R.; Rubilar, O.; Jiménez Castillo, M.I.; Lorente, J.A.; Serrano, M.J.; Raba, J.; Fernández Baldo, M.A. EGFR detection in extracellular vesicles of breast cancer patients through immunosensor based on silica-chitosan nanoplatform. Talanta 2019, 194, 243–252. [Google Scholar] [CrossRef]
- He, M.; Crow, J.; Roth, M.; Zeng, Y.; Godwin, A.K. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip 2014, 14, 3773. Available online: https://pubs.rsc.org/en/content/articlehtml/2014/lc/c4lc00662c (accessed on 26 February 2020). [CrossRef] [Green Version]
- Chen, C.; Skog, J.; Hsu, C.H.; Lessard, R.T.; Balaj, L.; Wurdinger, T.; Carter, B.S.; Breakefield, X.O.; Toner, M.; Irimia, D. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip 2010, 10, 505–511. [Google Scholar] [CrossRef] [Green Version]
- Ng, A.H.C.; Uddayasankar, U.; Wheeler, A.R. Immunoassays in microfluidic systems. Anal. Bioanal. Chem. 2010, 397, 991–1007. [Google Scholar] [CrossRef]
- Liang, L.G.; Kong, M.Q.; Zhou, S.; Sheng, Y.F.; Wang, P.; Yu, T.; Inci, F.; Kuo, W.P.; Li, L.J.; Demirci, U.; et al. An integrated double-filtration microfluidic device for isolation, enrichment and quantification of urinary extracellular vesicles for detection of bladder cancer. Sci. Rep. 2017, 7, 46224. Available online: https://www.nature.com/articles/srep46224#rightslink (accessed on 26 February 2020). [CrossRef]
- Peters, D.L.; Pretorius, P.J. Origin, translocation and destination of extracellular occurring DNA—A new paradigm in genetic behaviour. Clin. Chim. Acta 2011, 412, 806–811. [Google Scholar] [CrossRef]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, ra24–ra224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, D.; Huang, C.; Zheng, J.; Tang, J.; Li, J.; Yang, J.; Yang, R. Quantitative detection of exosomal microRNA extracted from human blood based on surface-enhanced Raman scattering. Biosens. Bioelectron. 2018, 101, 167–173. [Google Scholar] [CrossRef]
- Boriachek, K.; Umer, M.; Islam, M.N.; Gopalan, V.; Lam, A.K.; Nguyen, N.T.; Shiddiky, M.J.A. An amplification-free electrochemical detection of exosomal miRNA-21 in serum samples. Analyst 2018, 143, 1662–1669. [Google Scholar] [CrossRef] [PubMed]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 26, 3–23743. [Google Scholar] [CrossRef] [PubMed]
- Chapin, S.C.; Doyle, P.S. Ultrasensitive multiplexed microRNA quantification on encoded gel microparticles using rolling circle amplification. Anal. Chem. 2011, 83, 7179–7185. [Google Scholar] [CrossRef] [Green Version]
- Goldman, J.M.; Zhang, L.A.; Manna, A.; Armitage, B.A.; Ly, D.H.; Schneider, J.W. High affinity γpNA sandwich hybridization assay for rapid detection of short nucleic acid targets with single mismatch discrimination. Biomacromolecules 2013, 14, 2253–2261. [Google Scholar] [CrossRef]
- Graybill, R.M.; Bailey, R.C. Emerging Biosensing Approaches for microRNA Analysis. Anal. Chem. 2016, 88, 431–450. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Crow, J.; Lella, D.; Zhou, X.; Samuel, G.; Godwin, A.K.; Zeng, Y. Ultrasensitive quantification of tumor mRNAs in extracellular vesicles with an integrated microfluidic digital analysis chip. Lab Chip 2018, 18, 3790–3801. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, M.; Dreher, D.D.; Zeng, Y. Ultrasensitive microfluidic solid-phase ELISA using an actuatable microwell-patterned PDMS chip. Lab Chip 2013, 13, 4190. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Zhou, X.; Zeng, Y. Microfluidic exponential rolling circle amplification for sensitive microRNA detection directly from biological samples. Sens. Actuators B Chem. 2019, 279, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Slouka, Z.; Senapati, S.; Shah, S.; Lawler, R.; Shi, Z.; Stack, M.S.; Chang, H.C.; Integrated, D.C. voltage-driven nucleic acid diagnostic platform for real sample analysis: Detection of oral cancer. Talanta 2015, 145, 35–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramshani, Z.; Zhang, C.; Richards, K.; Chen, L.; Xu, G.; Stiles, B.L.; Hill, R.; Senapati, S.; Go, D.B.; Chang, H.-C. Extracellular vesicle microRNA quantification from plasma using an integrated microfluidic device. Commun. Biol. 2019, 2, 189. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sierra, J.; Marrugo-Ramírez, J.; Rodriguez-Trujillo, R.; Mir, M.; Samitier, J. Sensor-Integrated Microfluidic Approaches for Liquid Biopsies Applications in Early Detection of Cancer. Sensors 2020, 20, 1317. https://doi.org/10.3390/s20051317
Sierra J, Marrugo-Ramírez J, Rodriguez-Trujillo R, Mir M, Samitier J. Sensor-Integrated Microfluidic Approaches for Liquid Biopsies Applications in Early Detection of Cancer. Sensors. 2020; 20(5):1317. https://doi.org/10.3390/s20051317
Chicago/Turabian StyleSierra, Jessica, José Marrugo-Ramírez, Romen Rodriguez-Trujillo, Mònica Mir, and Josep Samitier. 2020. "Sensor-Integrated Microfluidic Approaches for Liquid Biopsies Applications in Early Detection of Cancer" Sensors 20, no. 5: 1317. https://doi.org/10.3390/s20051317
APA StyleSierra, J., Marrugo-Ramírez, J., Rodriguez-Trujillo, R., Mir, M., & Samitier, J. (2020). Sensor-Integrated Microfluidic Approaches for Liquid Biopsies Applications in Early Detection of Cancer. Sensors, 20(5), 1317. https://doi.org/10.3390/s20051317





