Recent Advances in Cell Adhesive Force Microscopy
Abstract
1. Introduction
2. Advances in Cell Adhesive Force Microscopy in Term of Spatial Resolution
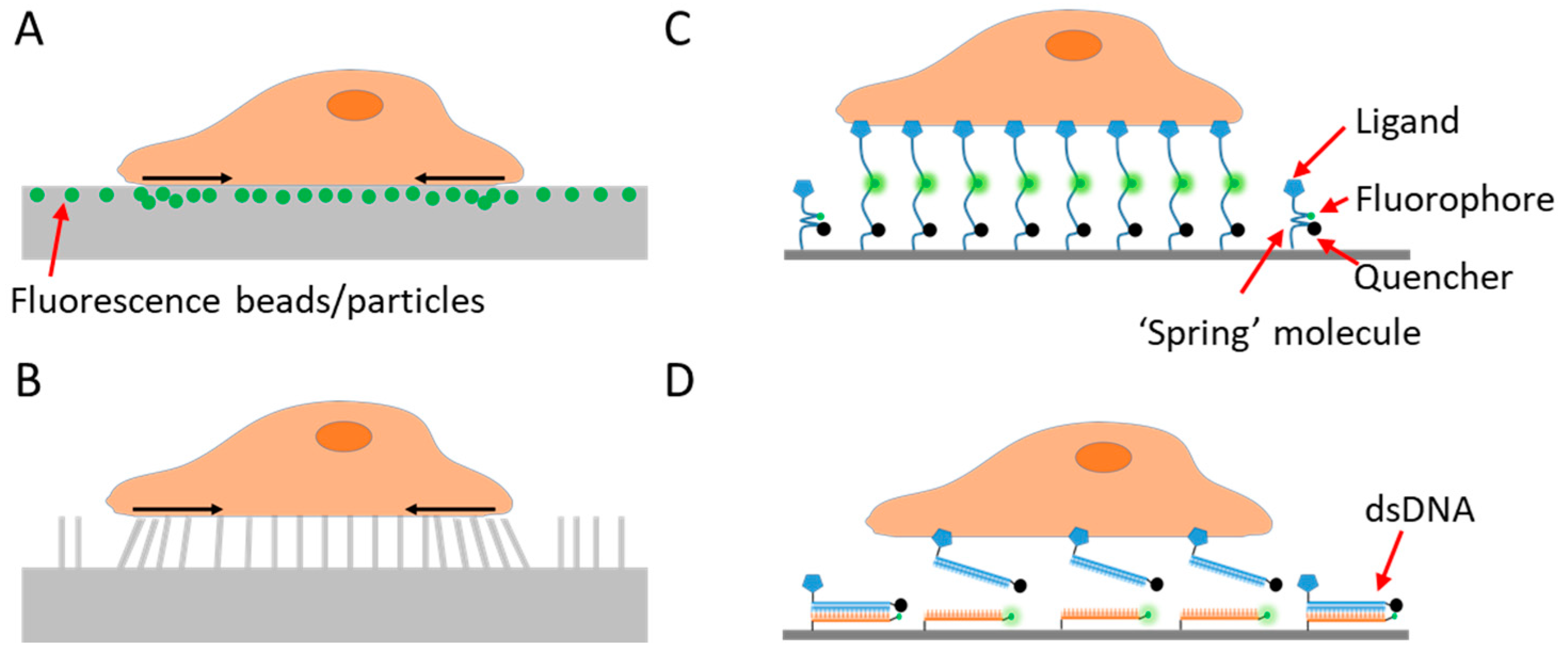
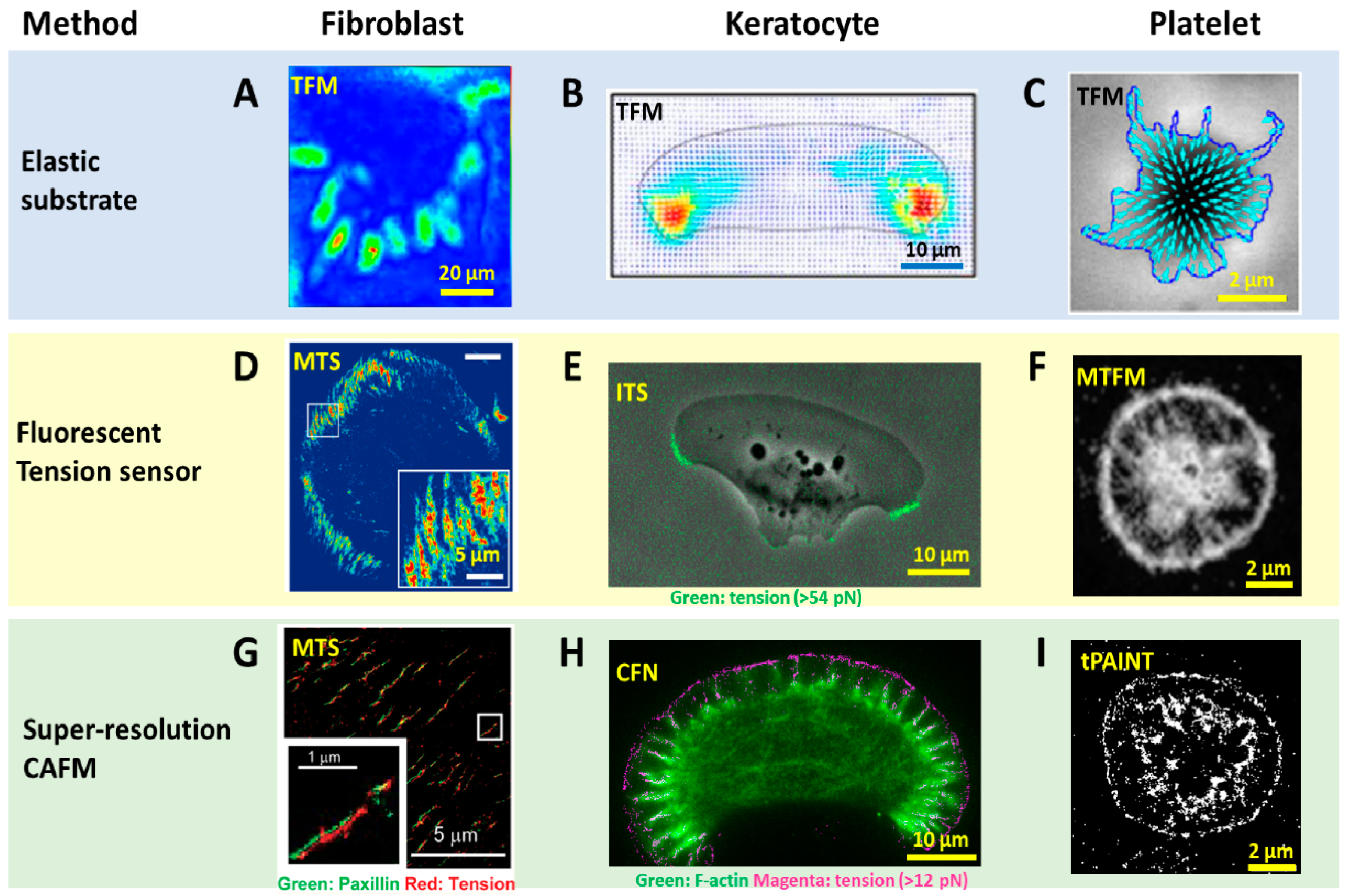
2.1. Traction Force Microscopy Pioneered Cell Adhesive Force Imaging
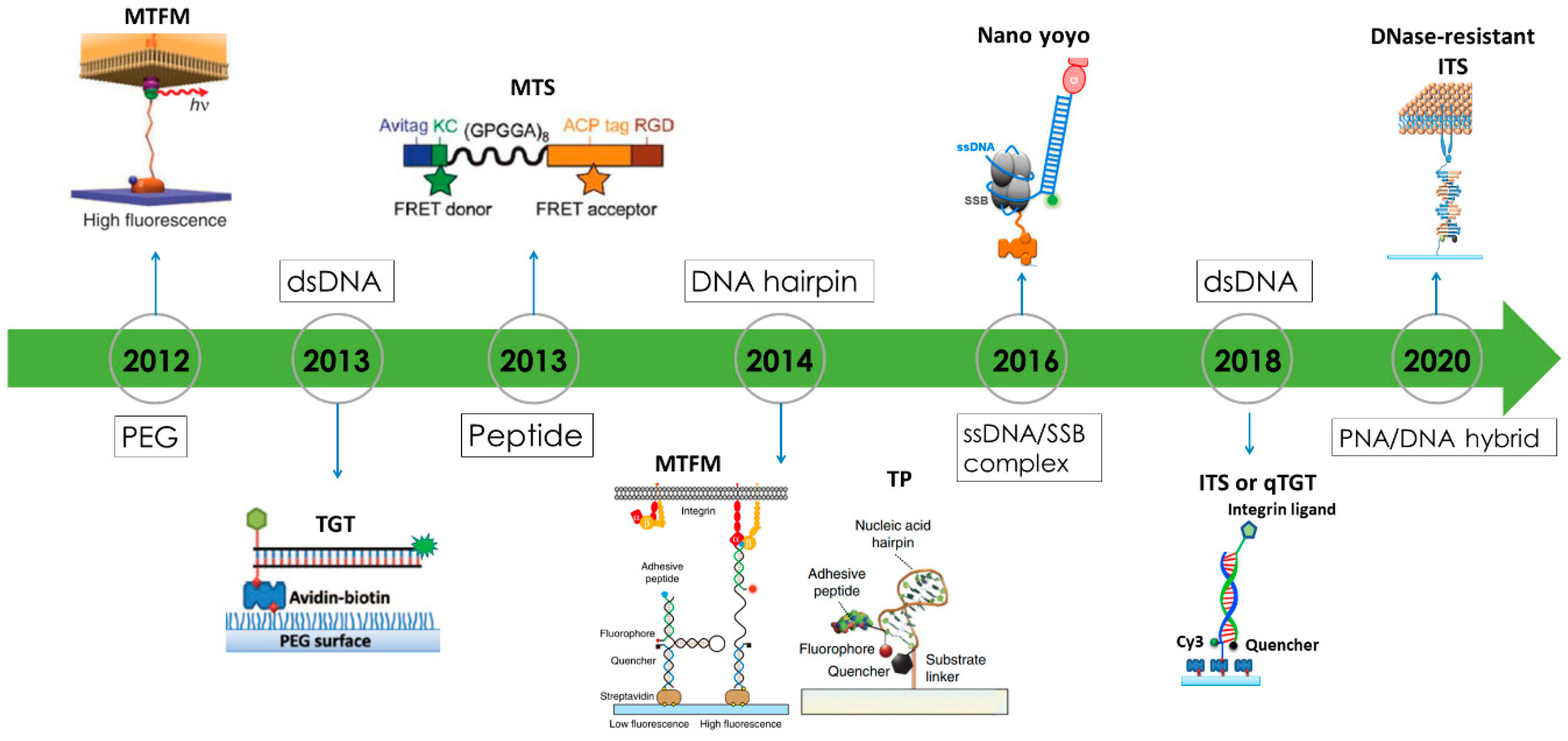
2.2. Fluorescent Tension Sensors Bring the Resolution of Force Imaging to Diffraction Limit
2.3. Super-Resolved Cell Adhesive Force Microscopies
3. Biomaterial Choices for Constructing Fluorescence Tension Sensors
4. Outlook
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CAFM | Cell adhesive force microscopy |
| CFN | Cellular force nanoscopy |
| FTS | Fluorescent tension sensor |
| ITS | Integrative tension sensor |
| MTFM | Molecular tension fluorescence microscopy |
| MTS | Molecular tension sensor |
| TFM | Traction force microscopy |
| TGT | Tension gauge tether |
| TP | Tension probe |
| t-PAINT | tension-point accumulation in nanoscale topology |
References
- Vogel, V.; Sheetz, M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006, 7, 265–275. [Google Scholar] [CrossRef] [PubMed]
- LeGoff, L.; Lecuit, T. Mechanical Forces and Growth in Animal Tissues. Cold Spring Harb. Perspect. Biol. 2015, 8, a019232. [Google Scholar] [CrossRef] [PubMed]
- Discher, D.E.; Janmey, P.; Wang, Y.-L. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science 2005, 310, 1139–1143. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Weaver, V.M. Mechanics, malignancy, and metastasis: The force journey of a tumor cell. Cancer Metastasis Rev. 2009, 28, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Geiger, B.; Spatz, J.P.; Bershadsky, A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009, 10, 21–33. [Google Scholar] [CrossRef]
- Rossy, J.; Laufer, J.M.; Legler, D.F. Role of Mechanotransduction and Tension in T Cell Function. Front. Immunol. 2018, 9, 2638. [Google Scholar] [CrossRef]
- Harris, A.K.; Wild, P.; Stopak, D. Silicone rubber substrata: A new wrinkle in the study of cell locomotion. Science 1980, 208, 177–179. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Schwarz, U.S.; Riveline, D.; Goichberg, P.; Tzur, G.; Sabanay, I.; Mahalu, D.; Safran, S.; Bershadsky, A.; Addadi, L.; et al. Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 2001, 3, 466–472. [Google Scholar] [CrossRef]
- Beningo, K.A.; Dembo, M.; Kaverina, I.; Small, J.V.; Wang, Y.-l. Nascent Focal Adhesions Are Responsible for the Generation of Strong Propulsive Forces in Migrating Fibroblasts. J. Cell Biol. 2001, 153, 881–888. [Google Scholar] [CrossRef]
- Tan, J.L.; Tien, J.; Pirone, D.M.; Gray, D.S.; Bhadriraju, K.; Chen, C.S. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc. Natl. Acad. Sci. USA 2003, 100, 1484. [Google Scholar] [CrossRef]
- Legant, W.R.; Miller, J.S.; Blakely, B.L.; Cohen, D.M.; Genin, G.M.; Chen, C.S. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat. Methods 2010, 7, 969. [Google Scholar] [CrossRef] [PubMed]
- Franck, C.; Maskarinec, S.A.; Tirrell, D.A.; Ravichandran, G. Three-Dimensional Traction Force Microscopy: A New Tool for Quantifying Cell-Matrix Interactions. PLoS ONE 2011, 6, e17833. [Google Scholar] [CrossRef] [PubMed]
- Plotnikov, S.V.; Pasapera, A.M.; Sabass, B.; Waterman, C.M. Force Fluctuations within Focal Adhesions Mediate ECM-Rigidity Sensing to Guide Directed Cell Migration. Cell 2012, 151, 1513–1527. [Google Scholar] [CrossRef] [PubMed]
- del Álamo, J.C.; Meili, R.; Álvarez-González, B.; Alonso-Latorre, B.; Bastounis, E.; Firtel, R.; Lasheras, J.C. Three-Dimensional Quantification of Cellular Traction Forces and Mechanosensing of Thin Substrata by Fourier Traction Force Microscopy. PLoS ONE 2013, 8, e69850. [Google Scholar] [CrossRef] [PubMed]
- Colin-York, H.; Shrestha, D.; Felce, J.H.; Waithe, D.; Moeendarbary, E.; Davis, S.J.; Eggeling, C.; Fritzsche, M. Super-Resolved Traction Force Microscopy (STFM). Nano Lett. 2016, 16, 2633–2638. [Google Scholar] [CrossRef]
- Stubb, A.; Laine, R.F.; Miihkinen, M.; Hamidi, H.; Guzmán, C.; Henriques, R.; Jacquemet, G.; Ivaska, J. Fluctuation-Based Super-Resolution Traction Force Microscopy. Nano Lett. 2020, 20, 2230–2245. [Google Scholar] [CrossRef]
- Polacheck, W.J.; Chen, C.S. Measuring cell-generated forces: A guide to the available tools. Nat. Methods 2016, 13, 415–423. [Google Scholar] [CrossRef]
- Meng, F.; Suchyna, T.M.; Sachs, F. A fluorescence energy transfer-based mechanical stress sensor for specific proteins in situ. FEBS J. 2008, 275, 3072–3087. [Google Scholar] [CrossRef]
- Grashoff, C.; Hoffman, B.D.; Brenner, M.D.; Zhou, R.; Parsons, M.; Yang, M.T.; McLean, M.A.; Sligar, S.G.; Chen, C.S.; Ha, T.; et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 2010, 466, 263–266. [Google Scholar] [CrossRef]
- Stabley, D.R.; Jurchenko, C.; Marshall, S.S.; Salaita, K.S. Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nat. Methods 2012, 9, 64–67. [Google Scholar] [CrossRef]
- Wang, X.; Ha, T. Defining Single Molecular Forces Required to Activate Integrin and Notch Signaling. Science 2013, 340, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yehl, K.; Narui, Y.; Salaita, K. Tension sensing nanoparticles for mechano-imaging at the living/nonliving interface. J. Am. Chem. Soc. 2013, 135, 5320–5323. [Google Scholar] [CrossRef] [PubMed]
- Morimatsu, M.; Mekhdjian, A.H.; Adhikari, A.S.; Dunn, A.R. Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Lett. 2013, 13, 3985–3989. [Google Scholar] [CrossRef] [PubMed]
- Blakely, B.L.; Dumelin, C.E.; Trappmann, B.; McGregor, L.M.; Choi, C.K.; Anthony, P.C.; Duesterberg, V.K.; Baker, B.M.; Block, S.M.; Liu, D.R.; et al. A DNA-based molecular probe for optically reporting cellular traction forces. Nat. Methods 2014, 11, 1229–1232. [Google Scholar] [CrossRef]
- Jurchenko, C.; Salaita, K.S. Lighting up the Force: Investigating Mechanisms of Mechanotransduction Using Fluorescent Tension Probes. Mol. Cell. Biol. 2015, 35, 2570–2582. [Google Scholar] [CrossRef]
- Matellan, C.; Del Río Hernández, A.E. Where No Hand Has Gone Before: Probing Mechanobiology at the Cellular Level. ACS Biomater. Sci. Eng. 2019, 5, 3703–3719. [Google Scholar] [CrossRef]
- Ma, V.P.-Y.; Salaita, K. DNA Nanotechnology as an Emerging Tool to Study Mechanotransduction in Living Systems. Small 2019, 15, 1900961. [Google Scholar] [CrossRef]
- Yasunaga, A.; Murad, Y.; Li, I.T.S. Quantifying molecular tension-classifications, interpretations and limitations of force sensors. Phys. Biol. 2020, 17, 011001. [Google Scholar] [CrossRef]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic Force Microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef]
- Ashkin, A. Optical trapping and manipulation of neutral particles using lasers. Proc. Natl. Acad. Sci. USA 1997, 94, 4853–4860. [Google Scholar] [CrossRef]
- Smith, S.B.; Finzi, L.; Bustamante, C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science 1992, 258, 1122. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.; Ritchie, K.; Merkel, R. Sensitive force technique to probe molecular adhesion and structural linkages at biological interfaces. Biophys. J. 1995, 68, 2580–2587. [Google Scholar] [CrossRef]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Dembo, M.; Wang, Y.L. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 1999, 76, 2307–2316. [Google Scholar] [CrossRef]
- Fu, J.; Wang, Y.K.; Yang, M.T.; Desai, R.A.; Yu, X.; Liu, Z.; Chen, C.S. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods 2010, 7, 733–736. [Google Scholar] [CrossRef]
- Pandey, P.; Hawkes, W.; Hu, J.; Megone, W.V.; Gautrot, J.; Anilkumar, N.; Zhang, M.; Hirvonen, L.; Cox, S.; Ehler, E.; et al. Cardiomyocytes Sense Matrix Rigidity through a Combination of Muscle and Non-muscle Myosin Contractions. Dev. Cell 2018, 44, 326–336.e323. [Google Scholar] [CrossRef]
- Sabass, B.; Gardel, M.L.; Waterman, C.M.; Schwarz, U.S. High resolution traction force microscopy based on experimental and computational advances. Biophys. J. 2008, 94, 207–220. [Google Scholar] [CrossRef]
- Hui, K.L.; Balagopalan, L.; Samelson, L.E.; Upadhyaya, A. Cytoskeletal forces during signaling activation in Jurkat T-cells. Mol. Biol. Cell 2015, 26, 685–695. [Google Scholar] [CrossRef]
- Wiegand, T.; Fratini, M.; Frey, F.; Yserentant, K.; Liu, Y.; Weber, E.; Galior, K.; Ohmes, J.; Braun, F.; Herten, D.P.; et al. Forces during cellular uptake of viruses and nanoparticles at the ventral side. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Glazier, R.; Brockman, J.M.; Bartle, E.; Mattheyses, A.L.; Destaing, O.; Salaita, K. DNA mechanotechnology reveals that integrin receptors apply pN forces in podosomes on fluid substrates. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Liu, Y.; Medda, R.; Liu, Z.; Galior, K.; Yehl, K.; Spatz, J.P.; Cavalcanti-Adam, E.A.; Salaita, K. Nanoparticle tension probes patterned at the nanoscale: Impact of integrin clustering on force transmission. Nano Lett 2014, 14, 5539–5546. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.J.; Chang, A.C.; Anderson, S.M.; Miller, C.M.; Prahl, L.S.; Odde, D.J.; Dunn, A.R. Regulation and dynamics of force transmission at individual cell-matrix adhesion bonds. Sci. Adv. 2020, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.C.; Mekhdjian, A.H.; Morimatsu, M.; Denisin, A.K.; Pruitt, B.L.; Dunn, A.R. Single Molecule Force Measurements in Living Cells Reveal a Minimally Tensioned Integrin State. ACS Nano 2016, 10, 10745–10752. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; O’Brien, C.; Mudiyanselage, A.P.K.K.K.; Li, N.W.; Bagheri, Y.; Wu, R.; Sun, Y.B.; You, M.X. Visualizing Intercellular Tensile Forces by DNA-Based Membrane Molecular Probes. J. Am. Chem. Soc. 2017, 139, 18182–18185. [Google Scholar] [CrossRef]
- Blanchard, A.T.; Bazrafshan, A.S.; Yi, J.; Eisman, J.T.; Yehl, K.M.; Bian, T.; Mugler, A.; Salaita, K. Highly Polyvalent DNA Motors Generate 100+ pN of Force via Autochemophoresis. Nano Lett. 2019, 19, 6977–6986. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X. Integrins outside focal adhesions transmit tensions during stable cell adhesion. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef]
- Jo, M.H.; Cottle, W.T.; Ha, T. Real-Time Measurement of Molecular Tension during Cell Adhesion and Migration Using Multiplexed Differential Analysis of Tension Gauge Tethers. ACS Biomater. Sci. Eng. 2019, 5, 3856–3863. [Google Scholar] [CrossRef]
- Wang, X.; Sun, J.; Xu, Q.; Chowdhury, F.; Roein-Peikar, M.; Wang, Y.; Ha, T. Integrin Molecular Tension within Motile Focal Adhesions. Biophys. J. 2015, 109, 2259–2267. [Google Scholar] [CrossRef]
- Wang, Y.; LeVine, D.N.; Gannon, M.; Zhao, Y.; Sarkar, A.; Hoch, B.; Wang, X. Force-activatable biosensor enables single platelet force mapping directly by fluorescence imaging. Biosens. Bioelectron. 2018, 100, 192–200. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Y.; Sarkar, A.; Wang, X. Keratocytes Generate High Integrin Tension at the Trailing Edge to Mediate Rear De-adhesion during Rapid Cell Migration. iScience 2018, 9, 502–512. [Google Scholar] [CrossRef]
- Murad, Y.; Li, I.T.S. Quantifying Molecular Forces with Serially Connected Force Sensors. Biophys. J. 2019, 116, 1282–1291. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Rahil, Z.; Li, I.T.S.; Chowdhury, F.; Leckband, D.E.; Chemla, Y.R.; Ha, T. Constructing modular and universal single molecule tension sensor using protein G to study mechano-sensitive receptors. Sci. Rep. 2016, 6, 21584. [Google Scholar] [CrossRef] [PubMed]
- Li, I.T.S.; Ha, T.; Chemla, Y.R. Mapping cell surface adhesion by rotation tracking and adhesion footprinting. Sci. Rep. 2017, 7, 44502. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Pal, K.; Tu, Y.; Wang, X. Cellular Force Nanoscopy with 50 nm Resolution Based on Integrin Molecular Tension Imaging and Localization. J. Am. Chem. Soc. 2020, 142, 6930–6934. [Google Scholar] [CrossRef]
- Brockman, J.M.; Su, H.; Blanchard, A.T.; Duan, Y.; Meyer, T.; Quach, M.E.; Glazier, R.; Bazrafshan, A.; Bender, R.L.; Kellner, A.V.; et al. Live-cell super-resolved PAINT imaging of piconewton cellular traction forces. Nat. Methods 2020, 17, 1018–1024. [Google Scholar] [CrossRef]
- Gardel, M.L.; Sabass, B.; Ji, L.; Danuser, G.; Schwarz, U.S.; Waterman, C.M. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J. Cell Biol. 2008, 183, 999–1005. [Google Scholar] [CrossRef]
- Legant, W.R.; Choi, C.K.; Miller, J.S.; Shao, L.; Gao, L.; Betzig, E.; Chen, C.S. Multidimensional traction force microscopy reveals out-of-plane rotational moments about focal adhesions. Proc. Natl. Acad. Sci. USA 2013, 110, 881–886. [Google Scholar] [CrossRef]
- Barnhart, E.; Lee, K.C.; Allen, G.M.; Theriot, J.A.; Mogilner, A. Balance between cell-substrate adhesion and myosin contraction determines the frequency of motility initiation in fish keratocytes. Proc. Natl. Acad. Sci. USA 2015, 112, 5045–5050. [Google Scholar] [CrossRef]
- Schwarz Henriques, S.; Sandmann, R.; Strate, A.; Koster, S. Force field evolution during human blood platelet activation. J. Cell Sci. 2012, 125, 3914–3920. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiu, Y.; Blanchard, A.T.; Chang, Y.; Brockman, J.M.; Ma, V.P.-Y.; Lam, W.A.; Salaita, K. Platelet integrins exhibit anisotropic mechanosensing and harness piconewton forces to mediate platelet aggregation. Proc. Natl. Acad. Sci. USA 2018, 115, 325. [Google Scholar] [CrossRef]
- Morimatsu, M.; Mekhdjian, A.H.; Chang, A.C.; Tan, S.J.; Dunn, A.R. Visualizing the Interior Architecture of Focal Adhesions with High-Resolution Traction Maps. Nano Lett. 2015, 15, 2220–2228. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, H. Förster resonance energy transfer—A spectroscopic nanoruler: Principle and applications. J. Photochem. Photobiol. C Photochem. Rev. 2011, 12, 20–30. [Google Scholar] [CrossRef]
- Rust, M.J.; Bates, M.; Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 2006, 3, 793–795. [Google Scholar] [CrossRef] [PubMed]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef] [PubMed]
- Cox, S.; Rosten, E.; Monypenny, J.; Jovanovic-Talisman, T.; Burnette, D.T.; Lippincott-Schwartz, J.; Jones, G.E.; Heintzmann, R. Bayesian localization microscopy reveals nanoscale podosome dynamics. Nat. Methods 2012, 9, 195–200. [Google Scholar] [CrossRef]
- Oesterhelt, F.; Rief, M.; Gaub, H.E. Single molecule force spectroscopy by AFM indicates helical structure of poly(ethylene-glycol) in water. New J. Phys. 1999, 1, 6. [Google Scholar] [CrossRef]
- Becker, N.; Oroudjev, E.; Mutz, S.; Cleveland, J.P.; Hansma, P.K.; Hayashi, C.Y.; Makarov, D.E.; Hansma, H.G. Molecular nanosprings in spider capture-silk threads. Nat. Mater. 2003, 2, 278–283. [Google Scholar] [CrossRef]
- Galior, K.; Liu, Y.; Yehl, K.; Vivek, S.; Salaita, K. Titin-Based Nanoparticle Tension Sensors Map High-Magnitude Integrin Forces within Focal Adhesions. Nano Lett. 2016, 16, 341–348. [Google Scholar] [CrossRef]
- Li, H.B.; Carrion-Vazquez, M.; Oberhauser, A.F.; Marszalek, P.E.; Fernandez, J.M. Point mutations alter the mechanical stability of immunoglobulin modules. Nat. Struct. Biol. 2000, 7, 1117–1120. [Google Scholar]
- Albrecht, C.; Blank, K.; Lalic-Mülthaler, M.; Hirler, S.; Mai, T.; Gilbert, I.; Schiffmann, S.; Bayer, T.; Clausen-Schaumann, H.; Gaub, H.E. DNA: A Programmable Force Sensor. Science 2003, 301, 367. [Google Scholar] [CrossRef]
- Jones, M.R.; Seeman, N.C.; Mirkin, C.A. Programmable materials and the nature of the DNA bond. Science 2015, 347, 6224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ge, C.; Zhu, C.; Salaita, K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat. Commun. 2014, 5, 5167. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Blanchfield, L.; Ma, V.P.-Y.; Andargachew, R.; Galior, K.; Liu, Z.; Evavold, B.; Salaita, K. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc. Natl. Acad. Sci. USA 2016, 113, 5610. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, F.; Li, I.T.S.; Ngo, T.T.M.; Leslie, B.J.; Kim, B.C.; Sokoloski, J.E.; Weiland, E.; Wang, X.; Chemla, Y.R.; Lohman, T.M.; et al. Defining Single Molecular Forces Required for Notch Activation Using Nano Yoyo. Nano Lett. 2016, 16, 3892–3897. [Google Scholar] [CrossRef]
- Zhao, Y.; Sarkar, A.; Wang, X. Peptide nucleic acid based tension sensor for cellular force imaging with strong DNase resistance. Biosens. Bioelectron. 2020, 150, 111959. [Google Scholar] [CrossRef]
- Boutry, C.M.; Beker, L.; Kaizawa, Y.; Vassos, C.; Tran, H.; Hinckley, A.C.; Pfattner, R.; Niu, S.; Li, J.; Claverie, J.; et al. Biodegradable and flexible arterial-pulse sensor for the wireless monitoring of blood flow. Nat. Biomed. Eng. 2019, 3, 47–57. [Google Scholar] [CrossRef]
| Detection Range | Spatial Resolution | References | ||
|---|---|---|---|---|
| TFM | ~1 nN | 1–5 μm | [7,34,35,36,37,38] | |
| Super-resolution TFM | <1 nN | 40–80 nm * | [15,16] | |
| FTS-based cell adhesive force microcopy | PEG | 0–20 pN | <1 μm | [22,23,39,40,41] |
| peptide | 1–100 pN | <1 μm | [42,43] | |
| DNA hairpin | 7–14 pN | <1 μm | [24,40,44,45] | |
| dsDNA | 12–54 pN | <1 μm | [46,47,48,49,50,51,52,53] | |
| CFN | 12–54 pN | 50 nm | [54] | |
| tPAINT | 7–14 pN | 25 nm | [55] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, Y.; Wang, X. Recent Advances in Cell Adhesive Force Microscopy. Sensors 2020, 20, 7128. https://doi.org/10.3390/s20247128
Tu Y, Wang X. Recent Advances in Cell Adhesive Force Microscopy. Sensors. 2020; 20(24):7128. https://doi.org/10.3390/s20247128
Chicago/Turabian StyleTu, Ying, and Xuefeng Wang. 2020. "Recent Advances in Cell Adhesive Force Microscopy" Sensors 20, no. 24: 7128. https://doi.org/10.3390/s20247128
APA StyleTu, Y., & Wang, X. (2020). Recent Advances in Cell Adhesive Force Microscopy. Sensors, 20(24), 7128. https://doi.org/10.3390/s20247128




