Estimating the Growing Stem Volume of Coniferous Plantations Based on Random Forest Using an Optimized Variable Selection Method
Abstract
:1. Introduction
2. Materials and Methods
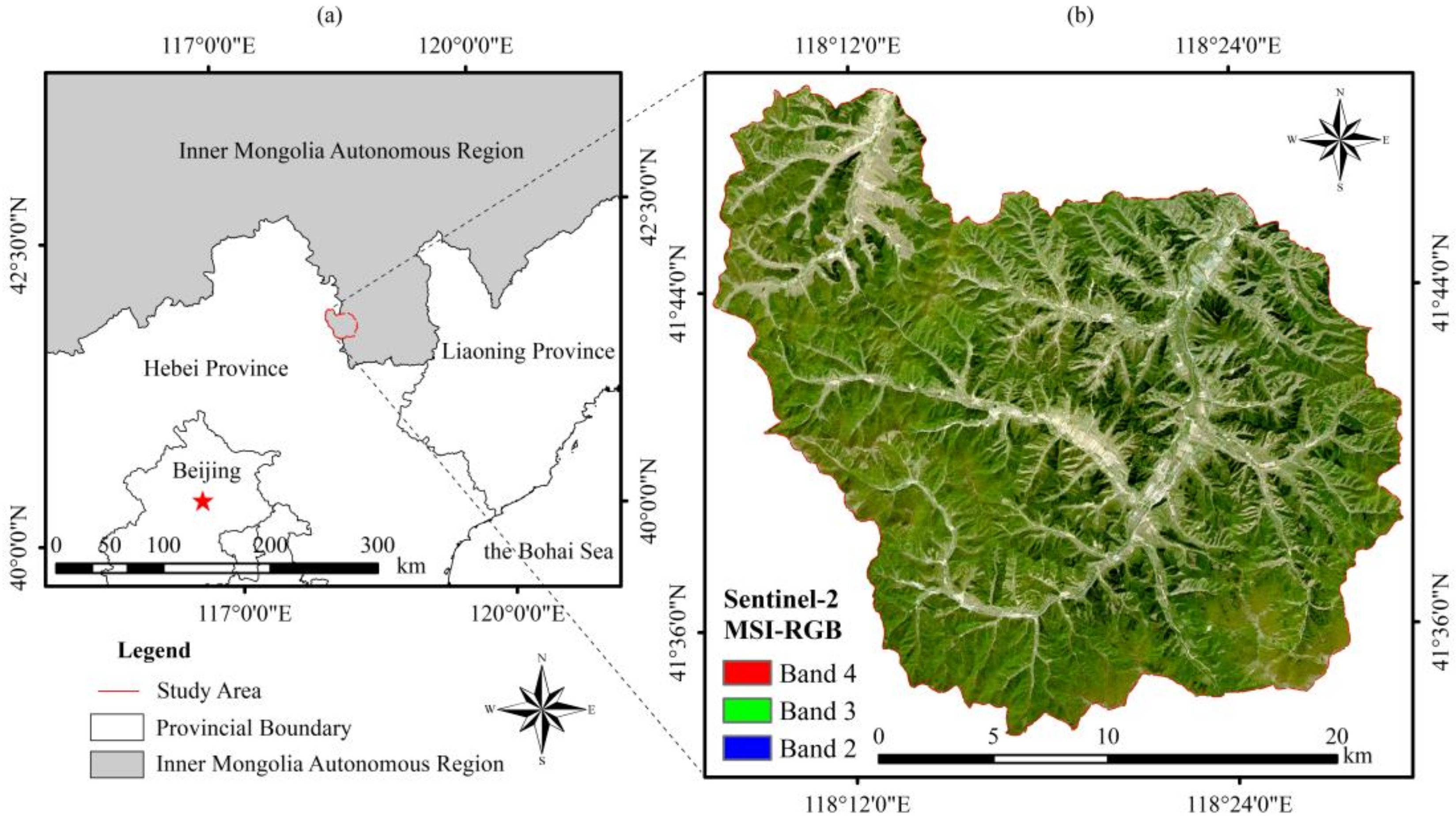
2.1. General Description of the Study Area
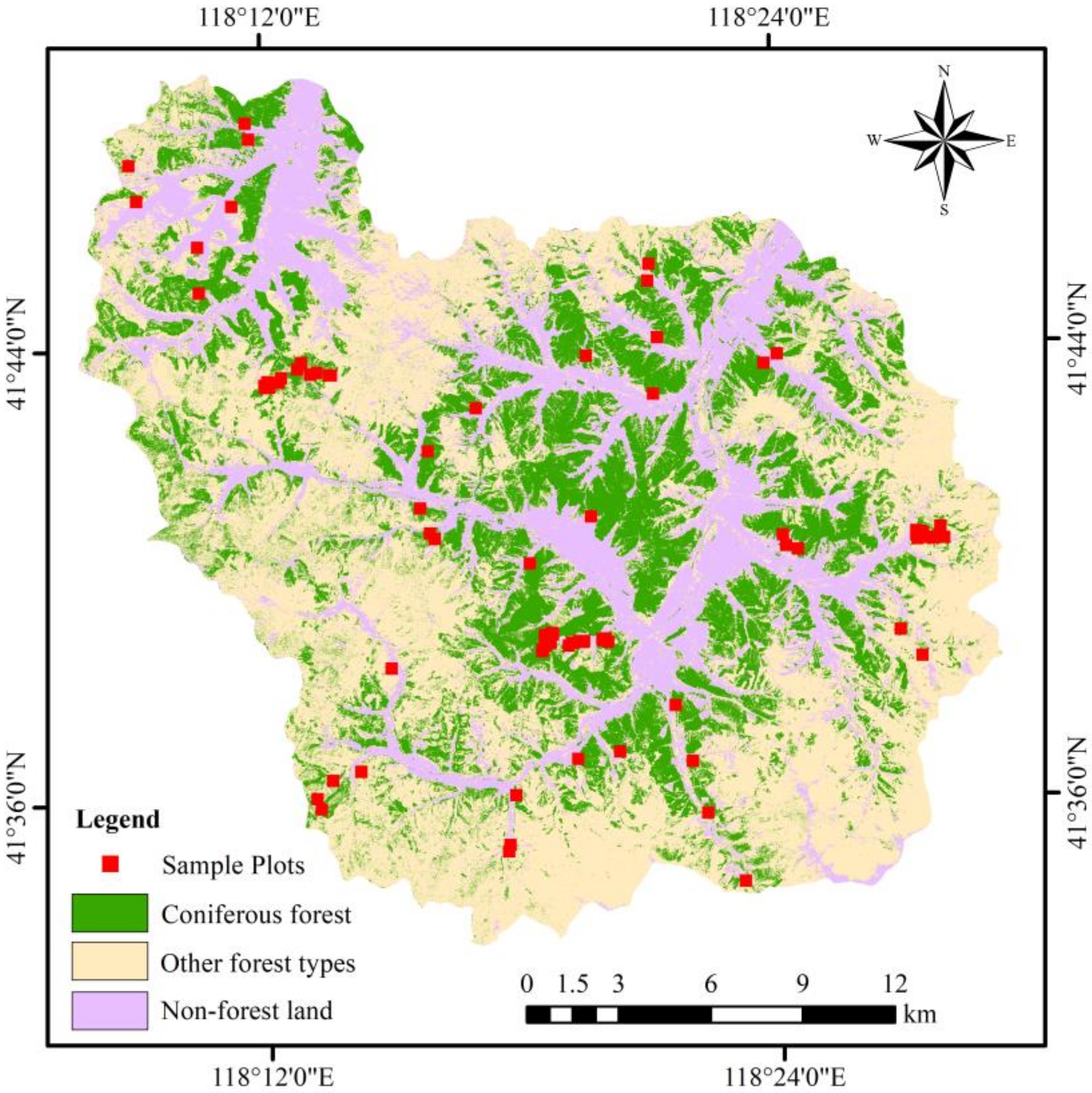
2.2. Sampling Design and GSV Survey
2.3. Remote Sensing Data and Preprocessing
2.4. Extraction and Selection of Spectral Variables and Topographic Factors
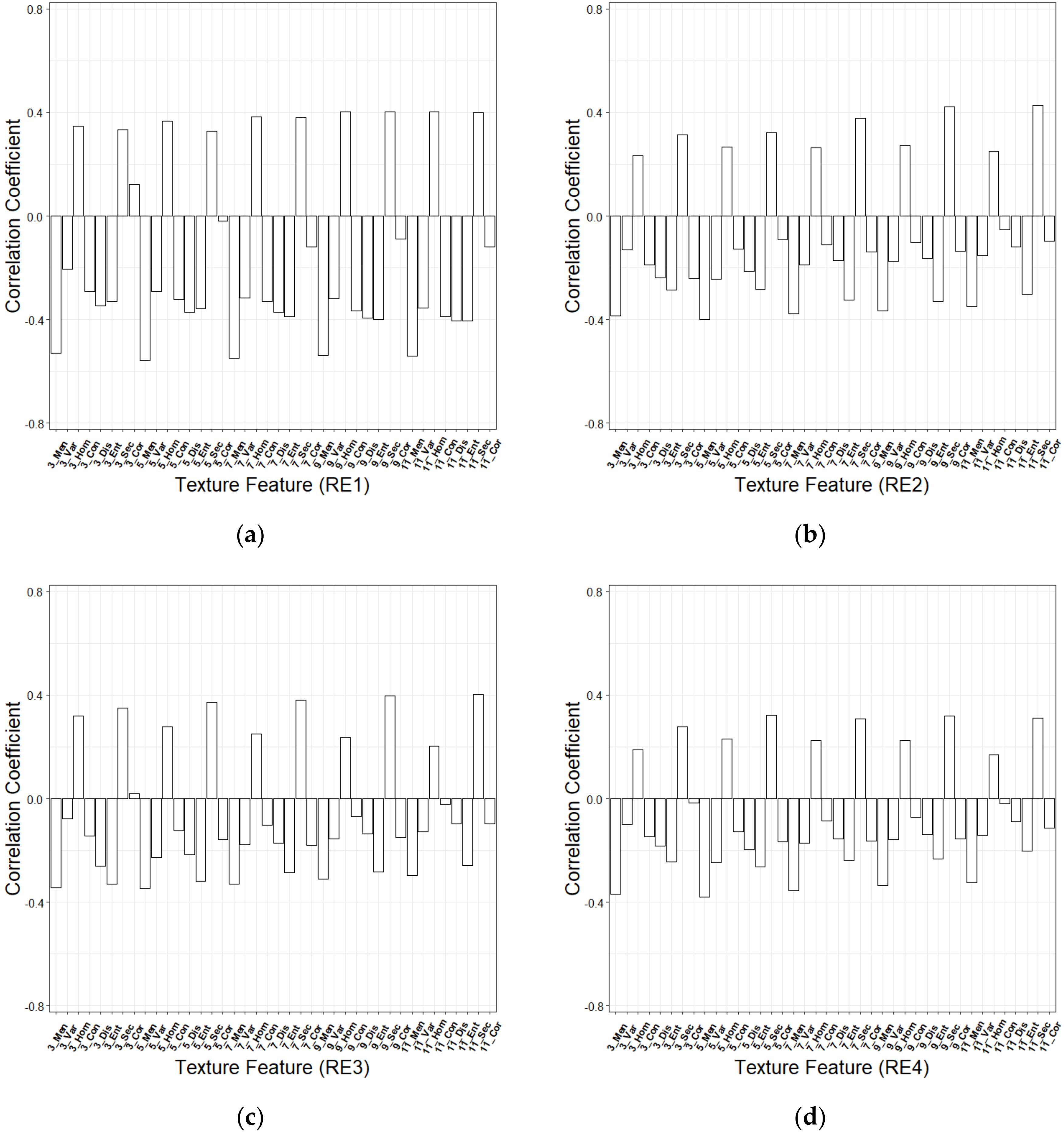
2.5. Extraction of the Texture Feature of Red-Edge Bands
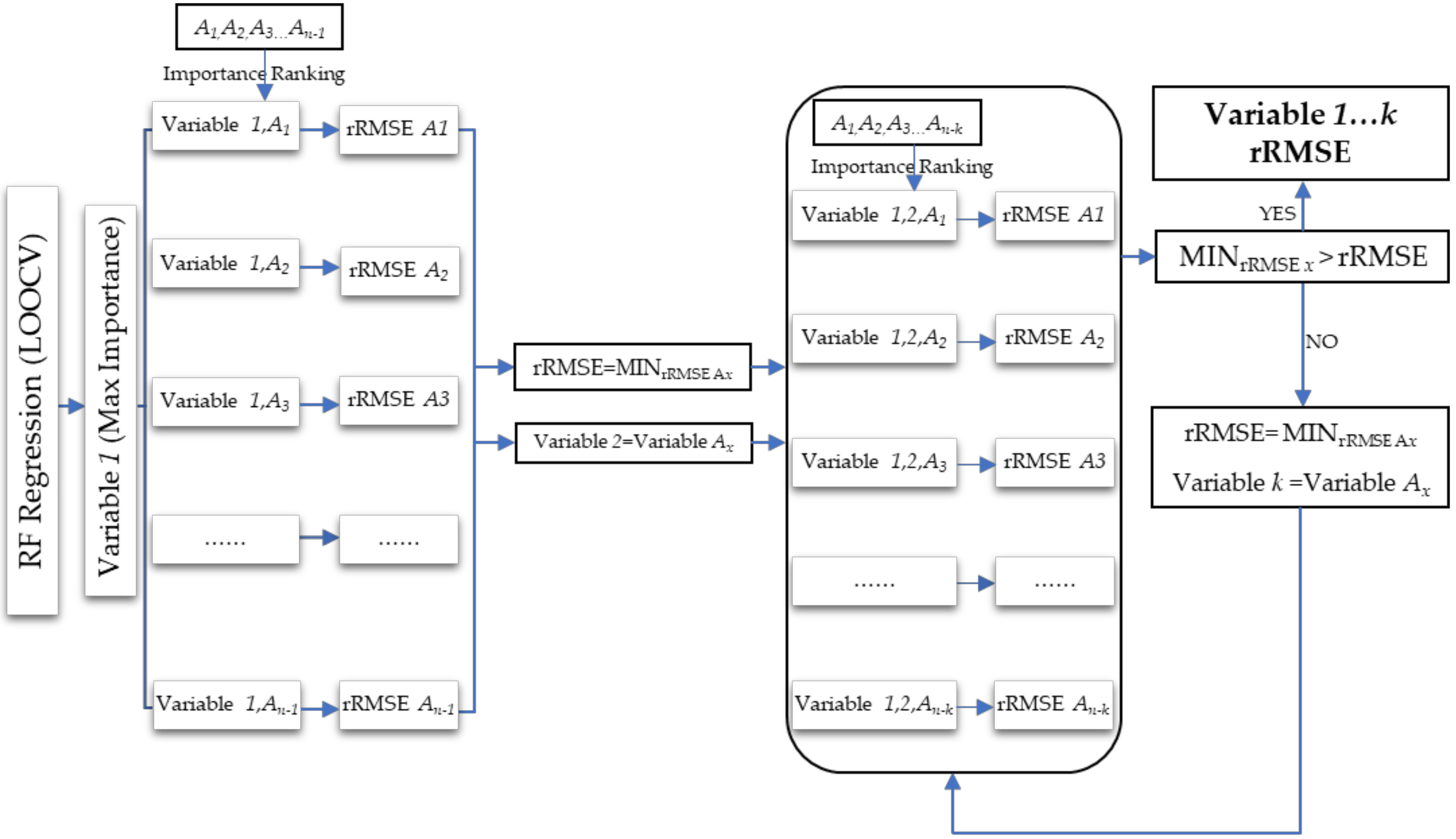
2.6. Random Forest Regression for GSV Estimation
2.7. Accuracy Assessment of GSV Estimation
3. Results
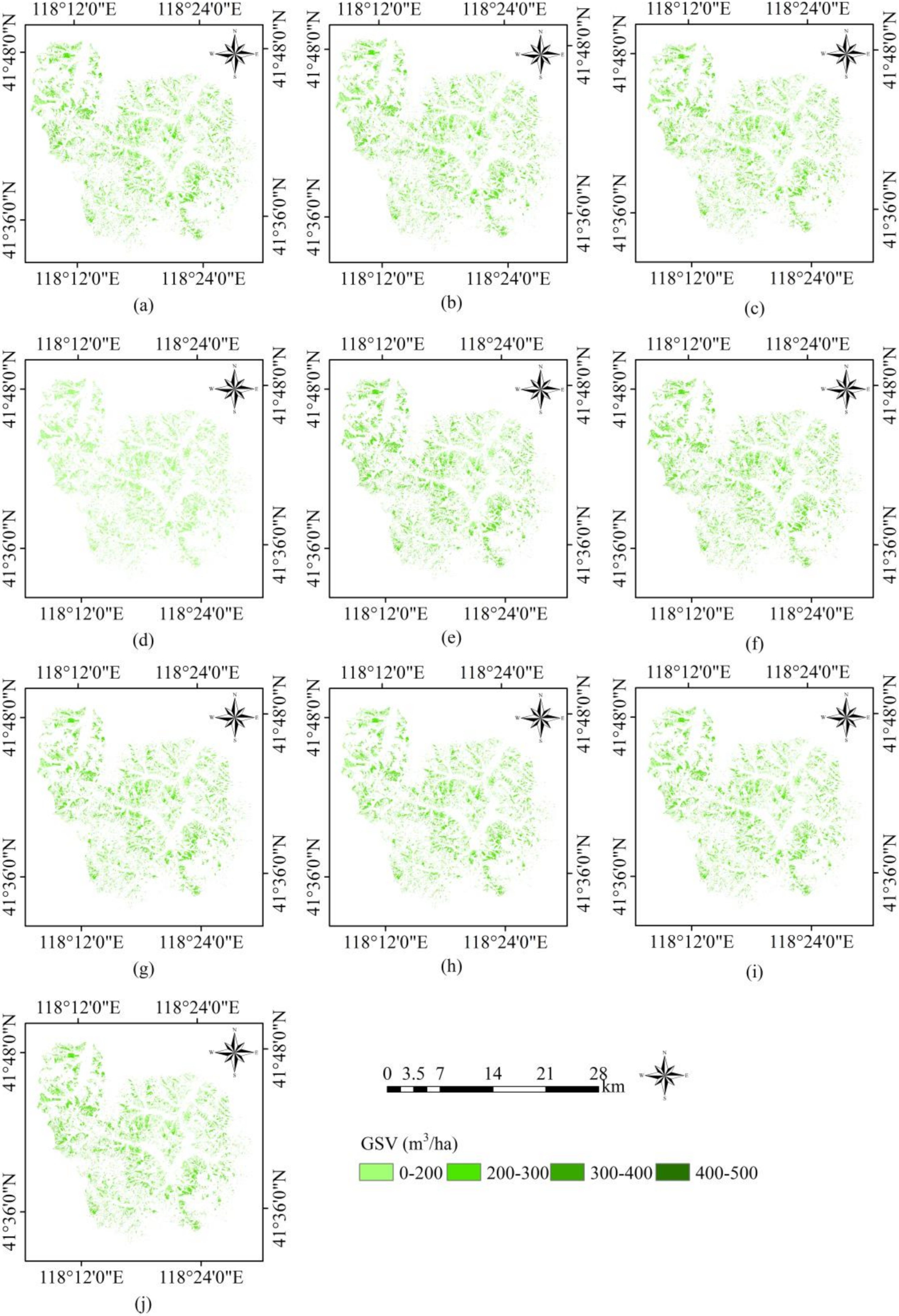
3.1. GSV Estimation and Mapping
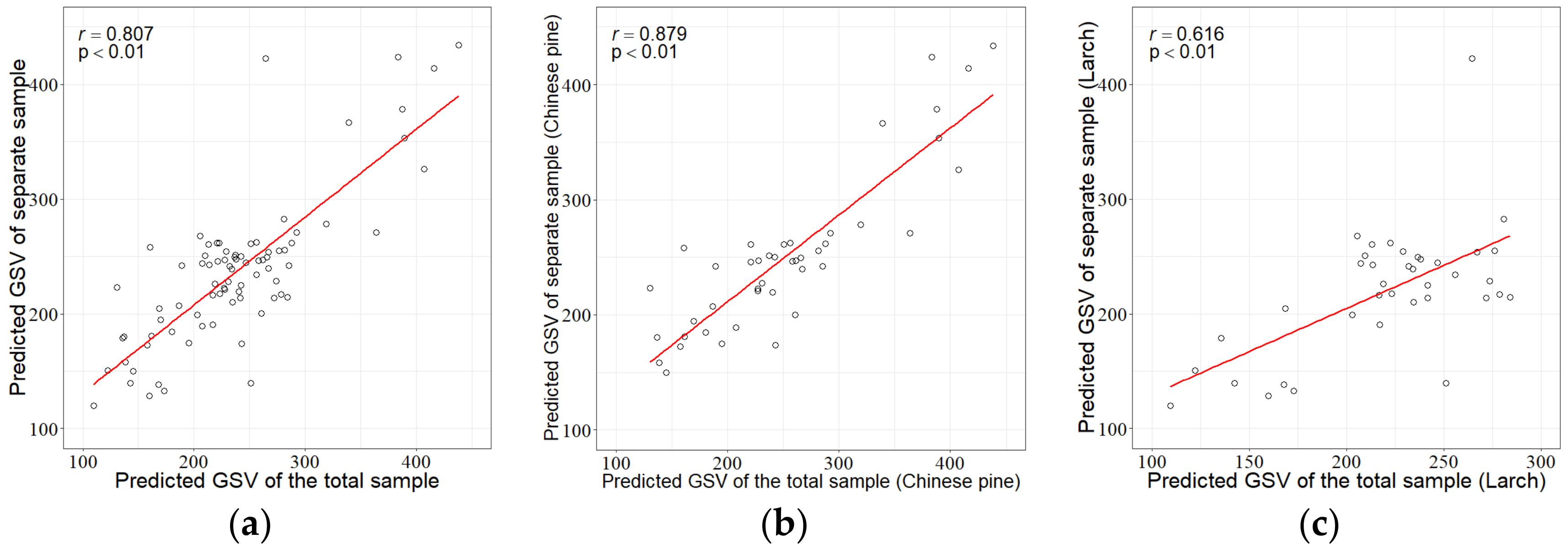
3.2. Uncertainty Analysis
4. Discussion
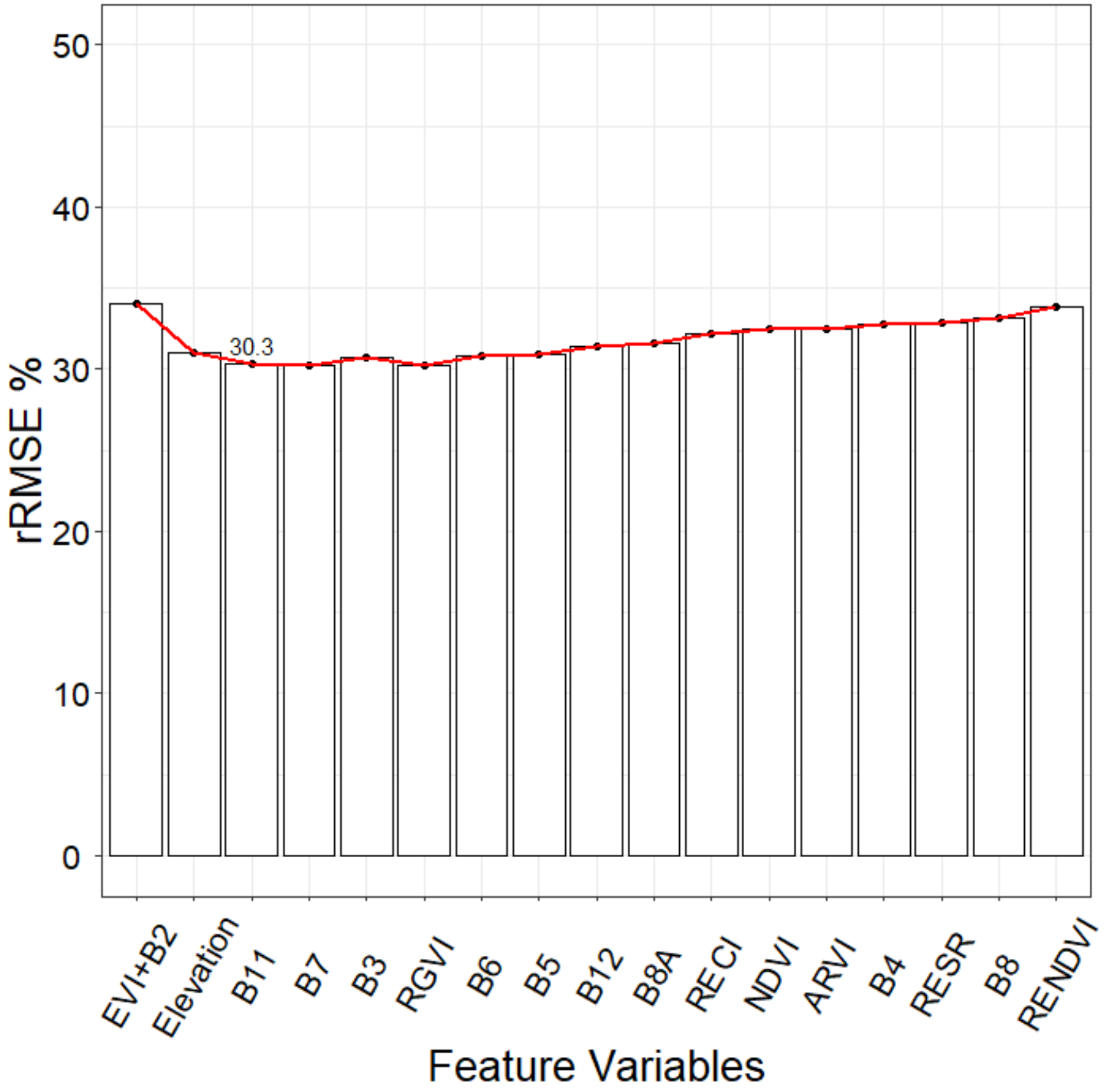
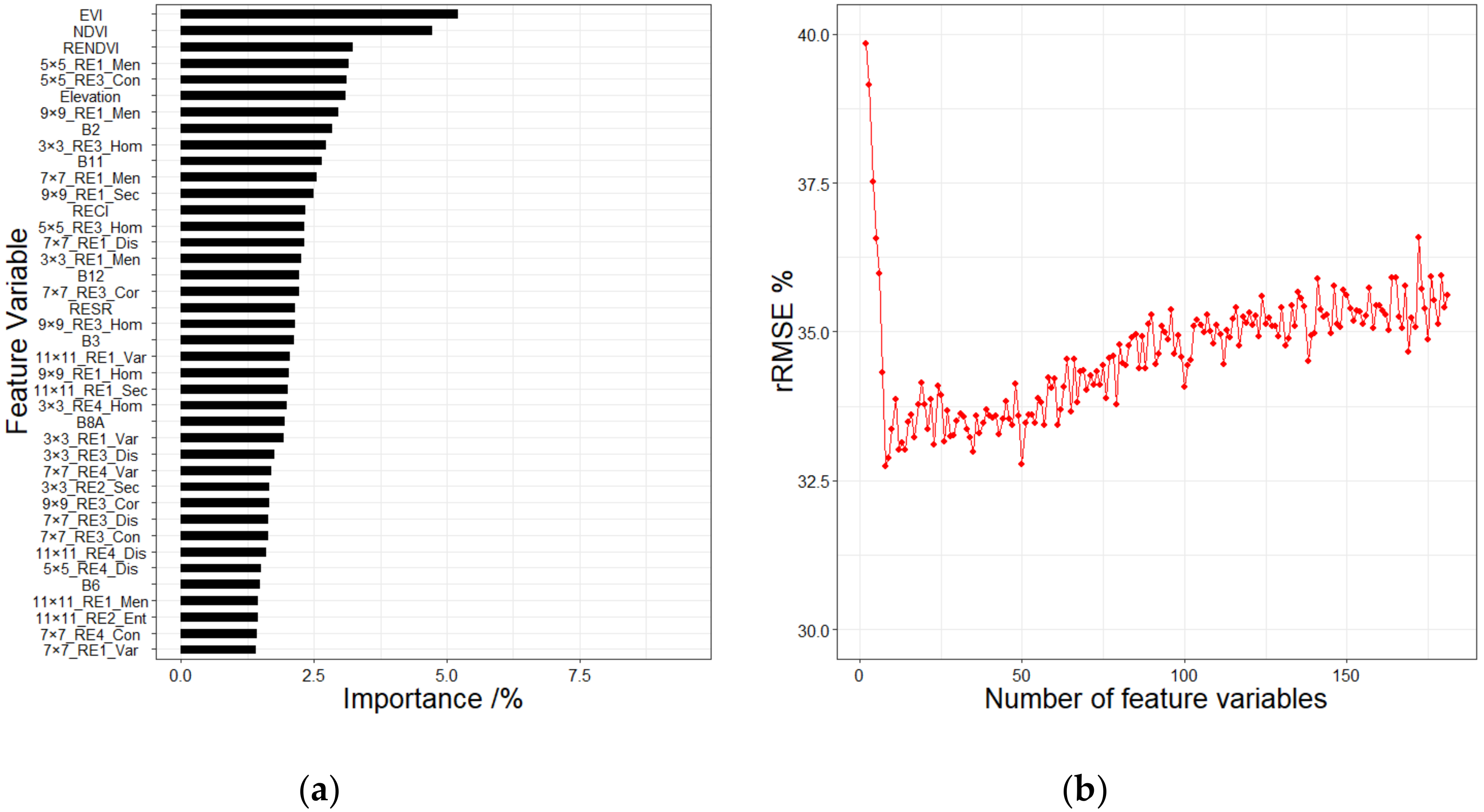
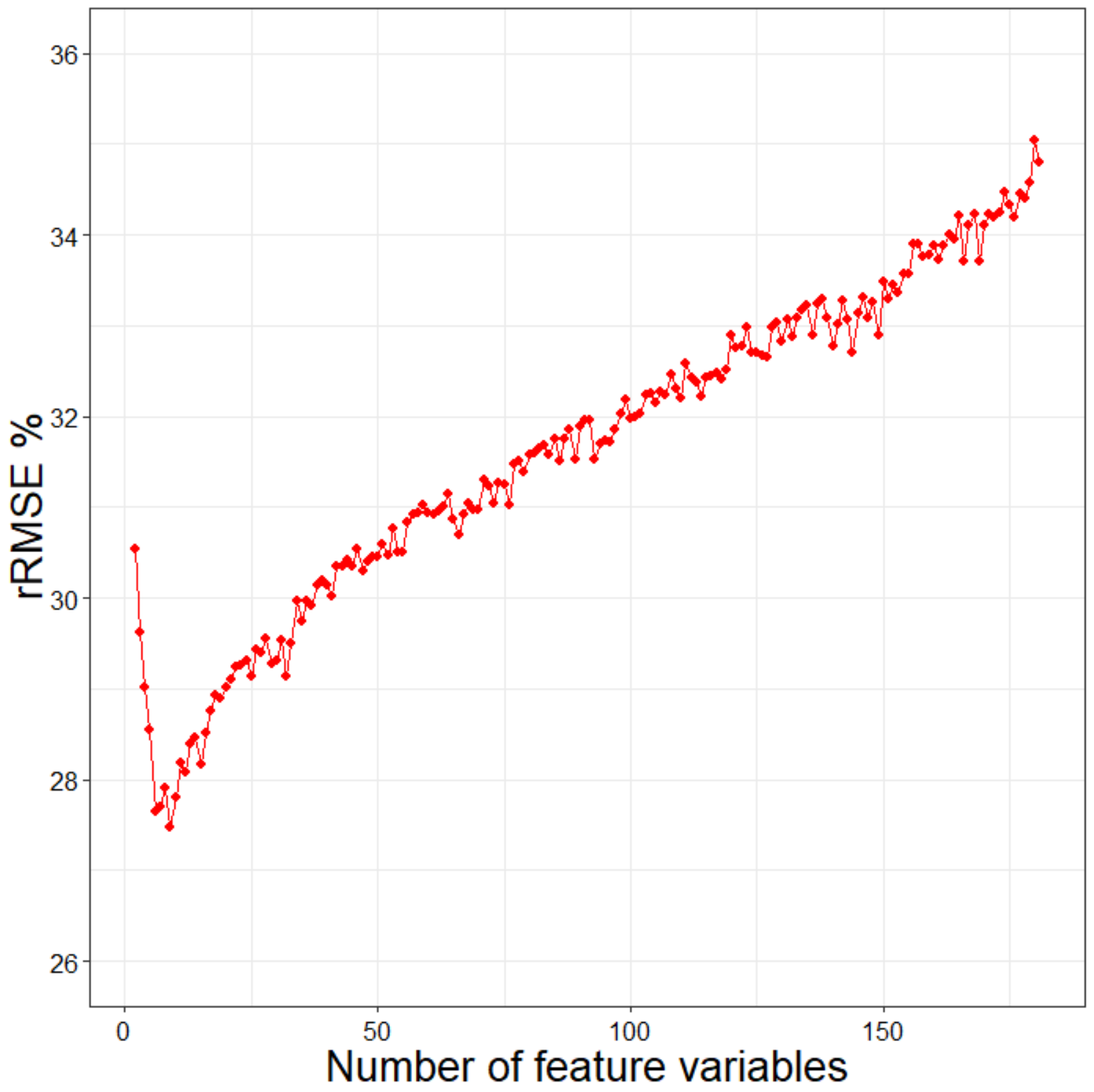
4.1. Feature Variable Selection Method
4.2. Uncertainty Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gower, S.T. Patterns and mechanisms of the forest carbon cycle. Ann. Rev. Environ. Resour. 2003, 28, 169–204. [Google Scholar] [CrossRef]
- Pedro, M.S.; Rammer, W.; Seidl, R. Tree species diversity mitigates disturbance impacts on the forest carbon cycle. Oecologia 2015, 177, 619–630. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Wang, C.; Lin, H.; Long, J.; Zhao, L.; Fu, H.; Liu, Z. Forest Growing Stock Volume Estimation in Subtropical Mountain Areas Using PALSAR-2 L-Band PolSAR Data. Forests 2019, 10, 276. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.; Kim, S.; Hong, S.; Kim, K.; Kim, E.; Im, J.; Heo, J. Effects of national forest inventory plot location error on forest carbon stock estimation using k-nearest neighbor algorithm. ISPRS J. Photogramm. Remote Sens. 2013, 81, 82–92. [Google Scholar] [CrossRef]
- Condés, S.; McRoberts, R.E. Updating national forest inventory estimates of growing stock volume using hybrid inference. For. Ecol. Manag. 2017, 400, 48–57. [Google Scholar] [CrossRef]
- Di Cosmo, L.; Gasparini, P.; Tabacchi, G. A national-scale, stand-level model to predict total above-ground tree biomass from growing stock volume. For. Ecol. Manag. 2016, 361, 269–276. [Google Scholar] [CrossRef]
- Brigot, G.; Colin-Koeniguer, E.; Plyer, A.; Janez, F. Adaptation and Evaluation of an Optical Flow Method Applied to Coregistration of Forest Remote Sensing Images. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 2923–2939. [Google Scholar] [CrossRef] [Green Version]
- Klinge, M.; Böhner, J.; Erasmi, S. Modelling forest lines and forest distribution patterns with remote sensing data in a mountainous region of semi-arid Central Asia. Biogeosci. Discuss. 2014, 11, 14667–14698. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Fehrmann, L.; Guan, F.; Forrester, D.I.; Guisasola, R.; Kleinn, C. Inventory-based estimation of forest biomass in Shitai County, China: A comparison of five methods. Ann. For. Res. 2016, 59, 269–280. [Google Scholar] [CrossRef] [Green Version]
- Isaacson, B.N.; SerbiniD, S.; Townsend, P.A. Detection of relative differences in phenology of forest species using Landsat and MODIS. Landsc. Ecol. 2012, 27, 529–543. [Google Scholar] [CrossRef]
- Amiro, B.D.; Chen, J.M. Forest-fire-scar aging using SPOT-VEGETATION for Canadian ecoregions. Can. J. For. Res. 2003, 33, 1116–1125. [Google Scholar] [CrossRef]
- Guo, X.-Y.; Zhang, H.-Y.; Wu, Z.; Zhao, J.-J.; Zhang, Z.-X. Comparison and Evaluation of Annual NDVI Time Series in China Derived from the NOAA AVHRR LTDR and Terra MODIS MOD13C1 Products. Sensors 2017, 17, 1298. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.S.; Tang, Y.; Qin, L.; Dai, H.S.; Zhao, Q.C.; Yang, Y. On-board radiometric calibration for thermal emission band of FY-3C/MERSI. Int. J. Remote Sens. 2019, 40, 1–14. [Google Scholar] [CrossRef]
- Pu, R.; Gong, P.; Yu, Q. Comparative Analysis of EO-1 ALI and Hyperion, and Landsat ETM+ Data for Mapping Forest Crown Closure and Leaf Area Index. Sensors 2008, 8, 3744–3766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacquemoud, S. Inversion of the PROSPECT + SAIL canopy reflectance model from AVIRIS equivalent spectra: Theoretical study. Remote Sens. Environ. 1993, 44, 281–292. [Google Scholar] [CrossRef]
- Zhong, Y.; Zhao, J.; Zhang, L. A Hybrid Object-Oriented Conditional Random Field Classification Framework for High Spatial Resolution Remote Sensing Imagery. IEEE Trans. Geosci. Remote Sens. 2014, 52, 7023–7037. [Google Scholar] [CrossRef]
- Chrysafis, I.; Mallinis, G.; Gitas, I.Z.; Tsakiri-Strati, M. Estimating Mediterranean forest parameters using multi seasonal Landsat 8 OLI imagery and an ensemble learning method. Remote Sens. Environ. 2017, 199, 154–166. [Google Scholar] [CrossRef]
- Chaozong, X.; Yuxing, Z.; Wei, W. A relief-based forest cover change extraction using GF-1 images. IEEE Geosci. Remote Sens. Symp. 2014, 4212–4215. [Google Scholar] [CrossRef]
- Inman, D.; Khosla, R.; Reich, R.M.; Westfall, D.G. Active remote sensing and grain yield in irrigated maize. Precis. Agric. 2007, 8, 241–252. [Google Scholar] [CrossRef]
- Lin, C.; Thomson, G.; Popescu, S. An IPCC-Compliant Technique for Forest Carbon Stock Assessment Using Airborne LiDAR-Derived Tree Metrics and Competition Index. Remote Sens. 2016, 8, 528. [Google Scholar] [CrossRef] [Green Version]
- Hakkenberg, C.R.; Zhu, K.; Peet, R.K.; Song, C. Mapping multi-scale vascular plant richness in a forest landscape with integrated LiDAR and hyperspectral remote-sensing. Ecology 2018, 99, 474–487. [Google Scholar] [CrossRef] [PubMed]
- Thiel, C.; Schmullius, C. The potential of ALOS PALSAR backscatter and InSAR coherence for forest growing stock volume estimation in Central Siberia. Remote Sens. Environ. 2016, 173, 258–273. [Google Scholar] [CrossRef]
- Mahdianpari, M.; Salehi, B.; Mohammadimanesh, F.; Motagh, M. Random forest wetland classification using ALOS-2 L-band, RADARSAT-2 C-band, and TerraSAR-X imagery. ISPRS J. Photogramm. Remote Sens. 2017, 130, 13–31. [Google Scholar] [CrossRef]
- Majasalmi, T.; Rautiainen, M. The potential of Sentinel-2 data for estimating biophysical variables in a boreal forest: A simulation study. Remote Sens. Lett. 2016, 7, 427–436. [Google Scholar] [CrossRef]
- Mura, M.; Bottalico, F.; Giannetti, F.; Bertani, R.; Giannini, R.; Mancini, M.; Orlandini, S.; Travaglini, D.; Chirici, G. Exploiting the capabilities of the Sentinel-2 multi spectral instrument for predicting growing stock volume in forest ecosystems. Int. J. Appl. Earth Obs. Geoinf. 2018, 66, 126–134. [Google Scholar] [CrossRef]
- Chrysafis, I.; Mallinis, G.; Siachalou, S.; Patias, P. Assessing the relationships between growing stock volume and Sentinel-2 imagery in a Mediterranean forest ecosystem. Remote Sens. Lett. 2017, 8, 508–517. [Google Scholar] [CrossRef]
- Hu, Y.; Xu, X.; Wu, F.; Sun, Z.; Xia, H.; Meng, Q.; Huang, W.; Zhou, H.; Gao, J.; Li, W.; et al. Estimating Forest Stock Volume in Hunan Province, China, by Integrating in Situ Plot Data, Sentinel-2 Images, and Linear and Machine Learning Regression Models. Remote Sens. 2020, 12, 186. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Li, L.; Lu, D.; Li, D. Exploring Bamboo Forest Aboveground Biomass Estimation Using Sentinel-2 Data. Remote Sens. 2018, 11, 7. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Lu, D.; Li, G.; Wang, G.; Chen, Q.; Liu, L.; Li, D. Comparative Analysis of Modeling Algorithms for Forest Aboveground Biomass Estimation in a Subtropical Region. Remote Sens. 2018, 10, 627. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Ge, H.; Lu, D.; Zhang, M.; Lai, Z.; Yao, R. Comparative Study on Variable Selection Approaches in Establishment of Remote Sensing Model for Forest Biomass Estimation. Remote Sens. 2019, 11, 1437. [Google Scholar] [CrossRef] [Green Version]
- Speiser, J.L.; Miller, M.E.; Tooze, J.A.; Ip, E.H. A comparison of random forest variable selection methods for classification prediction modeling. Expert Syst. Appl. 2019, 134, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Smith, A.R.; Kutia, M.; Wang, G.; Liu, H.; Sun, H. A Modified KNN Method for Mapping the Leaf Area Index in Arid and Semi-Arid Areas of China. Remote Sens. 2020, 12, 1884. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Liknes, G.C.; Domke, G.M. Using a remote sensing-based, percent tree cover map to enhance forest inventory estimation. For. Ecol. Manag. 2014, 331, 12–18. [Google Scholar] [CrossRef]
- Marselis, S.M.; Yebra, M.; Jovanovic, T.; Van Dijk, A.I. Deriving comprehensive forest structure information from mobile laser scanning observations using automated point cloud classification. Environ. Model. Softw. 2016, 82, 142–151. [Google Scholar] [CrossRef]
- García-Gutiérrez, J.; Martínez-álvarez, F.; Troncoso, A.; Riquelme, J.C. A comparison of machine learning regression techniques for lidar-derived estimation of forest variables. Neurocomputing 2015, 167, 24–31. [Google Scholar] [CrossRef]
- Chirici, G.; Barbati, A.; Corona, P.; Marchetti, M.; Travaglini, D.; Maselli, F.; Bertini, R. Non-parametric and parametric methods using satellite images for estimating growing stock volume in alpine and Mediterranean forest ecosystems. Remote Sens. Environ. 2008, 112, 2686–2700. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Yao, W.; Choi, S.; Park, T.; Myneni, R.B. A Comparative Study of Predicting DBH and Stem Volume of Individual Trees in a Temperate Forest Using Airborne Waveform LiDAR. IEEE Geosci. Remote Sens. Lett. 2015, 12, 2267–2271. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Haapanen, R.; Tuominen, S. Data Combination and Feature Selection for Multi-source Forest Inventory. Photogramm. Eng. Remote Sens. 2008, 74, 869–880. [Google Scholar] [CrossRef]
- Eitel, J.U.H.; Vierling, L.A.; Litvak, M.E.; Long, D.S.; Schulthess, U.; Ager, A.A.; Krofcheck, D.J.; Stoscheck, L. Broadband, red-edge information from satellites improves early stress detection in a New Mexico conifer woodland. Remote Sens. Environ. 2011, 115, 3640–3646. [Google Scholar] [CrossRef]
- Candra, E.D.; Hartono; Wicaksono, P. Above Ground Carbon Stock Estimates of Mangrove Forest Using Worldview-2 Imagery in Teluk Benoa, Bali; IOP Publishing: Bristol, UK, 2016; Volume 47, p. 12014. [Google Scholar]
- Zarco-Tejada, P.; Hornero, A.; Hernández-Clemente, R.; Beck, P. Understanding the temporal dimension of the red-edge spectral region for forest decline detection using high-resolution hyperspectral and Sentinel-2a imagery. ISPRS J. Photogramm. Remote Sens. 2018, 137, 134–148. [Google Scholar] [CrossRef] [PubMed]
- A Sims, D.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Viña, A.; Arkebauer, T.J.; Rundquist, D.; Keydan, G.; Leavitt, B. Remote estimation of leaf area index and green leaf biomass in maize canopies. Geophys. Res. Lett. 2003, 30. [Google Scholar] [CrossRef] [Green Version]
- Wallis, C.I.; Homeier, J.; Peña, J.; Brandl, R.; Farwig, N.; Bendix, J. Modeling tropical montane forest biomass, productivity and canopy traits with multispectral remote sensing data. Remote Sens. Environ. 2019, 225, 77–92. [Google Scholar] [CrossRef]
- Li, Y.; Li, C.; Li, M.; Liu, Z. Influence of Variable Selection and Forest Type on Forest Aboveground Biomass Estimation Using Machine Learning Algorithms. Forest 2019, 10, 1073. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Cao, C.; Dang, Y.; Xiang, H.; Zhao, J.; Zhang, Y.; Wang, X.; Guo, H. Retrieval of forest growing stock volume by two different methods using Landsat TM images. Int. J. Remote Sens. 2014, 35, 29–43. [Google Scholar] [CrossRef]
- Myroniuk, V.; Kutia, M.; Sarkissian, A.J.; Bilous, A.M.; Liu, S. Regional-Scale Forest Mapping over Fragmented Landscapes Using Global Forest Products and Landsat Time Series Classification. Remote Sens. 2020, 12, 187. [Google Scholar] [CrossRef] [Green Version]
- Chelgani, S.C.; Matin, S.; Makaremi, S. Modeling of free swelling index based on variable importance measurements of parent coal properties by random forest method. Measurement 2016, 94, 416–422. [Google Scholar] [CrossRef]
- Cutler, M.E.J.; Boyd, D.S.; Foody, G.M.; Vetrivel, A. Estimating tropical forest biomass with a combination of SAR image texture and Landsat TM data: An assessment of predictions between regions. ISPRS J. Photogramm. Remote Sens. 2012, 70, 66–77. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Han, N.; Li, X.; Du, H.; Mao, F.; Cui, L.; Liu, T.; Xing, L. Spatiotemporal Estimation of Bamboo Forest Aboveground Carbon Storage Based on Landsat Data in Zhejiang, China. Remote Sens. 2018, 10, 898. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Pattey, E.; Jégo, G. Assessment of vegetation indices for regional crop green LAI estimation from Landsat images over multiple growing seasons. Remote Sens. Environ. 2012, 123, 347–358. [Google Scholar] [CrossRef]
- Calvão, T.; Palmeirim, J.M. Mapping Mediterranean scrub with satellite imagery: Biomass estimation and spectral behaviour. Int. J. Remote Sens. 2004, 25, 3113–3126. [Google Scholar] [CrossRef]
- Zhang, M.; Du, H.; Zhou, G.; Li, X.; Mao, F.; Dong, L.; Zheng, J.; Liu, H.; Huang, Z.; He, S. Estimating Forest Aboveground Carbon Storage in Hang-Jia-Hu Using Landsat TM/OLI Data and Random Forest Model. Forest 2019, 10, 1004. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Li, Y.; Li, M. Improving Forest Aboveground Biomass (AGB) Estimation by Incorporating Crown Density and Using Landsat 8 OLI Images of a Subtropical Forest in Western Hunan in Central China. Forest 2019, 10, 104. [Google Scholar] [CrossRef] [Green Version]
- Willmott, C.J.; Ackleson, S.G.; Davis, R.E.; Feddema, J.J.; Klink, K.M.; LeGates, D.R.; O’Donnell, J.; Rowe, C.M. Statistics for the evaluation and comparison of models. J. Geophys. Res. Space Phys. 1985, 90, 8995–9005. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Liu, Z.; Lin, H.; Wang, G.; Sun, H.; Long, J.; Zhang, M. Estimating the Growing Stem Volume of Chinese Pine and Larch Plantations based on Fused Optical Data Using an Improved Variable Screening Method and Stacking Algorithm. Remote Sens. 2020, 12, 871. [Google Scholar] [CrossRef] [Green Version]
- Xie, B.; Cao, C.; Xu, M.; Bashir, B.; Singh, R.P.; Huang, Z.; Lin, X. Regional Forest Volume Estimation by Expanding LiDAR Samples Using Multi-Sensor Satellite Data. Remote Sens. 2020, 12, 360. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E.F. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2016, 9, 63–105. [Google Scholar] [CrossRef]
- Zhou, J.-J.; Zhou, Z.; Zhao, Q.; Han, Z.; Wang, P.; Xu, J.; Dian, Y. Evaluation of Different Algorithms for Estimating the Growing Stock Volume of Pinus massoniana Plantations Using Spectral and Spatial Information from a SPOT6 Image. Forest 2020, 11, 540. [Google Scholar] [CrossRef]


| Tree Specie | Plot Number | Range of Values | Mean | Standard Deviation | Coefficient of Variation (%) |
|---|---|---|---|---|---|
| Chinese pine | 43 | 91.97–514.96 | 245.52 | 110.59 | 45.0 |
| Larch | 38 | 86.17–466.23 | 217.21 | 91.84 | 42.3 |
| Total | 81 | 86.17–514.96 | 232.24 | 102.59 | 44.2 |
| Image Identification | Product Level | Acquisition Date |
|---|---|---|
| S2A_MSIL1C_20170922T025541_N0205_R032_T50TNM_20170922T030440 | L1C | 22 September, 2017 |
| S2A_MSIL1C_20170922T025541_N0205_R032_T50TPM_20170922T030440 | L1C | 22 September, 2017 |
| Sentinel-2 Bands | Central Wavelength (nm) | Bandwidth (nm) | Spatial Resolution (m) |
|---|---|---|---|
| Band 2-Blue | 496.6 | 98 | 10 |
| Band 3-Green | 560.0 | 45 | 10 |
| Band 4-Red | 664.5 | 38 | 10 |
| Band 5-Vegetation Red Edge | 703.9 | 19 | 20 |
| Band 6-Vegetation Red Edge | 740.2 | 18 | 20 |
| Band 7-Vegetation Red Edge | 782.5 | 28 | 20 |
| Band 8-NIR | 835.1 | 145 | 10 |
| Band 8A-Vegetation Red Edge | 864.8 | 33 | 20 |
| Band 11-SWIR | 1613.7 | 143 | 20 |
| Band 12-SWIR | 2202.4 | 242 | 20 |
| Feature Variable | Description | Reference |
|---|---|---|
| Band Reflectivity | B2-BLUE, B3-GREEN, B4-RED, B5-Red Edge1, B6-Red Edge2, B7-Red Edge3, B8-NIR, B8A-Red Edge4, B11-SWIR1, B12-SWIR2 | [39] |
| Vegetation Index | Normalized Difference Vegetation index (NDVI) | [39] |
| Enhanced Vegetation index (EVI) | [39] | |
| Red-green vegetation index (RGVI) | [39] | |
| Atmospherically resistant vegetation index (ARVI) | [39] | |
| Red Edge Normalized Difference Vegetation index (RENDVI) | [44] | |
| Red Edge Chlorophyll Index (RECI) | [45] | |
| Red Edge Simple Ratio (RESR) | [45] | |
| Topographic Factor | Elevation | [46] |
| Slope | [46] | |
| Aspect | [46] |
| Variable | Coefficient | Significance | VIF |
|---|---|---|---|
| Constant | −125.48 | 0.03 | - |
| B7 | −1162.73 | 0.00 | 1.10 |
| Elevation | 0.29 | 0.00 | 1.07 |
| RGVI | −111.09 | 0.04 | 1.60 |
| RENDVI | 379.13 | 0.00 | 1.65 |
| Texture Window | Red Edge Band | Texture Metric |
|---|---|---|
| 3 × 3, 5 × 5, 7 × 7, 9 × 9, 11 × 11 | Band 5-Vegetation Red Edge 1 (RE1), Band 6-Vegetation Red Edge 2 (RE2), Band 7-Vegetation Red Edge 3 (RE3), Band 8A-Vegetation Red Edge 4 (RE4) | Mean (Men) |
| Variance (Var) | ||
| Homogeneity (Hom) | ||
| Contrast (Con) | ||
| Dissimilarity (Dis) | ||
| Entropy (Ent) | ||
| Second moment (Sec) | ||
| Correlation (Cor) |
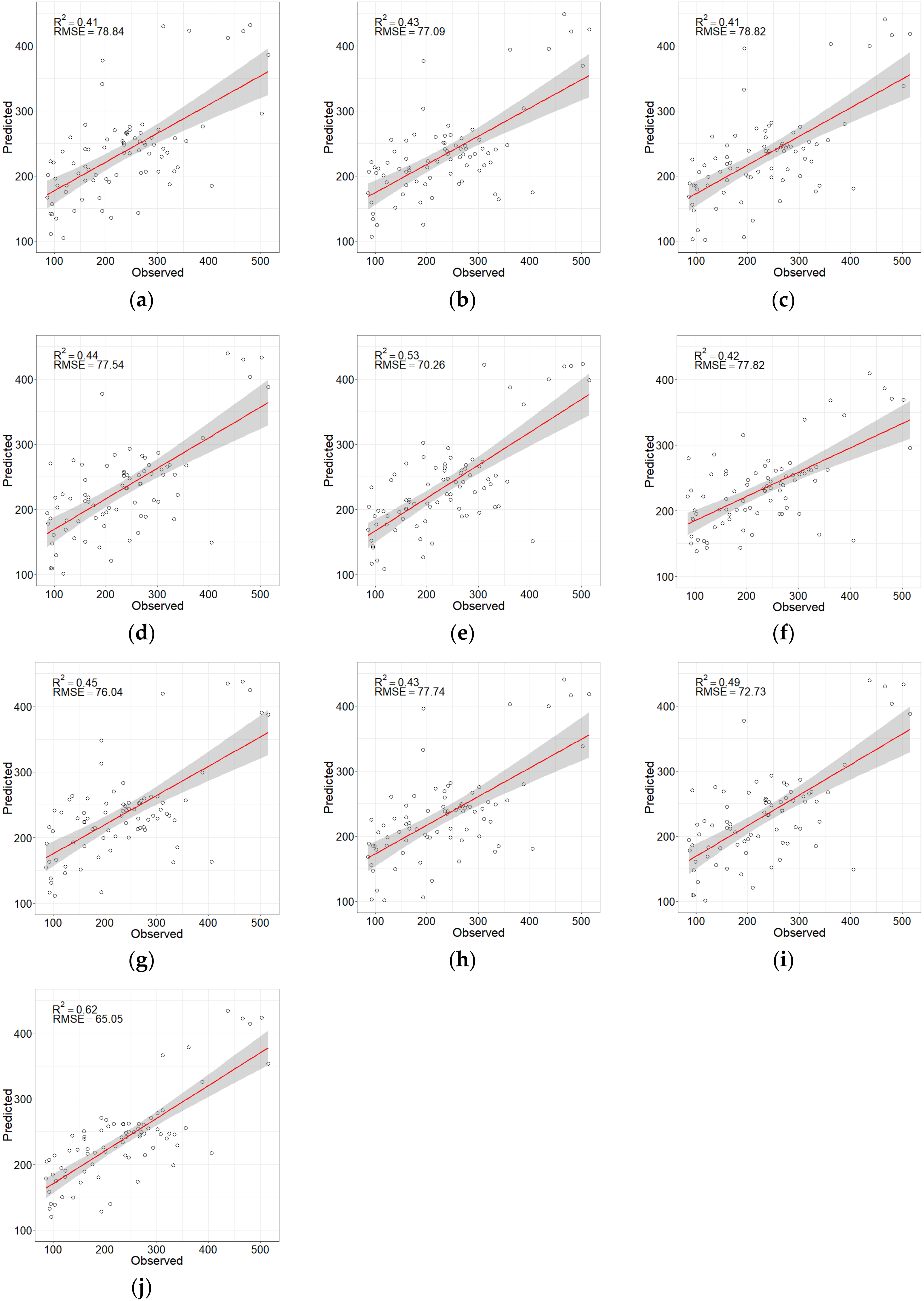
| Model | Method | Variables Combination | R2 | RMSE | rRMSE (%) | MAE | SDe |
|---|---|---|---|---|---|---|---|
| Without texture features | LSR | B7, Elevation, RENDVI, RGVI | 0.41 | 78.84 | 33.9 | 62.17 | 70.95 |
| RF | EVI, NDVI, Elevation, RENDVI, B12, B11, B4, B3, RECI, B2, B8A, B7, RGVI | 0.43 | 77.09 | 33.2 | 61.13 | 72.27 | |
| Boruta | EVI, NDVI, Elevation, RENDVI, B12, B11, B4, B3, RECI, B2, B8A, B7, RGVI, B5, B6, RESR | 0.41 | 78.72 | 33.9 | 63.06 | 73.43 | |
| VSURF | EVI, NDVI, B2 | 0.44 | 77.54 | 33.4 | 59.90 | 80.73 | |
| SRF | EVI, B2, Elevation, B11, B7 | 0.53 | 70.26 | 30.3 | 56.06 | 71.35 | |
| With texture features | LSR | 5 × 5_RE1_Men, Elevation, 11 × 11_RE2_Sec, 11 × 11_RE2_Ent, 9 × 9_RE2_Con, B7, 9 × 9_RE3_Cor | 0.42 | 77.82 | 33.5 | 59.04 | 58.18 |
| Boruta | NDVI, EVI, B11, B12, B2, B3, B4, Elevation, RECI, RENDVI, 3 × 3_RE1_Men, 5 × 5_RE1_Men, 7 × 7_RE1_Men, 9 × 9_RE1_Men, 11 × 11_RE1_Men, 3 × 3_RE3_Hom | 0.43 | 77.74 | 33.5 | 61.34 | 72.34 | |
| RF | EVI, NDVI, RENDVI, 5 × 5_RE1_Men, 5 × 5_RE3_Con, Elevation, 9 × 9_RE1_Men, B2 | 0.45 | 76.04 | 32.7 | 59.67 | 73.96 | |
| VSURF | EVI, NDVI, 5 × 5_RE1_Men, 9 × 9_RE1_Men, B12, Elevation, B2, B3, 9 × 9_RE1_Sec | 0.49 | 72.73 | 31.3 | 58.26 | 73.59 | |
| SRF | EVI, 11 × 11_RE1_Sec, B2, 5 × 5_RE1_Sec, Elevation, 11 × 11_RE3_Ent, 3 × 3_RE3_Var, 3 × 3_RE1_Hom, 9 × 9_RE4_Var | 0.62 | 65.05 | 28.0 | 52.69 | 65.04 |
| Model | Variable Selection Method | LSR | RF | Boruta | VSURF |
|---|---|---|---|---|---|
| Without texture features | LSR | ||||
| RF | −7.86 (0.00) | ||||
| Boruta | −0.42 (0.67) | 7.84 (0.00) | |||
| VSURF | 0.51 (0.61) | 8.38 (0.00) | 0.77 (0.44) | ||
| SRF | 2.01 (0.04) | 8.63 (0.00) | 2.63 (0.01) | 2.13 (0.00) | |
| With texture features | LSR | ||||
| RF | 0.08 (0.93) | ||||
| Boruta | −0.33 (0.74) | −0.82 (0.41) | |||
| VSURF | 0.42 (0.66) | 0.65 (0.52) | 1.82 (0.07) | ||
| SRF | 2.01 (0.04) | 2.40 (0.01) | 2.66 (0.00) | 2.00 (0.04) |
| Study Area | Feature Variable | Residual | VIF |
|---|---|---|---|
| Wangyedian Forest Farm | EVI | −0.09 | 3.222 |
| 11 × 11_RE1_Sec | −0.04 | 6.270 | |
| B2 | 0.12 | 3.244 | |
| 5 × 5_RE1_Sec | 0.01 | 5.308 | |
| Elevation | −0.17 | 1.064 | |
| 11 × 11_RE3_Ent | −0.04 | 6.115 | |
| 3 × 3_RE3_Var | −0.13 | 1.568 | |
| 3 × 3_RE1_Hom | −0.08 | 2.768 | |
| 9 × 9_RE4_Var | 0.04 | 2.430 |
| Sample Size | Tree Specie | R2 | RMSE | rRMSE (%) | MAE |
|---|---|---|---|---|---|
| Total or pooled sample plot set | Chinese pine | 0.73 | 62.40 | 25.4 | 51.05 |
| Larch | 0.44 | 67.92 | 31.3 | 54.53 | |
| Total | 0.62 | 65.05 | 28.0 | 56.69 | |
| Separate sample plot sets by species | Chinese pine | 0.68 | 63.20 | 24.9 | 50.06 |
| Larch | 0.53 | 65.67 | 29.8 | 49.45 | |
| Total | 0.62 | 64.37 | 27.7 | 49.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, F.; Kutia, M.; Sarkissian, A.J.; Lin, H.; Long, J.; Sun, H.; Wang, G. Estimating the Growing Stem Volume of Coniferous Plantations Based on Random Forest Using an Optimized Variable Selection Method. Sensors 2020, 20, 7248. https://doi.org/10.3390/s20247248
Jiang F, Kutia M, Sarkissian AJ, Lin H, Long J, Sun H, Wang G. Estimating the Growing Stem Volume of Coniferous Plantations Based on Random Forest Using an Optimized Variable Selection Method. Sensors. 2020; 20(24):7248. https://doi.org/10.3390/s20247248
Chicago/Turabian StyleJiang, Fugen, Mykola Kutia, Arbi J. Sarkissian, Hui Lin, Jiangping Long, Hua Sun, and Guangxing Wang. 2020. "Estimating the Growing Stem Volume of Coniferous Plantations Based on Random Forest Using an Optimized Variable Selection Method" Sensors 20, no. 24: 7248. https://doi.org/10.3390/s20247248
APA StyleJiang, F., Kutia, M., Sarkissian, A. J., Lin, H., Long, J., Sun, H., & Wang, G. (2020). Estimating the Growing Stem Volume of Coniferous Plantations Based on Random Forest Using an Optimized Variable Selection Method. Sensors, 20(24), 7248. https://doi.org/10.3390/s20247248








