Abstract
The mechanical properties of DNA have enabled it to be a structural and sensory element in many nanotechnology applications. While specific base-pairing interactions and secondary structure formation have been the most widely utilized mechanism in designing DNA nanodevices and biosensors, the intrinsic mechanical rigidity and flexibility are often overlooked. In this article, we will discuss the biochemical and biophysical origin of double-stranded DNA rigidity and how environmental and intrinsic factors such as salt, temperature, sequence, and small molecules influence it. We will then take a critical look at three areas of applications of DNA bending rigidity. First, we will discuss how DNA’s bending rigidity has been utilized to create molecular springs that regulate the activities of biomolecules and cellular processes. Second, we will discuss how the nanomechanical response induced by DNA rigidity has been used to create conformational changes as sensors for molecular force, pH, metal ions, small molecules, and protein interactions. Lastly, we will discuss how DNA’s rigidity enabled its application in creating DNA-based nanostructures from DNA origami to nanomachines.
1. Introduction
In the past two decades, DNA nanotechnology has put forth various remarkable structures and functions of DNA far beyond its role as the genetic material in living organisms. Owing to its outstanding properties (self-assembly, programmability, stability, resilience, etc.) DNA has been increasingly maneuvered in multiple different ways to propel the field of nanotechnology. DNA serves in multiple capacities, i.e., as aptamers, DNAzymes, molecular beacons, biosensors, nanoparticles, molecular machines, and nano-electronic components (e.g., nanowires, constituents of logic gates, etc.) [1]. One of the most fundamental features of DNA that has made many of its applications possible is its excellent mechanical rigidity against bending. Since the mechanical rigidity of DNA is sensitive to multiple intrinsic and extrinsic factors, it can be manipulated and employed for sensing applications.
A wide range of techniques has been used to determine dsDNA stiffness, persistence length (Lp), curvature, and geometry. Peters et al., have comprehensively reviewed many techniques used for probing DNA curvature and flexibility in vitro as well as in vivo [2]. Some of the popular techniques include computational simulations [3,4,5,6,7,8,9,10], electrophoretic mobility assays [11,12,13,14,15,16,17,18], cyclization analysis [4,19,20,21,22,23,24,25,26,27,28,29,30,31], tethered particle motion assays [32,33,34], optical tweezers [35,36,37,38,39], fluorescence spectroscopy [40,41,42,43,44,45,46,47], atomic force microscopy [48,49,50,51,52,53], single-molecule force spectroscopy [54,55], cryo-electron microscopy [56,57,58,59], scanning Tunneling microscopy [60,61,62], X-ray crystallography [63,64], small-angle X-ray scattering [65,66], NMR spectroscopy [67,68,69,70,71,72,73], transient electric birefringence [74,75], anti-Brownian electrophoretic trap [76], chromosomal conformation capture (3C) assay [77,78,79,80], genetic and recombination assays [81,82,83,84,85,86,87,88,89,90,91,92,93], etc. Generally, a combination of two or more of the above methods is needed to gain a complete understanding of conformation and kinetics. To utilize the mechanical rigidity of DNA to our advantage, we must first understand the factors that influence it. Here, we are primarily interested in double-stranded DNA (dsDNA) due to its relatively high stiffness compared to single-stranded DNA (ssDNA). The persistence length (Lp) of dsDNA is ~50 nm (at 10 mM Na+), over 60 times greater than that of ssDNA (Lp~0.75 nm at 150 mM Na+) [94].
First, the prime mechanism underlying the dsDNA stiffness is the stacking interactions of adjacent base pairs that provide its structural integrity [75]. In a recent report, Chen et al., used computational methods to quantitatively show that base-stacking contributes significantly to the local and global rigidity of dsDNA as compared to other chemical interactions such as backbone, ribose puckering, and base-pairing in dsDNA [3]. Figure 1a illustrates the geometry of base-stacking interaction among other chemical interactions. Bending of the DNA directly perturbs the base-stacking geometry along the dsDNA, hence unfavourable. Therefore, factors that directly influence the base-stacking of dsDNA will directly impact DNA stiffness.
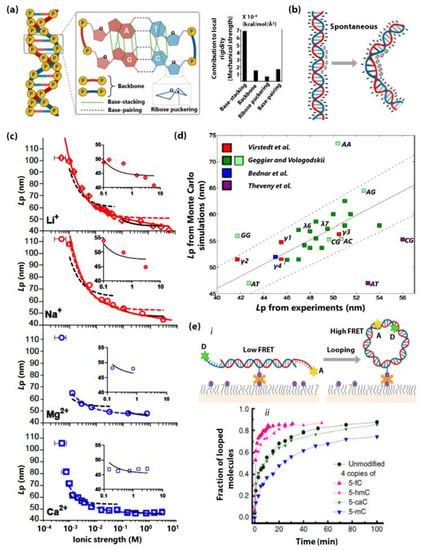
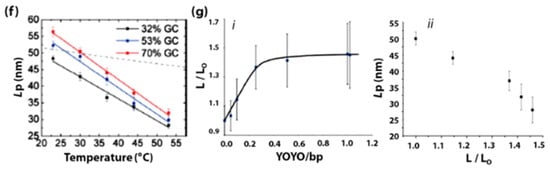
Figure 1.
(a) A schematic of dsDNA showing chemical interactions and structural features such as base-pairing, base-stacking, phosphate-backbone, sugar pucker, respectively, and their contribution to the local rigidity in terms of mechanical strength of dsDNA. Figure adapted and modified from [3] with permission from The Royal Society of Chemistry. (b) Cartoon representation of dsDNA spontaneously bending upon incorporating neutral phosphate analogs (shown as ‘0’) on one of the helical faces. Negatively charged phosphates are shown as ‘-’ [11]. (c) Dependence of Lp on the ionic strength of monovalent Li+, Na+ (top red) and divalent Mg2+, Ca2+ (bottom blue) ions. Figure reprinted with permission from [102]. Copyright (2019) by the American Physical Society. (d) A scatter plot of simulated and experimentally estimated poly(NN) dsDNA persistence length (Lp) data from different experiments pursued by various research groups (grouped by colour). Figure adapted with permission from [103]. Copyright 2016 American Chemical Society. (e,i) shows the schematic of a single-molecule dsDNA cyclization experiment. Two complementary sticky ends of the dsDNA are tagged with a fluorescence resonance energy transfer (FRET) pair of fluorescent donor (D) and acceptor (A) dyes, and the fraction of the dsDNA looped is probed by monitoring the increase in FRET between A and D. (ii) shows the fraction of the dsDNA looped as a function of time. Figure adapted with permission from [23]. Copyright 2016, the authors. (f) Dependence of Lp of three dsDNA (differing in their G-C content) on temperature. The melting temperature of all three dsDNA used in this experiment are greater than 75 °C. Figure adapted with permission from [32]. Copyright 2014 American Chemical Society. (g,i) shows the change in dsDNA length upon YOYO intercalation. L and Lo is the contour length of dsDNA in the presence and absence of YOYO, respectively. The curve demonstrates the increase in fractional extension L/Lo upon an increase in YOYO concentration. (ii) shows the dependence of Lp on fractional extension L/Lo. The curve demonstrates that Lp decreases upon the increase of the YOYO/base-pair ratio. Figure adapted from [48] with permission from The Royal Society of Chemistry.
Second, the electrostatic phosphate-phosphate repulsions and environmental ionic strength also have substantial influence on the dsDNA stiffness. It is well known that asymmetric neutralization of phosphate charges induce bending of the helix axis [11,95,96,97,98,99]. Figure 1b is a cartoon showing that DNA spontaneously bends upon the incorporation of neutral phosphate analogs on one of the helical faces. Apart from the bending rigidity, the stiffness of the phosphate backbone also gives rise to dsDNA’s outstanding torsional rigidity (overwinding behaviour of dsDNA) [100,101]. The salt-dependence of the stiffness of dsDNA is closely related to the screening of the phosphate backbone charges as well. In 2019, Guilbaud et al., showed that the Lp of dsDNA decreases significantly with increasing ionic strength in monovalent and divalent metal ions (Figure 1c) [102]. This decrease in persistence length could be attributed to the reduction in the energetic cost of bending due to screening of phosphate backbone charges by surrounding cations. However, the Lp seems to be independent of the size of metal ions when kept at the same ionic concentration [102].
Third, the rigidity of dsDNA is sequence-dependent. In 2017, Jonathan et al., used computational simulations of random and λ-phage dsDNA fragments to report that the apparent Lp shows a standard deviation of 4 nm over the sequence. Additionally, they demonstrated that poly(A), poly(TA), and phased A-tract sequence motifs are exceptionally straight and stiff, tightly coiled and exceptionally soft, and exceptionally bent and stiff, respectively [103]. Figure 1d shows a scatter plot of simulated (Monte Carlo) and experimentally (includes data from atomic force microscopy, electron microscopy, and cyclization experiments) estimated poly(NN) dsDNA persistence lengths (Lp) from different experiments from various studies. It is worth discussing here that nucleotide base modifications affect dsDNA stiffness. For example, 5-formylcytosine, 5-hydroxymethylcystosine and 5-hydroxymethyluracil are known to enhance the dsDNA flexibility while 5-methylcytosine is known to decrease it [54,104,105]. Figure 1e shows the fraction of the looped molecules as a function of time for unmodified dsDNA as well as dsDNA containing four copies of 5-formylcytosine (5-fC), 5-hydroxy-methylcystosine (5-hmC), 5-carboxylcystosine (5-caC), and 5-methylcytosine (5-mC). Higher flexibility of dsDNA leverages higher looping probability and faster looping kinetics. Figure 1e, shows the schematic of a single-molecule dsDNA looping experiment.
Fourth, dsDNA stiffness is strongly influenced by temperature. Through temperature-controlled single-particle tethered motion experiments, it has been shown that dsDNA stiffness is sharply reduced when temperature is increased (Figure 1f) [32]. The melting temperature of all three dsDNA used in Figure 1g are greater than 75 °C, while the experiment range is between 23–52 °C. This indicates that during the experiment the global conformation of all three DNA remains double-stranded. The authors have attributed this to local effects such as temperature-enhanced formation of kinks (due to unstacking of adjascent base pairs) and small melting bubbles (due to disruption of base pairing and/or base stacking). In line with the above discussion, it is worth emphasizing that basepair mismatches or melting bubbles significantly effect the rigidity of dsDNA [26,106,107,108]. In 2004, Yan et al., suggested that formation of melting bubbles by internal strand-sepraration provide a flexible-hinge that facilitates the formation of smooth bends and thus loops of dsDNA less than 150 bp [26]. In 2006, Yuan et al., demonstrated that distributed melting bubbles induce bigger bending angles and higher reduction in the stiffness of dsDNA as compared to the centrally located melting bubbles of comparable overall size [108]. In 2009, Forties et al., published a robust model that predicts the impact of small bubbles formed due to dsDNA melting (temperature-dependent) or DNA mismatches (sequence-dependent) on dsDNA flexibility [107].
Lastly, DNA-binding molecules such as bis-intercalators (e.g., YOYO-1) are known to alter dsDNA rigidity. Figure 1g demonstrates that as the concentration of YOYO-1 increases (in comparison to base-pair concentration), the fractional extension L/Lo of dsDNA increases (L and Lo are the contour lengths of dsDNA with and without YOYO-1), while the Lp decreases [48].
2. DNA-Springs as Regulators
2.1. Allosteric Regulation of Enzyme Activity
Regulation of protein or nucleic acid enzyme activity by factors such as pH, salt, temperature, and co-factor molecules have been widely investigated and understood. However, the effects of mechanical perturbation on enzyme activity remain under-explored. In the past decade, DNA-based molecular springs have been used to regulate the mechanical compliance of biomolecules by exerting mechanical strain to alter their conformation [109,110,111,112,113,114,115]. DNA spring is a bent dsDNA that stretches any molecule that bridges its ends, like a bow under tension. The concept of DNA springs was first introduced by Tyagi and Kramer in 1996 [116] (Figure 2a) where the conformational change in an DNA hairpin A (blue/yellow) was induced by a complementary DNA strand B (red). Here, hybridization of the loop of hairpin A (yellow) and strand B (red) into duplex A/B opens up the stem of hairpin A. As the stiffness of the double-stranded loop of hairpin A is stronger than that of the single-stranded loop of hairpin A, it acts as a spring and exerts force on the stem of hairpin A unzipping the double-stranded stem apart into single-stranded components. A more detailed discussion of the unzipping of hairpin stem due to formation of double-stranded DNA springs in hairpin loop is made in Section 3.1. Detection of this conformational change was achieved using fluorescence quenching using a fluorophore-quencher pair. The total fluorescence increases upon the formation of A/B duplex as the rigid dsDNA duplex keeps the fluorophore-quencher pair apart. DNA springs have since been employed in three different ways to probe the mechanical compliance of enzymes: (i) by influencing the spatial accessibility of molecules that regulate the enzyme activity, and by applying force directly on (ii) the enzyme or (iii) the substrate.
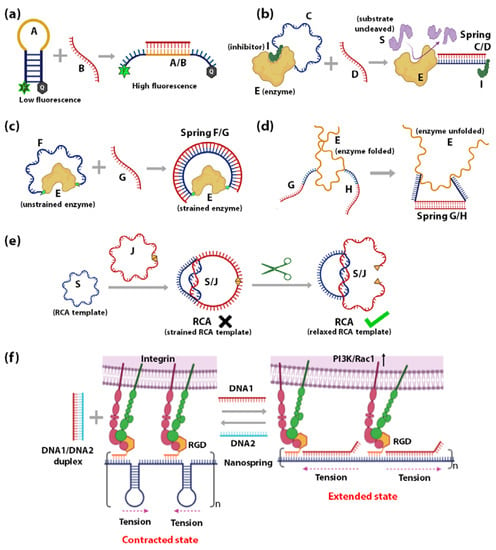
Figure 2.
(a) Schematic representation of a DNA spring where the increased stiffness of A/B duplex does not allow the two ends of strand A to self-hybridize [116]. Schematic representation of a DNA spring regulating the catalytic activity of enzyme E (b) by altering the spatial proximity of the enzyme-inhibitor I with the help of the stiffness of C/D duplex [117] (c) by exerting mechanical force (generated due to the stiffness of F/G duplex) on the enzyme itself [115] (d) by pulling apart the ribozyme E tertiary structure causing it to misfold by the formation of G/H duplex [118]. (e) Schematic representation of a topological constraint J regulating the activity of an RCA DNA polymerase by exerting strain on the RCA template S by S/J duplex formation [119]. (f) Schematic representation of a DNA nanospring regulating mechanical tension by contracting membrane-bound integrins [120].
- (i)
- Influencing the accessibility of regulatory molecules: The first strategy using DNA springs to regulate enzymes is by controlling the accessibility of enzyme regulatory factors, e.g., inhibitors. Ghadiri et al., reported the allosteric regulation of a protein enzyme Cereus Neutral Protease 5 (CNP) by a DNA spring using this strategy (Figure 2b) [117]. In this study, the two ends of DNA strand C (blue) were conjugated to CNP (shown as E) and its inhibitor (shown as I). The flexible nature of ssDNA C allows the inhibitor I to remain bound to the enzyme E until the complementary ssDNA D (red) hybridizes to strand C. The rigid dsDNA duplex C/D causes the dissociation of the inhibitor from the enzyme and keeps it away. CNP’s catalytic activity recovers noticeably upon the unbinding of the inhibitor I caused by the binding of strand B.
- (ii)
- Mechanical force regulation of enzymes: The second strategy uses DNA springs to exert force on enzymes to modulate their mechanochemistry. Choi et al., demonstrated this concept in 2005 (Figure 2c) [115] by covalently attaching two ends of a flexible 60-nucleotide ssDNA strand F (blue) to the two lobes of a maltose-binding protein (MBP). MBP retains its high binding affinity to maltose when F remains in the ssDNA state, as it does not apply external force to the MBP. However, when a complementary strand G binds to F, an external force up to 10 pN is applied on the MBP from the stiffness of the F/G duplex. This mechanical stress makes it energetically unfavourable for MBP to undergo the conformational changes required for maltose binding. As the length of strand G increases above 30 bases, the mechanical stress exerted on MBP increases, leading to further diminished maltose-binding affinity. Through this mechanism, regulation of MBP-maltose affinity was achieved through the external forces generated by a DNA spring. A similar concept was applied to achieve reversible allosteric regulation of enzyme guanylate kinase (GK) [109] and cAMP-dependent Protein Kinase A (PKA) [110]. The binding of GK to its substrates adenosine triphosphate (ATP) and guanosine monophosphate (GMP) requires a conformational change, which was reversibly inhibited by mechanical tension applied through a dsDNA spring. On the other hand, PKA was activated by the mechanical tension applied through a dsDNA spring, such that it demonstrated activity even in the absence of cyclic adenosine monophosphate (cAMP), the co-factor typically required for its activation. In 2007, Silverman et al., extended the application of DNA springs to the allosteric regulation of ribozymes (Figure 2d) [118]. Here, the hammerhead ribozyme’s mechanical control was attained by attaching two complementary ssDNA G and H to two different portions of the ribozyme. When G and H hybridize, they pull apart their corresponding attachment points, causing the tertiary structure of ribozyme E to unfold and lose function.
- (iii)
- Mechanical force regulation of substrates: The third strategy is to exert strain on the substrate such that it affects the efficiency of the enzyme. Based on this strategy, a circular ssDNA was used as a topological constraint to regulate the Rolling Circle Amplification (RCA) activity of Φ29 DNA Polymerase enzyme [119]. This was illustrated by Liu et al., where a circular RCA ssDNA template (shown as S, blue) was mechanically strained by a strong linking duplex formed with another circular ssDNA (shown as J, red) as in the case of DNA catenanes (Figure 2e). The strained template cannot undergo RCA until the spring/constraint is cleaved, and the strain is released. This system has been employed to demonstrate specific target-triggered RCA for detecting a specific E. coli strain with detection limits of 10 cells/mL.
2.2. Regulation of Live Cells
DNA-springs can be made to regulate the activity of not only individual biomolecules but also live cells. In 2017, Zhang et al., demonstrated the use of a DNA nano-spring for the reversible control of integrin clustering and cell membrane receptor function (Figure 2f) [120]. Here, a DNA nanospring was demonstrated to redirect the normal morphology of the cell to having multiple cell protrusions and even alter the mRNA expression levels of integrin-related genes. In this study, DNA nano-springs consist of long repeats of hairpin-forming sequences (generated by RCA) were used as a scaffold for assembling RGD-DNA sequences (shown in orange). RGD is a tripeptide (Arg-Gly-Asp) with a high affinity for cell adhesion through integrin. Upon the addition of external DNA sequences (shown as DNA1 and DNA2), the hairpins can undergo cycles of unhybridization/rehybridization. This stretches and contracts the distance between RGD and thus the distance between and force through the integrins. When the nanospring is contracted, the clustered integrins stimulate the cells to form focal adhesions. While in the extended state, increased distance between integrins triggers activation of PI3K/Rac1 signalling, causing membrane remodelling and generation of numerous cell protrusions. Regulation of cell adhesion has also been achieved through the rational design of DNA tethers with various mechanical stability levels. In these studies, complementary strands of dsDNA were conjugated to the surface and cell-adhesion receptors (e.g., RGD or Selectin), where they act like nano-springs with a tension tolerance. When external forces increased beyond a certain threshold, the DNA springs would break. This system has resulted in a series of studies leading to quantifying integrin adhesion force, notch receptor activation, and leukocyte rolling adhesions [121,122,123,124,125,126,127]. Lastly, regulation of subcellular membrane remodelling was achieved with the help of DNA origami nano-springs as reported in 2019 by Grome et al. [128]. Here, DNA nano-springs of varying structural stiffness were polymerized on the surface of the liposomes and were found to significantly influence membrane binding, membrane remodelling, as well as vesicle tubulation.
3. Mechanical Rigidity-Facilitated DNA Sensors
3.1. DNA Springs as Force-Sensors
Using DNA as springs for sensing nanoscale forces was introduced by Shroff et al., in 2005. Herein a FRET-based force sensor capable of sensing a force range of 0–20 pN was demonstrated using a circular ssDNA B (red) covalently attached to a FRET donor D (green) and acceptor A (yellow) (Figure 3a) [129]. Change in FRET occurs when the circular ssDNA B converts to dsDNA through hybridization with a complementary ssDNA C (blue) of different lengths (Figure 3a). The change in FRET measures the force applied through the DNA-spring on the ssDNA. The idea of hybridization-induced stress and strain on circular DNA was further used to assess the force sensitivity and nanomechanics of several systems. In 2013, Fields et al., illustrated a similar concept to control strain in a DNA hairpin system (Figure 3b) [130]. A DNA-vise was constructed from a hairpin B with a loop of 30–50 nucleotides and a stem of 49 base pairs. Hairpin B was tagged with a FRET-pair at its loop-stem junction, such that the unzipping of the stem can be monitored by the decrease of FRET. As more nucleotides within the hairpin loop hybridize with longer complementary ssDNA C (l), the stress increases to unzip the hairpin from the loop side. Within a range of l values (l < lbuckle), increasing stiffness of B/C duplex results in increasing unzipping of the stem of hairpin B as monitored by the decrease of FRET. However, at lengths above an Euler-buckling threshold (l > lbuckle) the base-pairing free energy to re-establish the full hairpin stem overcomes the bending energy of the DNA-spring, causing FRET to return to a high state.
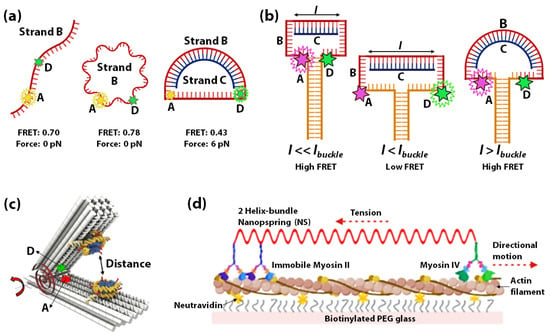
Figure 3.
Schematic representation of force exerted by DNA spring (B/C duplex) on (a) the single-stranded part of circular ssDNA B [129] and (b) the stem of hairpin B [130]. Increasing force increases the distance between the fluorophores D and A, resulting in a decrease in FRET (by unzipping in case of (b)). (c) Schematic representation of the DNA origami-based force-spectrometer consists of a spring-loaded hinge bearing two attached nucleosomes and a FRET pair of acceptor (A) and donor (D) fluorophores. The red torsional spring depicts the torque generated by the hinge. The nanospring is used for sensing distance (and interactive forces) between nucleosomes. Figure adapted with permission from [132]. Copyright 2016, the authors. (d) Schematic representation of a two-helix bundle DNA nanospring tethered to immobilized myosin II on one end and myosin VI on the other. Myosin VI is shown to more unidirectionally along actin filament against the load of the DNA nanospring [133].
Lately, the increased complexity of 3D DNA origami is being used for constructing origami-based nano-springs to leverage extremely sensitive DNA-based force-sensors. In 2013, Zhou et al., fabricated a DNA origami compliant joint structure, which acts as a tunable mechanical nano-spring [131]. The balance of tension in the flexible ssDNA components at the joint plays a key role in determining its geometry and mechanical properties. This study demonstrated the possibility of creating more elaborate DNA-origami sensors based on the mechanical flexibility of DNA. A DNA origami-based force-spectrometer was reported in 2016 by Funke et al. [132] where the fluctuations of the DNA spring-loaded hinge sensor allowed the measurement of inter-nucleosome distance at sub-nanometer resolution. Figure 3c shows the schematic of the DNA origami-based force spectrometer, which comprises of a spring-loaded hinge and two nucleosomes attached on opposite sides of the hinge. The torque generated by the hinge is shown by a red torsional spring. A FRET-pair was attached to the opposite sides of the spring-loaded hinge to gauge the distance between the nucleosomes. It is known that nucleosomes condense into arrays, which indicates that attractive forces exist between individual nucleosomes. The DNA origami-based force spectrometer, shown in Figure 3c, was used to measure the interaction strength between two nucleosomes directly. In this system, the strength of nucleosome-nucleosome interactions resulted in inter-nucleosome distance changes. These changes in distances were reflected by changes in FRET between the acceptor and donor fluorophores. In 2016, Iwaki et al., developed a programmable DNA origami-based nanospring that enabled the monitoring of force-induced transitions between two structurally distinct states of the mechanosensory motor protein myosin VI with nanometer-precision [133]. The DNA nanospring in this study comprises a 7,308 nucleotide ssDNA and more than 150 species of short ssDNA (<50 nucleotides) self-assembled into a two-helix bundle that is strained with negative superhelicity to form a coil structure. The DNA nanospring was attached to immobilized myosin II on one end and myosin VI on the other end. Figure 3d shows myosinVI moving unidirectionally along an actin filament against the load of the nanospring. This system helped study the stepping dynamics of Myosin VI under tension using fluorescently labelled DNA nanospring and TIRF microscopy. Lastly, DNA spring has been used in mechanical regulation and monitoring of biomolecules in live cells, taking advantage of the difference in rigidity of dsDNA vs ssDNA, for developing tools to study cell mechanics [134,135,136,137].
3.2. pH-Sensitive DNA Spring
In 2010, Wang et al., constructed a G-quadruplex/i-motif-based DNA nanospring powered by protons, which exhibited highly sensitive pH-dependent spring-like structural changes (Figure 4) [138]. The nanospring consists of two building blocks called subunit I and II, each of which is made up of two circular ssDNA C connected via ssDNA A and B. The subunits I and II can be connected by strand D in a linear array. The ssDNA A, B, and D consist of four G-rich stretches and form G-quadruplexes. The formation of G-quadruplexes reduces the distance between the circular DNA C rendering the nanospring in a compressed state. Upon adding a C-rich strand E, the strands A, B and D hybridize to strand E. Formation of the duplexes A/E, B/E, and D/E increases the distance between circular DNA C, imposing an extended state of the nanospring. This is due to the fact that in the single-stranded form the strands A, B and D have low rigidity and are floppy, while in the double-stranded form the A/E, B/E, and D/E are rigid due to their relatively increased persistence length. In the presence of slightly acidic conditions (~pH 6.0 or less), the cytosines are partially protonated, and the C-rich strand E un-hybridizes from strands A, B and D and folds into a closed i-motif structure, freeing the G-rich ssDNA strands A, B and D to form G-quadruplexes again. When the pH rises back to the alkaline conditions (~pH 8.0) the cytosines get deprotonated, leading to destabilization of the i-motif. This leaves the strand E free to hybridize back with strands A, B and D, which brings the nanospring back into the extended state. The above-explained DNA nanospring is pH-responsive, and the extended or compressed state can indicate a change between acidic and alkaline pH.
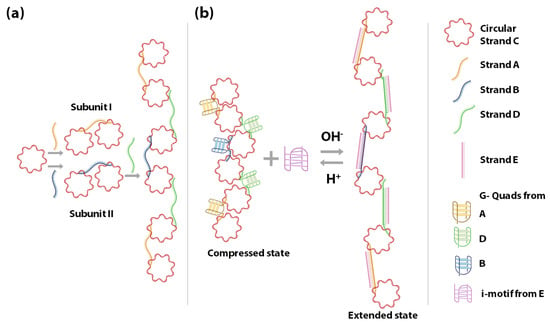
Figure 4.
Schematic representation of the proton-fueled DNA nanospring. (a) shows the constituent DNA strands and the construction of the nanospring. (b) shows the operation of the nanospring with the help of the C-rich strand E, and its compressed and extended state in response to acidic and alkaline pH, respectively [138].
3.3. DNA-Bows as Metal Ion/Small-Molecule Sensors
The interactions of DNA with metal ions and small molecules facilitate fundamental cellular processes [139,140,141,142] such as genomic stability [139,143,144,145,146,147], DNA-carcinogen interactions [146,147,148,149,150,151], drug development [152,153,154,155], and DNA-based metal-biosensors [156]. As mentioned earlier, the rigidity of DNA is affected by the type and concentration of metal ions. This mechanical energy stored in bent-DNA molecules has been utilized by Freeland et al., to quantify the interaction between DNA and metal ions (Mn+) and small organic molecules (Mols) using gel electrophoresis [17,18]. Their cost-effective and straightforward strategy utilizes the mechanical energy stored in bent DNA molecules called DNA-bows (Figure 5a) [17,157,158,159]. Each bow consists of two DNA strands with one part as a bent double-stranded fragment (F1) and the other as a mechanically stretched single-stranded fragment (F2) (Figure 5a). Depending on the environmental conditions, DNA-bows may decrease their bending elastic energy by transitioning to relaxed DNA-dimers or relaxed DNA-multimers (R), which show different electrophoretic mobilities (Figure 5b,c) for quantification. Freeland et al., reported that the equilibrium between the ss-DNA (S), DNA-bows (B) and multimers (R) is perturbed by the presence of metal ions and small organic molecules in a concentration-dependent manner, allowing for the quantitative determination of their interaction strength with DNA (Figure 5). This was indeed measured for various metal salts (MgCl2, MgSO4, KCl, CaCl2, Al(NO3)3, Zn(NO3)2, and AgNO3) and small organic molecules (guanidine, putrescine, spermidine, ethidium bromide, SYBR safe, ganciclovir, and thiamine). (Figure 5d,e) Undoubtedly, DNA-bows demonstrated the potential for developing sensitive and economical metal ions sensors to screen metal ion-aptamers, DNA-targeting drugs, and DNA-protein interactions in general.
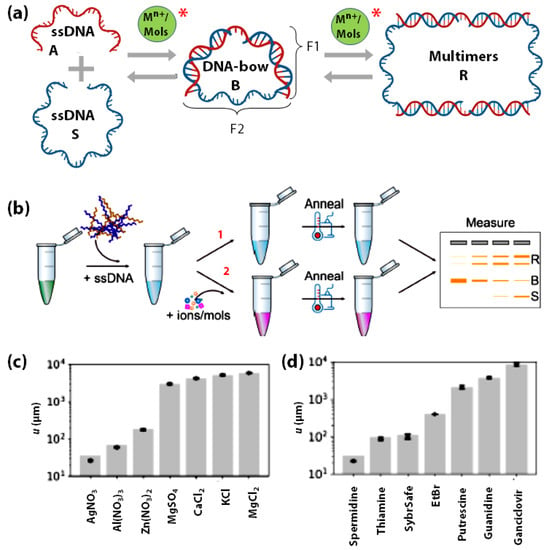
Figure 5.
Schematic of (a) the constitution of DNA-bows from single-stranded DNA, and the perturbation (depicted by *) of the equilibrium between ss-DNA, DNA-bows, and multimers due to their interaction with metal ions (Mn+) or small organic molecules (Mols), (b) the procedure followed for sensing DNA-metal ion/small molecule interaction via gel electrophoresis, where ‘1’ indicates the negative control with no Mn+/’Mols’ added while ‘2’ depicts the actual experiment with different concentrations of Mn+/’Mols’ added. Fitted u-values for quantifying the strength of DNA interactions with (c) metal ion salts and (d) small organic molecules. (b,c) are adapted with permission from [18]. Copyright 2020, by the authors.
3.4. DNA Stiffness Assisted Temperature and Osmolarity Sensing
The sensitivity of DNA towards changes in temperature and osmolarity are well known. In prokaryotes, the Histone-like nucleoid structuring protein (H-NS) is an abundant protein that plays a vital role in regulating nucleoid structure [160,161,162], in gene expression [163,164,165,166,167,168,169], and in mediating cellular response to changes in metabolite pH, temperature, and osmolarity [161,170,171]. It exhibits preferential binding to A & T-rich sequences and other regions of high intrinsic curvature, along the backbone of double-stranded DNA [162,166,172]. H-NS consists of two distinct domains (C-terminal and N-terminal) connected by a flexible linker segment. While the C-terminal domain bears a unique DNA binding motif, the N-terminal domain consists of a coiled-coil motif that mediates H-NS oligomerization [72,172,173]. Multiple studies suggest that H-NS bends dsDNA upon binding [174] and strongly confer dsDNA compaction [175,176]. However, it has also been shown that at high concentrations, H-NS binds to DNA from end-to-end with no significant compaction [162]. In 2003, using single-molecule force spectroscopy [177,178], Stavans et al., reported that at physiological concentrations, H-NS binding to λ-DNA covers extensive DNA-tracts, causing the contour length of the λ-DNA to extend 2-fold and DNA stiffness (measured by Lp) to increase 3-fold (Figure 6a) [179]. The result indicated that each H-NS dimer occupies 15–20 base-pairs along the λ-DNA, which was further supported by a structural investigation of the H-NS/DNA complex by using X-ray crystallography [180].
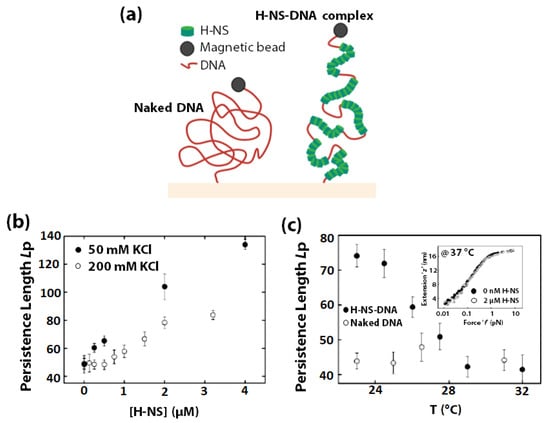
Figure 6.
The effect of osmolarity and temperature on H-NS/DNA complex was investigated through force-extension experiments. (a) Schematic representation of the extended configuration of H-NS-λ-DNA complexes compares to that of naked λ-DNA in the low-tension regime (≤10 pN). The setup for measuring the end-to-end extension of DNA as a function of stretching force (magnetic force) is shown where DNA is tethered to a magnetic bead on end and a glass surface on the other. Change in persistence length (Lp) of (b) H-NS-DNA as a function of increasing H-NS concentration in the presence of 50 mM (solid circles) and 200 mM (open circles) KCl, (c) H-NS-DNA complexes (solid circles) and naked DNA (open circles) with increasing temperature. Inset: Extension ‘z’ in nm (y-axis) of single H-NS-DNA complexes as a function of force (f) in pN (x-axis) measured at 37 °C for 0 (solid circles) and 2 µM (open circles) H-NS. (b,c) adapted with permission from [179]. Copyright 2003, Biophysical Society.
Stavans et al., showed that H-NS’s polymerization on DNA is sensitive to changes in osmolarity and temperature. The Lp of λ-DNA increases with increasing H-NS concentration and decreasing ionic strength (Figure 6b), and decreases with temperature (Figure 6c) up to 32 °C. However, at 37 °C, H-NS no longer induces any rigidity change of DNA. Not only do the above results provide remarkable insights into the mechanism of N-HS mediated gene silencing in vivo, but the system also demonstrated great potential as an in vivo osmolarity and temperature sensor for subcellular local environment.
3.5. DNA Bending Assisted Protein Sensing
Histone-like proteins in prokaryotic cells and transcription factors (TFs) are essential architectural and functional regulators of genetic materials in cells [181,182]. Understanding of their function and regulation requires sensitive strategies to detect their interactions with DNA [183,184,185,186,187,188]. In 2004, Shen et al., reported an elegant DNA-based nanomechanical sensor that involves bending of DNA upon binding to E. coli Integration Host Factor (IHF), a histone-like protein [189]. The device consists of a DNA double-helical shaft (consisting of the IHF binding site) connecting with two rigid DNA triple crossover (TX) motifs (Figure 7a). The two TX motifs are individually labelled with a FRET donor D (green) and acceptor A (yellow) [190].
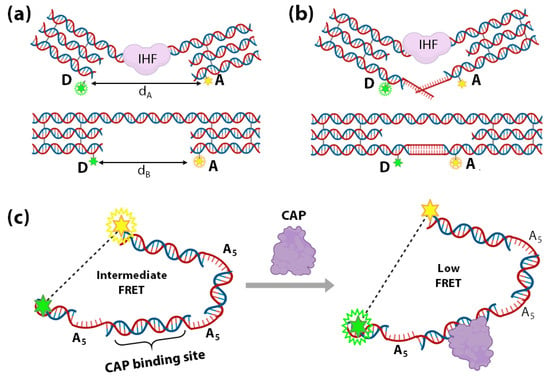
Figure 7.
(a,b) Schematic representation of a nanomechanical device with the double helices as rectangular boxes and the TX motifs as three infused rectangular boxes. The upper domain connects the two TX motifs with the binding site for IHF. Each TX motif is labelled with either FRET donor fluorescein (D) or FRET acceptor Cy3 (A). IHF (purple) binding distorts the connecting shaft in the lower panel, consequently increasing the distance between D and A (dB to dA) and thus decreasing FRET. (b) shows that the bottom domains are extended and connected by a cohesive tract [189]. DNA bending-based TF (CAP) sensor. (c) Left: Schematic of the sensor constituted by a dsDNA consisting of three A5 kinks and a CAP binding site (indicated by the blue dotted line). The donor (glowing green star) and the acceptor (glowing yellow star) fluorophores are depicted to be attached on either end of the sensor dsDNA, respectively. Right: Schematic of the pulling far apart of both the fluorophore-tagged ends of the dsDNA sensor from each other due to DNA bending induced by CAP binding [191].
The FRET signal decreases as the IHF binds and distorts its binding site, increasing the distance between the FRET-pair (Figure 7a). Shen et al., have further extended this system by connecting the TX motifs with a pair of complementary DNA strands (Figure 7b). If the IHF binding free energy exceeds the DNA hybridization free energy, IHF binding is energetically favoured (Figure 7b). Therefore, when the length of complementary base-pairs increases, the ability of IHF binding to the structure decreases as the overall free energy favours the DNA-bound structure more than IHF-bound structure. With this method, the device was used to estimate the binding free energy between IHF and DNA.
DNA-bending is one of the most significant mechanisms employed in TF-mediated gene modulation [192,193]. In 2012, Crawford et al., developed a DNA bending-based TF biosensor that can discriminate between the TF’s active and inactive forms. As a proof-of-concept, their system employs a FRET-based detection of low concentrations of Catabolite gene Activator Protein (CAP). The dsDNA sensor designed by Crawford et al., consists of three 5-adenine (A5) kinks (5 unpaired adenines) around a CAP-binding site (Figure 7c). The unpaired adenines in the A5 kinks confer specific sequence-directed bends, i.e., DNA bend angle of 73° ± 11° [194] bringing the fluorophore pair at the ends of the sensor DNA within the FRET range (Figure 7c). As shown in Figure 7c, the addition of CAP moves the ends farther apart, consequently reducing FRET in a concentration-dependent manner and enabling the detection of low concentrations of CAP. This system can be extrapolated appreciably to many more TFs, which marks significant progress in TF sensing.
4. DNA-Flexibility: A Game-Changer for DNA Nanostructures
The field of DNA origami is progressing rapidly and has produced nanostructures and nanomachines with fascinating applications. The 3-dimensional geometry of DNA origami nanostructures largely depends on the structural rigidity of the constituent DNA molecules [195]. In order to predict the shape and rigidity of origami in solution, multiple state-of-the-art computational modelling methods have been developed that can compute origami features like DNA bending, stretching, twisting, stiffness, and elasticity [196,197,198,199]. We will focus on two of the many frontiers in DNA origami research, namely the “circular DNA-based origami” and “DNA nanomachines”. With the examples discussed below, we aim to highlight how they heavily take advantage of the mechanical rigidity of DNA. We also highlight how small perturbations in the mechanical rigidity of DNA brings a significant change in the efficiency of functional DNA nanostructures.
4.1. Circular DNA as the Basis of Origami
One of the most captivating categories of bent or curved DNA is that of circular DNA. Although commercial synthesis of circular DNA is expensive for day-to-day experiments, various protocols are available for circularizing linear DNA. The propensity of a DNA to circularize is directly related to its sequence- and length-dependent mechanical rigidity. Cyclization propensity (calculated as the Jacobson-Stockmayer J factor) is a well-established method for experimentally and computationally probing a DNA fragment’s rigidity [4,130,200,201]. DNA nanotechnology has exploited the flexibility-based cyclization of DNA fragments to construct fascinating DNA origami nanostructures. These include circular interlocks, single- and multiple-ring DNA catenanes, Borromean rings, and rotaxanes [202,203,204,205]. Circular functional nucleic acids have been employed as rolling circle amplification templates, aptamers, enzymes two-input logic gates, biosensors and have proved to be a cornerstone for the advancement of DNA nanotechnology [206].
4.2. Mechanical Flexibility of DNA Can Tune the Efficiency of DNA-Based Nanomachines
Recent advances in artificially synthesized molecular machines have massively taken advantage of the structural and mechanical properties of DNA. DNA has been employed as a critical component of various types of synthetic molecular machines and of the molecular tracks which the machines navigate [207,208,209,210]. Nevertheless, these synthetic machines have yet not matched the biological macromolecular machines in terms of their vital characteristics, such as directionality, step size, processivity, speed, and chemical yield. Since DNA nanomechanics plays a crucial role in the systems overall free energy, it is intuitive to think that tuning the flexibility of the constitutive DNA may have a significant impact on the performance of such machines.
Tomov et al., have demonstrated the significance of DNA flexibility in the efficiency of functional DNA nanostructures through experiments and molecular simulations of a DNA bipedal motor’s walking dynamics [211,212,213,214,215]. For example, the foothold and the walker legs in their current design are shorter than the Lp of ds DNA and need to be made more flexible to achieve a larger step size (Figure 8a). However, the increase in such flexibility also enhances the search-space volume available to the walker, thus increasing the activation barrier to leg-placing and reducing the stepping yields. Additionally, they also pointed out that the walker’s efficiency is sensitive to the mechanical properties and curvature of the DNA origami track. They further pointed out that their design of the 2D origami sheet bears a curvature as the origami sheet’s effective designed pitch (10.67 bp/turn) is larger than that of real DNA (10.5 bp/turn). They suggested reducing this origami bending or curvature into a stiffer walking track will assist the walker in moving in the intended direction (Figure 8b).
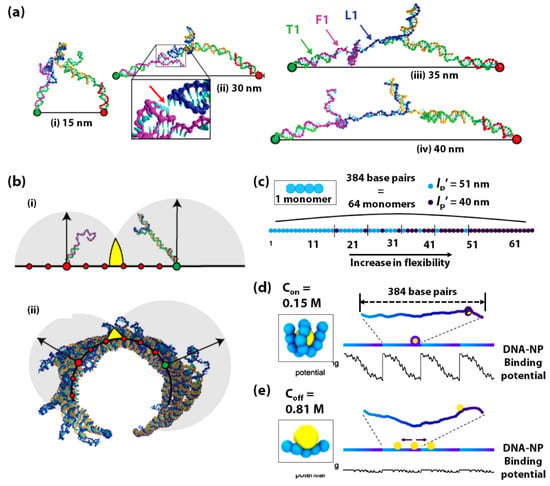
Figure 8.
(a) Brownian dynamics simulations of coarse-grained models of the transition state configurations of the DNA bipedal walker, illustrating the increasingly stretched nature of the walker at (i) 15 nm, (ii) 30 nm, (iii) 35 nm, and (iv) 40 nm step sizes, with T1, F1, and L1 being one of the two the ‘foothold strand’, ‘Fuel strand’, and ‘Leg strand’. The red arrow in (ii) indicates the position of a leg-placing base pair. (b) Schematic of the DNA bipedal walker (blue-orange) with (i) one foot up and the other down on a flat surface, and (ii) an axial view of the configuration of the 2D origami sheet curled up into a tubular structure, with the red circles denoting the short axis foothold positions. The step size (~32 nm) is denoted by the larger green and red circles, which is the same for both (i) and (ii). The gray circles in both (i) and (ii) represent the regions of space well-sampled (95% of configurations have their terminal base within the sphere) by the unbound section of the walker (blue) and the foothold-bound fuel (purple) when the secondary structure was forbidden in the simulations. Arrows represent the radii of the spheres, and the yellow colour indicates the overlap between the spheres. (a,b) are adapted with permission from [211]. Copyright 2017, the authors. (c) Schematic of the dsDNA fragment modelled by a chain of 64 monomers (1 monomer = 6 basepairs) with flexibility gradient. The local persistence length (Lp’) is set for each monomer at either 51 or 40 nm. The sequence-dependent dsDNA flexibility is gradually increased by increasing the fraction of monomers with Lp’ = 40 nm. Schematic of the long dsDNA containing four repeat fragments with the flexibility gradient, together with its DNA-NP binding potential energy along the entire dsDNA molecule at solution salt concentrations of (d) Con = 0.15 M and (e) Coff = 0.81 M, respectively. The inset in (d,e) shows the specific representative configuration of DNA-NP complex for which the bending energy is calculated. (c–e) are adapted with permission from [212]. Copyright 2019, the authors.
It is a known fact that while the average persistence length (Lp) of dsDNA is ~50 nm and increases significantly with increasing GC content, the local persistence length (Lp’) of dinucleotide steps ranges from 40–55 nm [216,217]. In a unique computational study, Park et al., have exploited the sequence-dependent flexibility of DNA to propel a DNA-based Brownian ratchet for directional transporting positively charged nanoparticles [212]. In this study, Brownian dynamics simulations of coarse-grained models have been implemented to construct a single, 130 nm long, dsDNA with its sequence-dependent local flexibility gradually increasing along its length at the physiological salt concentration (0.15 M) (Figure 8c). Further, a positively charged nanoparticle (NP) is simulated to bind to this DNA. The DNA-NP binding causes the DNA to bend (Figure 8d), which augments the effect of the variation in sequence-dependent DNA flexibility. This DNA flexibility gradient creates an asymmetric potential for the DNA-NP binding and fuels the directional and processive motion of the NP towards the higher flexibility region on the DNA. However, upon increasing the salt concentration to 0.81 M the DNA-NP binding does not induce significant bending (Figure 8e), and thus this binding is less dependent on DNA flexibility. In this case, there is a negligible gradient; therefore, the NP diffuses randomly in either direction on the dsDNA. The DNA-NP system designed in this study has this 130 nm long fragment repeated such that the asymmetric potential is repeated periodically in a single, long dsDNA molecule. By repeatedly switching the salt concentration between 0.15 M (Con) and 0.81 M (Coff) over several cycles, the directional and processive motion of NP is demonstrated.
In one of the latest reports, Suzuki et al., have taken advantage of serially repeated tension-adjustable modules to induce large reversible structural deformations in a DNA-origami nano arm (fabricated as an eight-helix DNA bundle) [218]. The tension-adjustable modules are composed of a stem (4 helices), stiff piers (>4 helices including the stem), and bridge strands. The deformation of the nano arm into increasingly complex shapes has been demonstrated by placing varying amounts of tension in its flexible DNA modules.
5. Summary and Future Directions
In summary, dsDNA’s mechanical rigidity has been used in a wide range of nanotechnology applications, from understanding and controlling biologically active molecules to probing cellular mechanics and creating nanomachines. The free energy required to bend a piece of dsDNA adds a new dimension in controlling the conformational states of the molecule in addition to the traditional base-pairing interactions. However, there are still only a few examples that fully utilize the mechanical bending of dsDNA in sensing applications at the current stage. We foresee that incorporating DNA bending as a functional element to the existing designs can create new generations of DNA-based nanodevices and sensors with bifunctional properties and extended sensitivity range in the near future.
Author Contributions
Writing—original draft preparation, R.S.; writing—review and editing, R.S., I.T.S.L., Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC), grant number RGPIN-2017-04407, New Frontiers in Research Fund, grant number NFRFE-2018-00969, and Michael Smith Foundation for Health Research, grant number SCH-2020-0559. This work was also supported by the Arkansas Biosciences Institute, grant numbers ABI-0189, ABI-0226, ABI-0277, ABI-0326, and ABI2021, and the National Science Foundation, grant number CBET-1826642.
Conflicts of Interest
Y.W. declares that a patent (pending) with him as an inventor has been filed for the concept and realization of the bent DNA bows as amplifiers and biosensors by the University of Arkansas. Other authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Seeman, N.C. DNA Nanotechnology at 40. Nano Lett. 2020, 20, 1477–1478. [Google Scholar] [CrossRef]
- Peters, J.P.; Maher, L.J. DNA Curvature and Flexibility in Vitro and in Vivo. Q. Rev. Biophys. 2010, 43, 23–63. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.T.; Yang, H.; Chu, J.W. Structure-Mechanics Statistical Learning Unravels the Linkage between Local Rigidity and Global Flexibility in Nucleic Acids. Chem. Sci. 2020, 11, 4969–4979. [Google Scholar] [CrossRef]
- Vologodskii, A.; Frank-Kamenetskii, M.D. Strong Bending of the DNA Double Helix. Nucleic Acids Res. 2013, 41, 6785–6792. [Google Scholar] [CrossRef]
- Drozdetski, A.V.; Anandakrishnan, R.; Onufriev, A. Estimating Persistence Length of DNA from Molecular Dynamics Simulations. Biophys. J. 2015, 108, 233a. [Google Scholar] [CrossRef][Green Version]
- Beveridge, D.L. Molecular Dynamics Simulations of DNA Curvature and Flexibility: Helix Phasing and Premelting. Biopolymers 2020, 73, 380–403. [Google Scholar] [CrossRef] [PubMed]
- Curuksu, J.; Zacharias, M.; Lavery, R.; Zakrzewska, K. Local and Global Effects of Strong DNA Bending Induced during Molecular Dynamics Simulations. Nucleic Acids Res. 2009, 37, 3766–3773. [Google Scholar] [CrossRef]
- Brinkers, S.; Dietrich, H.R.C.; De Groote, F.H.; Young, I.T.; Rieger, B. The Persistence Length of Double Stranded DNA Determined Using Dark Field Tethered Particle Motion. J. Chem. Phys. 2009, 130. [Google Scholar] [CrossRef]
- Wang, H.; Milstein, J.N. Simulation Assisted Analysis of the Intrinsic Stiffness for Short DNA Molecules Imaged with Scanning Atomic Force Microscopy. PLoS ONE 2015, 10, 1–11. [Google Scholar] [CrossRef][Green Version]
- Olson, W.K.; Zhurkin, V.B. Modeling DNA Deformations. Curr. Opin. Struct. Biol. 2000, 10, 286–297. [Google Scholar] [CrossRef]
- Strauss, J.K.; Maher, L.J. DNA Bending by Asymmetric Phosphate Neutralization. Science 1994, 266, 1829–1834. [Google Scholar] [CrossRef] [PubMed]
- Strutz, K. Intrinsic Curvature of Plasmid DNAs Analyzed by Polyacrylamide Gel Electrophoresis. Electrophoresis 1996, 17, 989–995. [Google Scholar] [CrossRef] [PubMed]
- Crothers, D.M.; Drak, J. Global Features of DNA Structure by Comparative Gel Electrophoresis. Methods Enzymol. Acad. Press 1992, 212, 46–71. [Google Scholar] [CrossRef]
- Hellman, L.M.; Fried, M.G. Electrophoretic Mobility Shift Assays. Nat. Methods 2005, 2, 557–558. [Google Scholar] [CrossRef]
- Jost, J.P.; Saluz, H.P. (Eds.) A Laboratory Guide to In Vitro Studies of Protein-DNA Interactions; Birkhäuser: Basel, Switzerland, 1991; Volume 5, pp. 245–257. [Google Scholar] [CrossRef]
- Harrington, R.E. Studies of DNA Bending and Flexibility Using Gel Electrophoresis. Electrophoresis 1993, 14, 732–746. [Google Scholar] [CrossRef]
- Freeland, J.; Khadka, P.; Wang, Y. Mechanical-Energy-Based Amplifiers for Probing Interactions of DNA with Metal Ions. Phys. Rev. E 2018, 98, 62403. [Google Scholar] [CrossRef]
- Freeland, J.; Zhang, L.; Wang, S.-T.; Ruiz, M.; Wang, Y. Bent DNA Bows as Amplifiers and Biosensors for Detecting DNA-Interacting Salts and Molecules. bioRxiv 2020. [Google Scholar] [CrossRef]
- Vafabakhsh, R.; Ha, T. Extreme Bendability of DNA Less than 100 Base Pairs Long Revealed by Single-Molecule Cyclization. Science 2012, 337, 1097–1101. [Google Scholar] [CrossRef]
- Jeong, J.; Kim, H.D. Determinants of Cyclization-Decyclization Kinetics of Short DNA with Sticky Ends. bioRxiv 2018. [Google Scholar] [CrossRef]
- Du, Q.; Vologodskaia, M.; Kuhn, H.; Frank-Kamenetskii, M.; Vologodskii, A. Gapped DNA and Cyclization of Short DNA Fragments. Biophys. J. 2005, 88, 4137–4145. [Google Scholar] [CrossRef]
- Le, T.T.; Kim, H.D. Studying DNA Looping by Single-Molecule FRET. J. Vis. Exp. 2014, 88, e51667. [Google Scholar] [CrossRef] [PubMed]
- Ngo, T.T.M.; Yoo, J.; Dai, Q.; Zhang, Q.; He, C.; Aksimentiev, A.; Ha, T. Effects of Cytosine Modifications on DNA Flexibility and Nucleosome Mechanical Stability. Nat. Commun. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dlakic, M.; Harrington, R.E. Bending and Torsional Flexibility of G/C-Rich Sequences as Determined by Cyclization Assays. J. Biol. Chem. 1995, 270, 29945–29952. [Google Scholar] [CrossRef] [PubMed]
- Shore, D.; Langowski, J.; Baldwin, R.L. DNA Flexibility Studied by Covalent Closure of Short Fragments into Circles. Proc. Natl. Acad. Sci. USA 1981, 78, 4833–4837. [Google Scholar] [CrossRef]
- Yan, J.; Marko, J.F. Localized Single-Stranded Bubble Mechanism for Cyclization of Short Double Helix DNA. Phys. Rev. Lett. 2004, 93, 3–6. [Google Scholar] [CrossRef]
- Cherstvy, A.G. DNA Cyclization: Suppression or Enhancement by Electrostatic Repulsions? J. Phys. Chem. B 2011, 115, 4286–4294. [Google Scholar] [CrossRef]
- Cloutier, T.E.; Widom, J. DNA Twisting Flexibility and the Formation of Sharply Looped Protein–DNA Complexes. Proc. Natl. Acad. Sci. USA 2005, 102, 3645LP–3650. [Google Scholar] [CrossRef]
- Crothers, D.M.; Drak, J.; Kahn, J.D.; Levene, S.D. DNA Bending, Flexibility, and Helical Repeat by Cyclization Kinetics. Methods Enzymol. 1992, 212, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Cloutier, T.E.; Widom, J. Spontaneous Sharp Bending of Double-Stranded DNA. Mol. Cell 2004, 14, 355–362. [Google Scholar] [CrossRef]
- Du, Q.; Smith, C.; Shiffeldrim, N.; Vologodskaia, M.; Vologodskii, A. Cyclization of Short DNA Fragments and Bending Fluctuations of the Double Helix. Proc. Natl. Acad. Sci. USA 2005, 102, 5397LP–5402. [Google Scholar] [CrossRef]
- Driessen, R.P.C.; Sitters, G.; Laurens, N.; Moolenaar, G.F.; Wuite, G.J.L.; Goosen, N.; Dame, R.T. Effect of Temperature on the Intrinsic Flexibility of DNA and Its Interaction with Architectural Proteins. Biochemistry 2014, 53, 6430–6438. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Mousley, B.; Gambino, S.; Helou, E.; Loparo, J.; Price, A.C. Tethered Particle Motion with Single DNA Molecules. Am. J. Phys. 2015, 83, 418–426. [Google Scholar] [CrossRef]
- Kovari, D.T.; Yan, Y.; Finzi, L.; Dunlap, D. Tethered Particle Motion: An Easy Technique for Probing DNA Topology and Interactions with Transcription Factors. Methods Mol. Biol. 2018, 1665, 317–340. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.D.; Yin, H.; Landick, R.; Gelles, J.; Block, S.M. Stretching DNA with Optical Tweezers. Biophys. J. 1997, 72, 1335–1346. [Google Scholar] [CrossRef]
- Gross, P.; Laurens, N.; Oddershede, L.B.; Bockelmann, U.; Peterman, E.J.G.; Wuite, G.J.L. Quantifying How DNA Stretches, Melts and Changes Twist under Tension. Nat. Phys. 2011, 7, 731–736. [Google Scholar] [CrossRef]
- Bockelmann, U.; Thomen, P.; Essevaz-Roulet, B.; Viasnoff, V.; Heslot, F. Unzipping DNA with Optical Tweezers: High Sequence Sensitivity and Force Flips. Biophys. J. 2002, 82, 1537–1553. [Google Scholar] [CrossRef]
- van Mameren, J.; Wozniak, A.; Ragona, S. Single-Molecule DNA Stretching Using Optical Tweezers. Micros. Today 2009, 17, 42–43. [Google Scholar] [CrossRef]
- Epstein, C.S.; Mann, A.J. Measurement of the DNA Spring Constant Using Optical Tweezers. Phys. Rev. E. 2003, 67, 051906. [Google Scholar]
- Kang, J.; Jung, J.; Kim, S.K. Flexibility of Single-Stranded DNA Measured by Single-Molecule FRET. Biophys. Chem. 2014, 195, 49–52. [Google Scholar] [CrossRef]
- Murphy, M.C.; Rasnik, I.; Cheng, W.; Lohman, T.M.; Ha, T. Probing Single-Stranded DNA Conformational Flexibility Using Fluorescence Spectroscopy. Biophys. J. 2004, 86, 2530–2537. [Google Scholar] [CrossRef]
- Dragan, A.I.; Privalov, P.L. Use of Fluorescence Resonance Energy Transfer (FRET) in Studying Protein-Induced DNA Bending. Methods Enzymol. 2008, 450, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.L.; Parkhurst, L.K.; Parkhurst, L.J. Changes in DNA Bending and Flexing Due to Tethered Cations Detected by Fluorescence Resonance Energy Transfer. Nucleic Acids Res. 2006, 34, 1028–1035. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Woźniak, A.K.; Schröder, G.F.; Grubmüller, H.; Seidela, C.A.M.; Oesterhelt, F. Single-Molecule FRET Measures Bends and Kinks in DNA. Proc. Natl. Acad. Sci. USA 2008, 105, 18337–18342. [Google Scholar] [CrossRef]
- Tóth, K.; Sauermann, V.; Langowski, J. DNA Curvature in Solution Measured by Fluorescence Resonance Energy Transfer. Biochemistry 1998, 37, 8173–8179. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Kim, H.D. Probing the Elastic Limit of DNA Bending. Nucleic Acids Res. 2014, 42, 10786–10794. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, S.V.; Sugimura, S.; Vivas, P.; Crothers, D.M.; Ansari, A. Direct Observation of DNA Bending/Unbending Kinetics in Complex with DNA-Bending Protein IHF. Proc. Natl. Acad. Sci. USA 2006, 103, 18515LP–18520. [Google Scholar] [CrossRef] [PubMed]
- Maaloum, M.; Muller, P.; Harlepp, S. DNA-Intercalator Interactions: Structural and Physical Analysis Using Atomic Force Microscopy in Solution. Soft Matter 2013, 9, 11233–11240. [Google Scholar] [CrossRef]
- Hansma, H.G.; Laney, D.E.; Bezanilla, M.; Sinsheimer, R.L.; Hansma, P.K. Applications for Atomic Force Microscopy of DNA. Biophys. J. 1995, 68, 1672–1677. [Google Scholar] [CrossRef]
- Mazur, A.K.; Maaloum, M. Atomic Force Microscopy Study of DNA Flexibility on Short Length Scales: Smooth Bending versus Kinking. Nucleic Acids Res. 2014, 42, 14006–14012. [Google Scholar] [CrossRef][Green Version]
- Beckwitt, E.C.; Kong, M.; Van Houten, B. Studying Protein-DNA Interactions Using Atomic Force Microscopy. Semin. Cell Dev. Biol. 2018, 73, 220–230. [Google Scholar] [CrossRef]
- Hansma, H. Surface Biology of DNA by Atomic Force Microscopy. Annu. Rev. Phys. Chem. 2001, 52, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Faas, F.G.A.; Rieger, B.; van Vliet, L.J.; Cherny, D.I. DNA Deformations near Charged Surfaces: Electron and Atomic Force Microscopy Views. Biophys. J. 2009, 97, 1148–1157. [Google Scholar] [CrossRef] [PubMed]
- Kahn, J.D. DNA, Flexibly Flexible. Biophys. J. 2014, 107, 282–284. [Google Scholar] [CrossRef][Green Version]
- Camunas-Soler, J.; Ribezzi-Crivellari, M.; Ritort, F. Elastic Properties of Nucleic Acids by Single-Molecule Force Spectroscopy. Annu. Rev. Biophys. 2016, 45, 65–84. [Google Scholar] [CrossRef] [PubMed]
- Demurtas, D.; Amzallag, A.; Rawdon, E.J.; Maddocks, J.H.; Dubochet, J.; Stasiak, A. Bending Modes of DNA Directly Addressed by Cryo-Electron Microscopy of DNA Minicircles. Nucleic Acids Res. 2009, 37, 2882–2893. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lukoyanova, N.; Miah, S.; Lucas, J.; Vaughan, C.K. Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10. Cell Rep. 2018, 24, 744–754. [Google Scholar] [CrossRef]
- Bednar, J.; Furrer, P.; Katritch, V.; Stasiak, A.; Dubochet, J.; Stasiak, A. Determination of DNA Persistence Length by Cryo-Electron Microscopy. Separation of the Static and Dynamic Contributions to the Apparent Persistence Length of DNA. J. Mol. Biol. 1995, 254, 579–594. [Google Scholar] [CrossRef]
- Bai, X.C.; Martin, T.G.; Scheres, S.H.W.; Dietz, H. Cryo-EM Structure of a 3D DNA-Origami Object. Proc. Natl. Acad. Sci. USA 2012, 109, 20012–20017. [Google Scholar] [CrossRef]
- Dunlop, D. Scanning Tunneling Microscopy of DNA. IEEE Eng. Med. Biol. Mag. 1996, 15, 46–50. [Google Scholar] [CrossRef]
- Marilley, M.; Pasero, P.; Humbert, A.; Granjeaud, S.; Dayez, M.; Pierrisnard, R.; Jordan, B. Scanning Tunneling Microscopy Study of a DNA Fragment of Known Size and Sequence. Microsc. Microanal. Microstruct. 1994, 5, 47–56. [Google Scholar] [CrossRef]
- Müller, T.; Gilson, E.; Schmidt, R.; Giraldo, R.; Sogo, J.; Gross, H.; Gasser, S. Imaging the Asymmetrical DNA Bend Induced by Repressor Activator Protein 1 with Scanning Tunneling Microscopy. J. Struct. Biol. 1994, 113, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Subirana, J.A. A Crystallographic Approach to DNA Bending: Prediction of Nusleosome Formation by DNA Triple Repeats and: Other Repetitive Sequences. FEBS Lett. 1997, 417, 352–354. [Google Scholar] [CrossRef]
- Kornyshev, A.A.; Lee, D.J.; Wynveen, A.; Leikin, S. Signatures of DNA Flexibility, Interactions and Sequence-Related Structural Variations in Classical X-Ray Diffraction Patterns. Nucleic Acids Res. 2011, 39, 7289–7299. [Google Scholar] [CrossRef] [PubMed]
- Mathew-Fenn, R.S.; Das, R.; Harbury, P.A.B. Remeasuring the Double Helix. Science 2008, 322, 446–449. [Google Scholar] [CrossRef]
- Mastroianni, A.J.; Sivak, D.A.; Geissler, P.L.; Alivisatos, A.P. Probing the Conformational Distributions of Subpersistence Length DNA. Biophys. J. 2009, 97, 1408–1417. [Google Scholar] [CrossRef]
- Vermeulen, A.; Zhou, H.; Pardi, A. Determining DNA Global Structure and DNA Bending by Application of NMR Residual Dipolar Couplings. J. Am. Chem. Soc. 2000, 122, 9638–9647. [Google Scholar] [CrossRef]
- Ben Imeddourene, A.; Xu, X.; Zargarian, L.; Oguey, C.; Foloppe, N.; Mauffret, O.; Hartmann, B. The Intrinsic Mechanics of B-DNA in Solution Characterized by NMR. Nucleic Acids Res. 2016, 44, 3432–3447. [Google Scholar] [CrossRef]
- Wu, Z.; Maderia, M.; Barchi, J.J.; Marquez, V.E.; Bax, A. Changes in DNA Bending Induced by Restricting Nucleotide Ring Pucker Studied by Weak Alignment NMR Spectroscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 24–28. [Google Scholar] [CrossRef]
- Campagne, S.; Gervais, V.; Milon, A. Nuclear Magnetic Resonance Analysis of Protein—DNA Interactions. J. R. Soc. Interface 2011, 8, 1065–1078. [Google Scholar] [CrossRef]
- Nathan, D.; Crothers, D.M. Bending and Flexibility of Methylated and Unmethylated EcoRI DNA. J. Mol. Biol. 2002, 316, 7–17. [Google Scholar] [CrossRef]
- Renzoni, D.; Esposito, D.; Pfuhl, M.; Hinton, J.C.D.; Higgins, C.F.; Driscoll, P.C.; Ladbury, J.E. Structural Characterization of the N-Terminal Oligomerization Domain of the Bacterial Chromatin-Structuring Protein, H-NS1 1Edited by A. R. Fersht. J. Mol. Biol. 2001, 306, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Meints, G.A.; Drobny, G.P. Dynamic Impact of Methylation at the M. HhaI Target Site: A Solid-State Deuterium NMR Study. Biochemistry 2001, 40, 12436–12443. [Google Scholar] [CrossRef]
- Hagerman, P.J. Investigation of the Flexibility of DNA Using Transient Electric Birefringence. Biopolymers 1981, 20, 1503–1535. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.B.; Hagerman, P.J. Origin of the Intrinsic Rigidity of DNA. Nucleic Acids Res. 2004, 32, 4055–4059. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.E.; Moemer, W.E. Method for Trapping and Manipulating Nanoscale Objects in Solution. Appl. Phys. Lett. 2005, 86, 1–3. [Google Scholar] [CrossRef]
- Dekker, J.; Rippe, K.; Dekker, M.; Kleckner, N. Capturing Chromosome Conformation. Science 2002, 295, 1306LP–1311. [Google Scholar] [CrossRef]
- Göndör, A.; Rougier, C.; Ohlsson, R. High-Resolution Circular Chromosome Conformation Capture Assay. Nat. Protoc. 2008, 3, 303–313. [Google Scholar] [CrossRef]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289LP–293. [Google Scholar] [CrossRef]
- Zhou, G.-L.; Xin, L.; Song, W.; Di, L.-J.; Liu, G.; Wu, X.-S.; Liu, D.-P.; Liang, C.-C. Active Chromatin Hub of the Mouse Alpha-Globin Locus Forms in a Transcription Factory of Clustered Housekeeping Genes. Mol. Cell. Biol. 2006, 26, 5096–5105. [Google Scholar] [CrossRef]
- Becker, N.A.; Kahn, J.D.; James Maher, L. Bacterial Repression Loops Require Enhanced DNA Flexibility. J. Mol. Biol. 2005, 349, 716–730. [Google Scholar] [CrossRef]
- Becker, N.A.; Kahn, J.D.; Maher, L.J., 3rd. Effects of Nucleoid Proteins on DNA Repression Loop Formation in Escherichia coli. Nucleic Acids Res. 2007, 35, 3988–4000. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Oehler, S.; Müller-Hill, B. Repression of Lac Promoter as a Function of Distance, Phase and Quality of an Auxiliary Lac Operator. J. Mol. Biol. 1996, 257, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Oehler, S.; Eismann, E.R.; Krämer, H.; Müller-Hill, B. The Three Operators of the Lac Operon Cooperate in Repression. EMBO J. 1990, 9, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Oehler, S.; Müller-Hill, B. High Local Concentration: A Fundamental Strategy of Life. J. Mol. Biol. 2010, 395, 242–253. [Google Scholar] [CrossRef]
- Becker, N.A.; Kahn, J.D.; Maher, L.J., 3rd. Eukaryotic HMGB Proteins as Replacements for HU in E. coli Repression Loop Formation. Nucleic Acids Res. 2008, 36, 4009–4021. [Google Scholar] [CrossRef]
- Bellomy, G.R.; Mossing, M.C.; Record, M.T.J. Physical Properties of DNA in Vivo as Probed by the Length Dependence of the Lac Operator Looping Process. Biochemistry 1988, 27, 3900–3906. [Google Scholar] [CrossRef]
- Garcia, H.G.; Grayson, P.; Han, L.; Inamdar, M.; Kondev, J.; Nelson, P.C.; Phillips, R.; Widom, J.; Wiggins, P.A. Biological Consequences of Tightly Bent DNA: The Other Life of a Macromolecular Celebrity. Biopolymers 2007, 85, 115–130. [Google Scholar] [CrossRef]
- Zhang, Y.; McEwen, A.E.; Crothers, D.M.; Levene, S.D. Statistical-Mechanical Theory of DNA Looping. Biophys. J. 2006, 90, 1903–1912. [Google Scholar] [CrossRef]
- Zhang, Y.; McEwen, A.E.; Crothers, D.M.; Levene, S.D. Analysis of In-Vivo LacR-Mediated Gene Repression Based on the Mechanics of DNA Looping. PLoS ONE 2006, 1. [Google Scholar] [CrossRef]
- Krämer, H.; Niemöller, M.; Amouyal, M.; Revet, B.; von Wilcken-Bergmann, B.; Müller-Hill, B. Lac Repressor Forms Loops with Linear DNA Carrying Two Suitably Spaced Lac Operators. EMBO J. 1987, 6, 1481–1491. [Google Scholar] [CrossRef]
- Law, S.M.; Bellomy, G.R.; Schlax, P.J.; Record, M.T.J. In Vivo Thermodynamic Analysis of Repression with and without Looping in Lac Constructs. Estimates of Free and Local Lac Repressor Concentrations and of Physical Properties of a Region of Supercoiled Plasmid DNA in Vivo. J. Mol. Biol. 1993, 230, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Mossing, M.C.; Record, M.T.J. Upstream Operators Enhance Repression of the Lac Promoter. Science 1986, 233, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.B.; Cui, Y.; Bustamante, C. Overstretching B-DNA: The Elastic Response of Individual Double-Stranded and Single-Stranded DNA Molecules. Science 1996, 271, 795–799. [Google Scholar] [CrossRef]
- Williams, L.D.; Maher, L.J. 3rd. Electrostatic Mechanisms of DNA Deformation. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 497–521. [Google Scholar] [CrossRef] [PubMed]
- Range, K.; Mayaan, E.; Maher, L.J.; York, D.M. The Contribution of Phosphate-Phosphate Repulsions to the Free Energy of DNA Bending. Nucleic Acids Res. 2005, 33, 1257–1268. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Strauss, J.K.; Roberts, C.; Nelson, M.G.; Switzer, C.; Maher, L.J. DNA Bending by Hexamethylene-Tethered Ammonium Ions. Proc. Natl. Acad. Sci. USA 1996, 93, 9515–9520. [Google Scholar] [CrossRef]
- Manning, G.S.; Ebralidse, K.K.; Mirzabekov, A.D.; Rich, A. An Estimate of the Extent of Folding of Nucleosomal DNA by Laterally Asymmetric Neutralization of Phosphate Groups. J. Biomol. Struct. Dyn. 1989, 6, 877–889. [Google Scholar] [CrossRef]
- Strauss-Soukup, J.K.; Vaghefi, M.M.; Hogrefe, R.I.; Maher, L.J. Effects of Neutralization Pattern and Stereochemistry on DNA Bending by Methylphosphonate Substitutions. Biochemistry 1997, 36, 8692–8698. [Google Scholar] [CrossRef]
- Kim, Y.J.; Kim, D.N. Structural Basis for Elastic Mechanical Properties of the DNA Double Helix. PLoS ONE 2016, 11, 1–11. [Google Scholar] [CrossRef]
- Gore, J.; Bryant, Z.; Nöllmann, M.; Le, M.U.; Cozzarelli, N.R.; Bustamante, C. DNA Overwinds When Stretched. Nature 2006, 442, 836–839. [Google Scholar] [CrossRef]
- Guilbaud, S.; Salomé, L.; Destainville, N.; Manghi, M.; Tardin, C. Dependence of DNA Persistence Length on Ionic Strength and Ion Type. Phys. Rev. Lett. 2019, 122, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.S.; Glowacki, J.; Grandchamp, A.E.; Manning, R.S.; Maddocks, J.H. Sequence-Dependent Persistence Lengths of DNA. J. Chem. Theory Comput. 2017, 13, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Severin, P.M.D.; Zou, X.; Gaub, H.E.; Schulten, K. Cytosine Methylation Alters DNA Mechanical Properties. Nucleic Acids Res. 2011, 39, 8740–8751. [Google Scholar] [CrossRef] [PubMed]
- Carson, S.; Wilson, J.; Aksimentiev, A.; Weigele, P.R.; Wanunu, M. Hydroxymethyluracil Modifications Enhance the Flexibility and Hydrophilicity of Double-Stranded DNA. Nucleic Acids Res. 2015, 44, 2085–2092. [Google Scholar] [CrossRef] [PubMed]
- Destainville, N.; Manghi, M.; Palmeri, J. Microscopic Mechanism for Experimentally Observed Anomalous Elasticity of DNA in Two Dimensions. Biophys. J. 2009, 96, 4464–4469. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Forties, R.A.; Bundschuh, R.; Poirier, M.G. The Flexibility of Locally Melted DNA. Nucleic Acids Res. 2009, 37, 4580–4586. [Google Scholar] [CrossRef]
- Yuan, C.; Rhoades, E.; Lou, X.W.; Archer, L.A. Spontaneous Sharp Bending of DNA: Role of Melting Bubbles. Nucleic Acids Res. 2006, 34, 4554–4560. [Google Scholar] [CrossRef][Green Version]
- Choi, B.; Zocchi, G.; Wu, Y.; Chan, S.; Jeanne Perry, L.; Perry, L. Allosteric Control through Mechanical Tension. Phys. Rev. Lett. 2005, 95, 78102. [Google Scholar] [CrossRef]
- Choi, B.; Zocchi, G. Mimicking CAMP-Dependent Allosteric Control of Protein Kinase A through Mechanical Tension. J. Am. Chem. Soc. 2006, 128, 8541–8548. [Google Scholar] [CrossRef]
- Tseng, C.-Y. UCLA Mechanical Control of Enzymes Using DNA Molecular Springs. Ph.D. Thesis, UCLA, Los Angeles, CA, USA, 2013. Available online: https://escholarship.org/uc/item/49d611tc (accessed on 27 September 2018).
- Zocchi, G. Controlling Proteins Through Molecular Springs. Annu. Rev. Biophys. 2009, 38, 75–88. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, A.; Qu, H.; Zocchi, G. Protein–DNA Chimeras: Synthesis of Two-Arm Chimeras and Non-Mechanical Effects of the DNA Spring. J. Phys. Condens. Matter 2009, 21, 335103. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.Y.; Zocchi, G. Equilibrium Softening of an Enzyme Explored with the DNA Spring. Appl. Phys. Lett. 2014, 104. [Google Scholar] [CrossRef]
- Choi, B.; Zocchi, G.; Canale, S.; Wu, Y.; Chan, S.; Perry, L.J. Artificial Allosteric Control of Maltose Binding Protein. Phys. Rev. Lett. 2005, 94, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Kramer, F.R. Molecular Beacon Probes That Fluoresce on Hybridiztion. Nat. Publ. Gr. 1996, 14, 303–308. [Google Scholar]
- Saghatelian, A.; Guckian, K.M.; Thayer, D.A.; Ghadiri, M.R. DNA Detection and Signal Amplification via an Engineered Allosteric Enzyme. J. Am. Chem. Soc. 2003, 125, 344–345. [Google Scholar] [CrossRef]
- Zelin, E.; Silverman, S.K. Allosteric Control of Ribozyme Catalysis by Using DNA Constraints. ChemBioChem 2007, 8, 1907–1911. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, Q.; Li, Z.; Gu, J.; Brennan, J.D.; Li, Y. Programming a Topologically Constrained DNA Nanostructure into a Sensor. Nat. Commun. 2016, 7, 1–7. [Google Scholar] [CrossRef]
- Zhang, K.; Deng, R.; Sun, Y.; Zhang, L.; Li, J. Reversible Control of Cell Membrane Receptor Function Using DNA Nano-Spring Multivalent Ligands. Chem. Sci. 2017, 8, 7098–7105. [Google Scholar] [CrossRef]
- Wang, X.; Ha, T. Defining Single Molecular Forces Required to Activate Integrin and Notch Signaling. Science 2013, 340, 991–994. [Google Scholar] [CrossRef]
- Wang, X.; Rahil, Z.; Li, I.T.S.; Chowdhury, F.; Leckband, D.E.; Chemla, Y.R.; Ha, T. Constructing Modular and Universal Single Molecule Tension Sensor Using Protein G to Study Mechano-Sensitive Receptors. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Chowdhury, F.; Li, I.T.S.; Leslie, B.J.; Doğanay, S.; Singh, R.; Wang, X.; Seong, J.; Lee, S.H.; Park, S.; Wang, N.; et al. Single Molecular Force across Single Integrins Dictates Cell Spreading. Integr. Biol. 2015, 7, 1265–1271. [Google Scholar] [CrossRef]
- Chowdhury, F.; Li, I.T.S.; Ngo, T.T.M.; Leslie, B.J.; Kim, B.C.; Sokoloski, J.E.; Weiland, E.; Wang, X.; Chemla, Y.R.; Lohman, T.M.; et al. Defining Single Molecular Forces Required for Notch Activation Using Nano Yoyo. Nano Lett. 2016, 16, 3892–3897. [Google Scholar] [CrossRef] [PubMed]
- Li, I.T.S.; Ha, T.; Chemla, Y.R. Mapping Cell Surface Adhesion by Rotation Tracking and Adhesion Footprinting. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Yasunaga, A.; Murad, Y.; Li, I.T.S. Quantifying Molecular Tension-Classifications, Interpretations and Limitations of Force Sensors. Phys. Biol. 2019, 17, 011001. [Google Scholar] [CrossRef]
- Murad, Y.; Li, I.T.S. Quantifying Molecular Forces with Serially Connected Force Sensors. Biophys. J. 2019, 116, 1282–1291. [Google Scholar] [CrossRef] [PubMed]
- Grome, M.W.; Zhang, Z.; Lin, C. Stiffness and Membrane Anchor Density Modulate DNA-Nanospring-Induced Vesicle Tubulation. ACS Appl. Mater. Interfaces 2019, 11, 22987–22992. [Google Scholar] [CrossRef] [PubMed]
- Shroff, H.; Reinhard, B.M.; Siu, M.; Agarwal, H.; Spakowitz, A.; Liphardt, J. Biocompatible Force Sensor with Optical Readout and Dimensions of 6 Nm 3. Nano Lett. 2005, 5, 1509–1514. [Google Scholar] [CrossRef] [PubMed]
- Fields, A.P.; Meyer, E.A.; Cohen, A.E. Euler Buckling and Nonlinear Kinking of Double-Stranded DNA. Nucleic Acids Res. 2013, 41, 9881–9890. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Marras, A.E.; Su, H.J.; Castro, C.E. DNA Origami Compliant Nanostructures with Tunable Mechanical Properties. ACS Nano 2014, 8, 27–34. [Google Scholar] [CrossRef]
- Funke, J.J.; Ketterer, P.; Lieleg, C.; Schunter, S.; Korber, P.; Dietz, H. Uncovering the Forces between Nucleosomes Using DNA Origami. Sci. Adv. 2016, 2. [Google Scholar] [CrossRef]
- Iwaki, M.; Wickham, S.F.; Ikezaki, K.; Yanagida, T.; Shih, W.M. A Programmable DNA Origami Nanospring That Reveals Force-Induced Adjacent Binding of Myosin VI Heads. Nat. Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Sachs, F. Orientation-Based FRET Sensor for Real-Time Imaging of Cellular Forces. J. Cell Sci. 2012, 125, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Sachs, F. Visualizing Dynamic Cytoplasmic Forces with a Compliance-Matched FRET Sensor. J. Cell Sci. 2011, 124, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Suchyna, T.M.; Lazakovitch, E.; Gronostajski, R.M.; Sachs, F. Real Time FRET Based Detection of Mechanical Stress in Cytoskeletal and Extracellular Matrix Proteins. Cell. Mol. Bioeng. 2011, 4, 148–159. [Google Scholar] [CrossRef]
- Guo, J.; Wang, Y.; Sachs, F.; Meng, F. Actin Stress in Cell Reprogramming. Proc. Natl. Acad. Sci. USA 2014, 111, E5252–E5261. [Google Scholar] [CrossRef]
- Wang, C.; Huang, Z.; Lin, Y.; Ren, J.; Qu, X. Artificial DNA Nano-Spring Powered by Protons. Adv. Mater. 2010, 22, 2792–2798. [Google Scholar] [CrossRef]
- Lipfert, J.; Doniach, S.; Das, R.; Herschlag, D. Understanding Nucleic Acid-Ion Interactions. Annu. Rev. Biochem. 2014, 83, 813–841. [Google Scholar] [CrossRef]
- Bui, V.C.; Nguyen, T.H. DNA Aggregation Induced by Mg2+ Ions under Different Conditions. J. Mol. Recognit. 2018, 31, 1–5. [Google Scholar] [CrossRef]
- Kim, K.-J.; Choi, S.; Sang Cho, Y.; Yang, S.-J.; Cho, Y.-S.; Kim, K.K. Magnesium Ions Enhance Infiltration of Osteoblasts in Scaffolds via Increasing Cell Motility. J. Mater. Sci. Mater. Med. 2017, 28, 96. [Google Scholar] [CrossRef]
- Draper, D.E.; Grilley, D.; Soto, A.M. Ions and RNA Folding. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 221–243. [Google Scholar] [CrossRef]
- Yang, L.; Arora, K.; Beard, W.A.; Wilson, S.H.; Schlick, T. Critical Role of Magnesium Ions in DNA Polymerase Beta’s Closing and Active Site Assembly. J. Am. Chem. Soc. 2004, 126, 8441–8453. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.; Tainer, J.A.; McCammon, J.A. Unraveling the Three-Metal-Ion Catalytic Mechanism of the DNA Repair Enzyme Endonuclease IV. Proc. Natl. Acad. Sci. USA 2007, 104, 1465–1470. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, A. Role of Magnesium in Genomic Stability. Mutat. Res. 2001, 475, 113–121. [Google Scholar] [CrossRef]
- Fei, B.-L.; Xu, W.-S.; Tao, H.-W.; Li, W.; Zhang, Y.; Long, J.-Y.; Liu, Q.-B.; Xia, B.; Sun, W.-Y. Effects of Copper Ions on DNA Binding and Cytotoxic Activity of a Chiral Salicylidene Schiff Base. J. Photochem. Photobiol. B. 2014, 132, 36–44. [Google Scholar] [CrossRef]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy Metal Toxicity and the Environment. Exp. Suppl. 2012, 101, 133–164. [Google Scholar] [CrossRef]
- Hartwig, A.; Asmuss, M.; Ehleben, I.; Herzer, U.; Kostelac, D.; Pelzer, A.; Schwerdtle, T.; Bürkle, A. Interference by Toxic Metal Ions with DNA Repair Processes and Cell Cycle Control: Molecular Mechanisms. Environ. Health Perspect. 2002, 110, 797–799. [Google Scholar] [CrossRef]
- Asmuss, M.; Mullenders, L.H.; Hartwig, A. Interference by Toxic Metal Compounds with Isolated Zinc Finger DNA Repair Proteins. Toxicol. Lett. 2000, 112–113, 227–231. [Google Scholar] [CrossRef]
- Anastassopoulou, J.; Theophanides, T. Magnesium-DNA Interactions and the Possible Relation of Magnesium to Carcinogenesis. Irradiation and Free Radicals. Crit. Rev. Oncol. Hematol. 2002, 42, 79–91. [Google Scholar] [CrossRef]
- Sigel, A.; Sigel, H.; Sigel, R.K.O. Metal. Ions in Toxicology: Effects, Interactions, Interdependencies. Met. Ions Life Sci. 2011, 8, vii–viii. [Google Scholar]
- Liang, L.; Shen, J.-W.; Wang, Q. Molecular Dynamics Study on DNA Nanotubes as Drug Delivery Vehicle for Anticancer Drugs. Colloids Surf. B. Biointerfaces 2017, 153, 168–173. [Google Scholar] [CrossRef]
- Palchaudhuri, R.; Hergenrother, P.J. DNA as a Target for Anticancer Compounds: Methods to Determine the Mode of Binding and the Mechanism of Action. Curr. Opin. Biotechnol. 2007, 18, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Hurley, L.H. DNA and Its Associated Processes as Targets for Cancer Therapy. Nat. Rev. Cancer 2002, 2, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Agudelo, D.; Bourassa, P.; Bérubé, G.; Tajmir-Riahi, H.-A. Intercalation of Antitumor Drug Doxorubicin and Its Analogue by DNA Duplex: Structural Features and Biological Implications. Int. J. Biol. Macromol. 2014, 66, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Saran, R.; Liu, J. Metal Sensing by DNA. Chem. Rev. 2017, 117, 8272–8325. [Google Scholar] [CrossRef]
- Qu, H.; Tseng, C.Y.; Wang, Y.; Levine, A.J.; Zocchi, G. The Elastic Energy of Sharply Bent Nicked DNA. EPL 2010, 90, 1–5. [Google Scholar] [CrossRef]
- Qu, H.; Wang, Y.; Tseng, C.Y.; Zocchi, G. Critical Torque for Kink Formation in Double-Stranded DNA. Phys. Rev. X 2011, 1, 1–11. [Google Scholar] [CrossRef]
- Qu, H.; Zocchi, G. The Complete Bending Energy Function for Nicked DNA. EPL Europhys. Lett. 2011, 94, 18003. [Google Scholar] [CrossRef]
- Ussery, D.W.; Hinton, J.C.D.; Jorji, B.J.A.M.; Granum, P.E.; Seirafi, A.; Stephen, R.J.; Tupper, A.E.; Berridge, G.; Sidebotham, J.M.; Higgins, C.F. The Chromatin-Associated Protein H-NS. Biochimie 1994, 76, 968–980. [Google Scholar] [CrossRef]
- Williams, R.M.; Rimsky, S. Molecular Aspects of the E. Coli Nucleoid Protein, H-NS: A Central Controller of Gene Regulatory Networks. FEMS Microbiol. Lett. 1997, 156, 175–185. [Google Scholar] [CrossRef]
- Tupper, A.E.; Owen-Hughes, T.A.; Ussery, D.W.; Santos, D.S.; Ferguson, D.J.; Sidebotham, J.M.; Hinton, J.C.; Higgins, C.F. The Chromatin-Associated Protein H-NS Alters DNA Topology in Vitro. EMBO J. 1994, 13, 258–268. [Google Scholar] [CrossRef]
- Hommais, F.; Evelyne, K.; Laurent-Winter, C.; Soutourina, O.; Malpertuy, A.; Le Caer, J.-P.; Danchin, A.; Bertin, P. Large-Scale Monitoring of Pleiotropic Regulation of Gene Expression by the Prokaryotic Nucleoid-Associated Protein, H-NS. Mol. Microbiol. 2001, 40, 20–36. [Google Scholar] [CrossRef] [PubMed]
- Caramel, A.; Schnetz, K. Lac and Lambda Repressors Relieve Silencing of the Escherichia Coli Bgl Promoter. Activation by Alteration of a Repressing Nucleoprotein Complex. J. Mol. Biol. 1998, 284, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, V.H.; Santana, F.J.; Calva, E.; Puente, J.L. Transcriptional Regulation of Type III Secretion Genes in Enteropathogenic Escherichia Coli: Ler Antagonizes H-NS-Dependent Repression. Mol. Microbiol. 2001, 39, 664–678. [Google Scholar] [CrossRef]
- Rimsky, S.; Zuber, F.; Buckle, M.; Buc, H. A Molecular Mechanism for the Repression of Transcription by the H-NS Protein. Mol. Microbiol. 2001, 42, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Falconi, M.; Colonna, B.; Prosseda, G.; Micheli, G.; Gualerzi, C.O. Thermoregulation of Shigella and Escherichia Coli EIEC Pathogenicity. A Temperature-Dependent Structural Transition of DNA Modulates Accessibility of VirF Promoter to Transcriptional Repressor H-NS. EMBO J. 1998, 17, 7033–7043. [Google Scholar] [CrossRef]
- Friedberg, D.; Umanski, T.; Fang, Y.; Rosenshine, I. Hierarchy in the Expression of the Locus of Enterocyte Effacement Genes of Enteropathogenic Escherichia Coli. Mol. Microbiol. 1999, 34, 941–952. [Google Scholar] [CrossRef]
- Göransson, M.; Sondén, B.; Nilsson, P.; Dagberg, B.; Foreman, K.; Emanuelsson, K.; Uhlin, B.E. Transcriptional Silencing and Thermoregulation of Gene Expression in Escherichia Coli. Nature 1990, 344, 682–685. [Google Scholar] [CrossRef]
- McLeod, S.M.; Johnson, R.C. Control of Transcription by Nucleoid Proteins. Curr. Opin. Microbiol. 2001, 4, 152–159. [Google Scholar] [CrossRef]
- Atlung, T.; Ingmer, H. H-NS: A Modulator of Environmentally Regulated Gene Expression. Mol. Microbiol. 1997, 24, 7–17. [Google Scholar] [CrossRef]
- Thei Dame, R.; Wyman, C.; Goosen, N. Structural Basis for Preferential Binding of H-NS to Curved DNA. Biochimie 2001, 83, 231–234. [Google Scholar] [CrossRef]
- Smyth, C.P.; Lundbäck, T.; Renzoni, D.; Siligardi, G.; Beavil, R.; Layton, M.; Sidebotham, J.M.; Hinton, J.C.; Driscoll, P.C.; Higgins, C.F.; et al. Oligomerization of the Chromatin-Structuring Protein H-NS. Mol. Microbiol. 2000, 36, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Spurio, R.; Falconi, M.; Brandi, A.; Pon, C.L.; Gualerzi, C.O. The Oligomeric Structure of Nucleoid Protein H-NS Is Necessary for Recognition of Intrinsically Curved DNA and for DNA Bending. EMBO J. 1997, 16, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Dame, R.T.; Wyman, C.; Goosen, N. H-NS Mediated Compaction of DNA Visualised by Atomic Force Microscopy. Nucleic Acids Res. 2000, 28, 3504–3510. [Google Scholar] [CrossRef]
- Spassky, A.; Rimsky, S.; Garreau, H.; Buc, H. H1a, an E. Coli DNA-Binding Protein Which Accumulates in Stationary Phase, Strongly Compacts DNA in Vitro. Nucleic Acids Res. 1984, 12, 5321–5340. [Google Scholar] [CrossRef] [PubMed]
- Ali, B.M.J.; Amit, R.; Braslavsky, I.; Oppenheim, A.B.; Gileadi, O.; Stavans, J. Compaction of Single DNA Molecules Induced by Binding of Integration Host Factor (IHF). Proc. Natl. Acad. Sci. USA 2001, 98, 10658LP–10663. [Google Scholar] [CrossRef] [PubMed]
- Strick, T.R.; Allemand, J.-F.; Bensimon, D.; Bensimon, A.; Croquette, V. The Elasticity of a Single Supercoiled DNA Molecule. Science 1996, 271, 1835LP–1837. [Google Scholar] [CrossRef] [PubMed]
- Amit, R.; Oppenheim, A.B.; Stavans, J. Increased Bending Rigidity of Single DNA Molecules by H-NS, a Temperature and Osmolarity Sensor. Biophys. J. 2003, 84, 2467–2473. [Google Scholar] [CrossRef]
- Arold, S.T.; Leonard, P.G.; Parkinson, G.N.; Ladbury, J.E. H-NS Forms a Superhelical Protein Scaffold for DNA Condensation. Proc. Natl. Acad. Sci. USA 2010, 107, 15728LP–15732. [Google Scholar] [CrossRef]
- Anuchin, A.M.; Goncharenko, A.V.; Demidenok, O.I.; Kaprelyants, A.S. Histone-like Proteins of Bacteria (Review). Appl. Biochem. Microbiol. 2011, 47, 580. [Google Scholar] [CrossRef]
- Branden, C.I.; Tooze, J. Introduction to Protein Structure; Garland Science: New York, NY, USA, 2012. [Google Scholar]
- Robertson, G.; Hirst, M.; Bainbridge, M.; Bilenky, M.; Zhao, Y.; Zeng, T.; Euskirchen, G.; Bernier, B.; Varhol, R.; Delaney, A.; et al. Genome-Wide Profiles of STAT1 DNA Association Using Chromatin Immunoprecipitation and Massively Parallel Sequencing. Nat. Methods 2007, 4, 651–657. [Google Scholar] [CrossRef]
- Wu, H.-M.; Crothers, D.M. The Locus of Sequence-Directed and Protein-Induced DNA Bending. Nature 1984, 308, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Kotlarz, D.; Fritsch, A.; Buc, H. Variations of Intramolecular Ligation Rates Allow the Detection of Protein-Induced Bends in DNA. EMBO J. 1986, 5, 799–803. [Google Scholar] [CrossRef] [PubMed]
- Finzi, L.; Dunlap, D.D. Single-Molecule Approaches to Probe the Structure, Kinetics, and Thermodynamics of Nucleoprotein Complexes That Regulate Transcription. J. Biol. Chem. 2010, 285, 18973–18978. [Google Scholar] [CrossRef] [PubMed]
- Lymperopoulos, K.; Crawford, R.; Torella, J.P.; Heilemann, M.; Hwang, L.C.; Holden, S.J.; Kapanidis, A.N. Single-Molecule DNA Biosensors for Protein and Ligand Detection. Angew. Chem.—Int. Ed. 2010, 49, 1316–1320. [Google Scholar] [CrossRef] [PubMed]
- Kapanidis, A.N.; Laurence, T.A.; Lee, N.K.; Margeat, E.; Kong, X.; Weiss, S. Alternating-Laser Excitation of Single Molecules. Acc. Chem. Res. 2005, 38, 523–533. [Google Scholar] [CrossRef]
- Shen, W.; Bruist, M.F.; Goodman, S.D.; Seeman, N.C. A Protein-Driven DNA Device That Measures the Excess Binding Energy of Proteins That Distort DNA. Angew. Chem.—Int. Ed. 2004, 43, 4750–4752. [Google Scholar] [CrossRef]
- Stryer, L.; Haugland, R.P. Energy Transfer: A Spectroscopic Ruler. Proc. Natl. Acad. Sci. USA 1967, 58, 719–726. [Google Scholar] [CrossRef]
- Crawford, R.; Kelly, D.J.; Kapanidis, A.N. A Protein Biosensor That Relies on Bending of Single DNA Molecules. Chem. Phys. Chem. 2012, 13, 918–922. [Google Scholar] [CrossRef]
- van der Vliet, P.C. Bending of DNA by Transcription Factors. BioEssays 1993, 15, 25–32. [Google Scholar] [CrossRef]
- Tang, G.Q.; Deshpande, A.P.; Patel, S.S. Transcription Factor-Dependent DNA Bending Governs Promoter Recognition by the Mitochondrial RNA Polymerase. J. Biol. Chem. 2011, 286, 38805–38813. [Google Scholar] [CrossRef]
- Dornberger, U.; Hillisch, A.; Gollmick, F.A.; Fritzsche, H.; Diekmann, S. Solution Structure of a Five-Adenine Bulge Loop within a DNA Duplex. Biochemistry 1999, 38, 12860–12868. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA Origami: Scaffolds for Creating Higher Order Structures. Chem. Rev. 2017, 117. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.; Aksimentiev, A. In Situ Structure and Dynamics of DNA Origami Determined through Molecular Dynamics Simulations. Proc. Natl. Acad. Sci. USA 2013, 110, 20099–20104. [Google Scholar] [CrossRef] [PubMed]
- Pan, K.; Kim, D.N.; Zhang, F.; Adendorff, M.R.; Yan, H.; Bathe, M. Lattice-Free Prediction of Three-Dimensional Structure of Programmed DNA Assemblies. Nat. Commun. 2014, 5, 1–7. [Google Scholar] [CrossRef]
- Kim, D.N.; Kilchherr, F.; Dietz, H.; Bathe, M. Quantitative Prediction of 3D Solution Shape and Flexibility of Nucleic Acid Nanostructures. Nucleic Acids Res. 2012, 40, 2862–2868. [Google Scholar] [CrossRef]
- Douglas, S.M.; Marblestone, A.H.; Teerapittayanon, S.; Vazquez, A.; Church, G.M.; Shih, W.M. Rapid Prototyping of 3D DNA-Origami Shapes with CaDNAno. Nucleic Acids Res. 2009, 37, 5001–5006. [Google Scholar] [CrossRef]
- Zhang, Y.; Crothers, D.M. High-Throughput Approach for Detection of DNA Bending and Flexibility Based on Cyclization. Proc. Natl. Acad. Sci. USA 2003, 100, 3161–3166. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Bishop, A.R.; Rasmussen, K.O.; Alexandrov, B.S. The Role of Structural Parameters in DNA Cyclization. BMC Bioinformatics 2016, 17, 1–10. [Google Scholar] [CrossRef]
- Gubu, A.; Wang, J.; Jin, H.; Tang, X. Synthesis and “DNA Interlocks” Formation of Small Circular Oligodeoxynucleotides. ACS Appl. Mater. Interfaces 2020, 12, 12584–12590. [Google Scholar] [CrossRef]
- Schmidt, T.L.; Heckel, A. Construction of a Structurally Defined Double-Stranded DNA Catenane. Nano Lett. 2011, 11, 1739–1742. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Sun, W.; Seeman, N.C. Assembly of Borromean Rings from DNA. Nature 1997, 386, 137–138. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, D.; Schmidt, T.L.; Hannam, J.S.; Purohit, C.S.; Heckel, A.; Famulok, M. A Double-Stranded DNA Rotaxane. Nat. Nanotechnol. 2010, 5, 436–442. [Google Scholar] [CrossRef]
- Li, J.; Mohammed-Elsabagh, M.; Paczkowski, F.; Li, Y. Circular Nucleic Acids: Discovery, Functions and Applications. Chem. Bio. Chem. 2020, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C. From Genes to Machines: DNA Nanomechanical Devices. Trends Biochem. Sci. 2005, 30, 119–125. [Google Scholar] [CrossRef]
- Krishnan, Y.; Simmel, F.C. Nucleic Acid Based Molecular Devices. Angew. Chem. Int. Ed. Engl. 2011, 50, 3124–3156. [Google Scholar] [CrossRef]
- Ramezani, H.; Dietz, H. Building Machines with DNA Molecules. Nat. Rev. Genet. 2020, 21, 5–26. [Google Scholar] [CrossRef]
- Wang, F.; Willner, B.; Willner, I. DNA-Based Machines. Top. Curr. Chem. 2014, 354, 279–338. [Google Scholar] [CrossRef]
- Khara, D.C.; Schreck, J.S.; Tomov, T.E.; Berger, Y.; Ouldridge, T.E.; Doye, J.P.K.; Nir, E. DNA Bipedal Motor Walking Dynamics: An Experimental and Theoretical Study of the Dependency on Step Size. Nucleic Acids Res. 2017, 46, 1553–1561. [Google Scholar] [CrossRef]
- Park, S.; Song, J.; Kim, J.S. In Silico Construction of a Flexibility-Based DNA Brownian Ratchet for Directional Nanoparticle Delivery. Sci. Adv. 2019, 5, eaav4943. [Google Scholar] [CrossRef]
- Tomov, T.E.; Tsukanov, R.; Liber, M.; Masoud, R.; Plavner, N.; Nir, E. Rational Design of DNA Motors: Fuel Optimization through Single-Molecule Fluorescence. J. Am. Chem. Soc. 2013, 135, 11935–11941. [Google Scholar] [CrossRef] [PubMed]
- Tomov, T.E.; Tsukanov, R.; Glick, Y.; Berger, Y.; Liber, M.; Avrahami, D.; Gerber, D.; Nir, E. DNA Bipedal Motor Achieves a Large Number of Steps Due to Operation Using Microfluidics-Based Interface. ACS Nano 2017, 11, 4002–4008. [Google Scholar] [CrossRef] [PubMed]
- Liber, M.; Tomov, T.E.; Tsukanov, R.; Berger, Y.; Nir, E. A Bipedal DNA Motor That Travels Back and Forth between Two DNA Origami Tiles. Small 2015, 11, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Geggier, S.; Vologodskii, A. Sequence Dependence of DNA Bending Rigidity. Proc. Natl. Acad. Sci. USA 2010, 107, 15421LP–15426. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.-M.; Reifenberger, J.G.; Cao, H.; Dorfman, K.D. Sequence-Dependent Persistence Length of Long DNA. Phys. Rev. Lett. 2017, 119, 227802. [Google Scholar] [CrossRef]
- Suzuki, Y.; Kawamata, I.; Mizuno, K.; Murata, S. Large Deformation of a DNA-Origami Nanoarm Induced by the Cumulative Actuation of Tension-Adjustable Modules. Angew. Chem. Int. Ed. 2020, 59, 6230–6234. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).