2.1. Curve Similarity Model
2.1.1. Original Definition
One curve is typically represented by a function. A definition must be provided to study the problem of the curve similarity. In geometry, there is a strict definition for shape similarity, which is an accurate similarity. In engineering applications, due to the large number of error factors, the definition of fuzzy curve similarity is adopted. Taking a 2D plane curve as an example, a kind of curve similarity is defined as follows:
Definition 1 ([
32])
. Given functions f1(x) and f2(x), is the distance between two functions, and also known as the function similarity distance or the curve similarity distance, where [C1, C2] is the function definition domain or the definition domain.
Definition 2 ([
32])
. For a given threshold ε, if d(f1,f2) < ε, then f1(x) and f2(x) are similar, otherwise they are not. As mentioned above in the definitions of curve similarity distance (CSD) and curve similarity, the two functions have the same definition domains, that is to say, two curves must be aligned first, which is very limited in practical application. In most cases, the definition domains of two functions are different, and it is necessary to perform a truncation, translation, stretching, or even rotation transformation on a function to calculate the similarity distance.
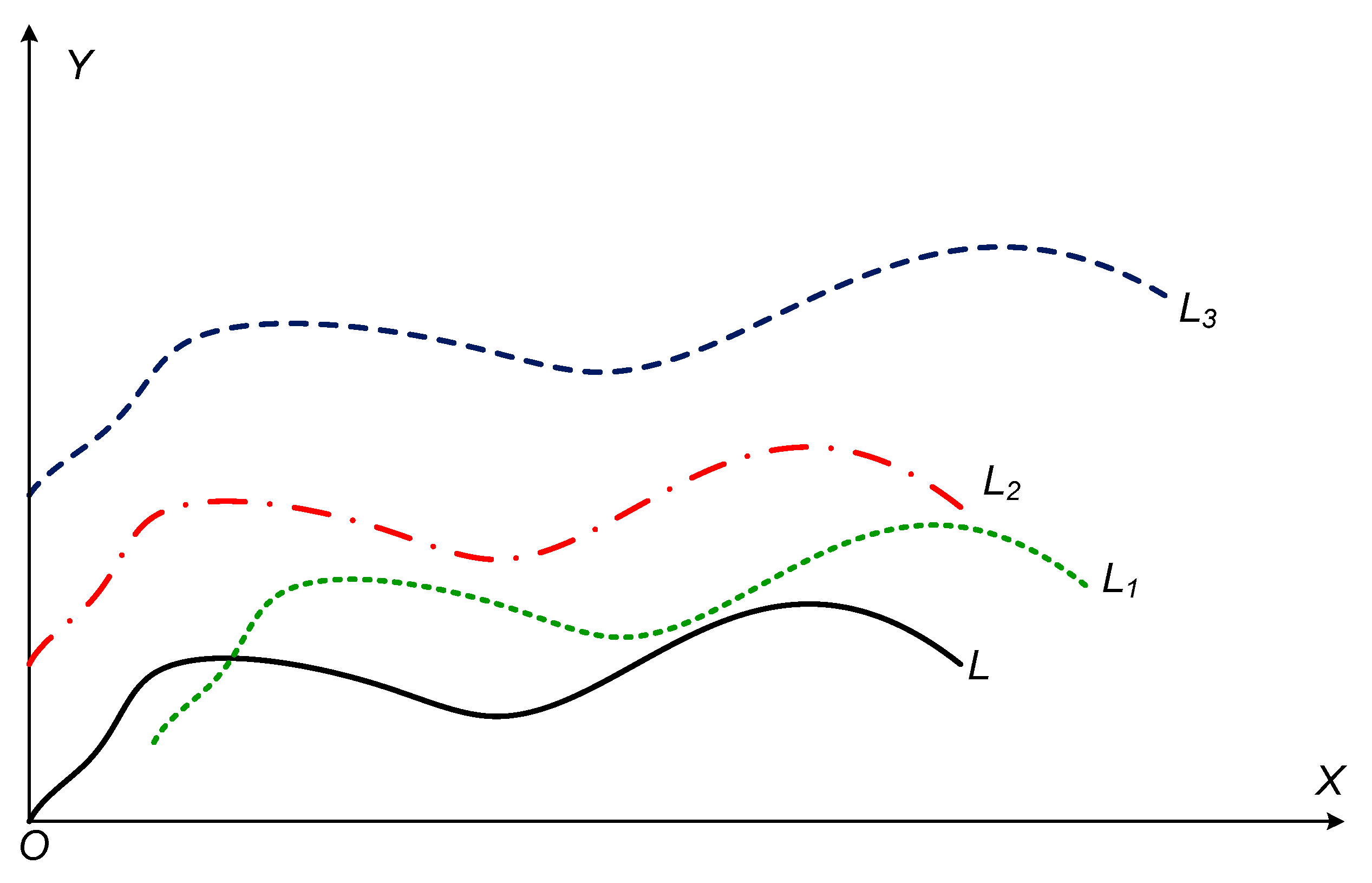
As shown in
Figure 1, given a curve
L, the curves
L1 to
L3 are new curves obtained after applying different transformations such as translation or stretching. If the above calculation method is adopted, the distances between
L1 to
L3 and
L are different. Similar problems are available in the calculation of DTW and Fréchet distances of the curves.
From the perspective of transformation, the above several curves are similar, and the distance is 0. Therefore, when to measure the similarity between curves, the transformation of the curve should be taken into consideration. Firstly, a curve similarity distance definition based on curve transformation may be suggested.
2.1.2. Improvement Definition
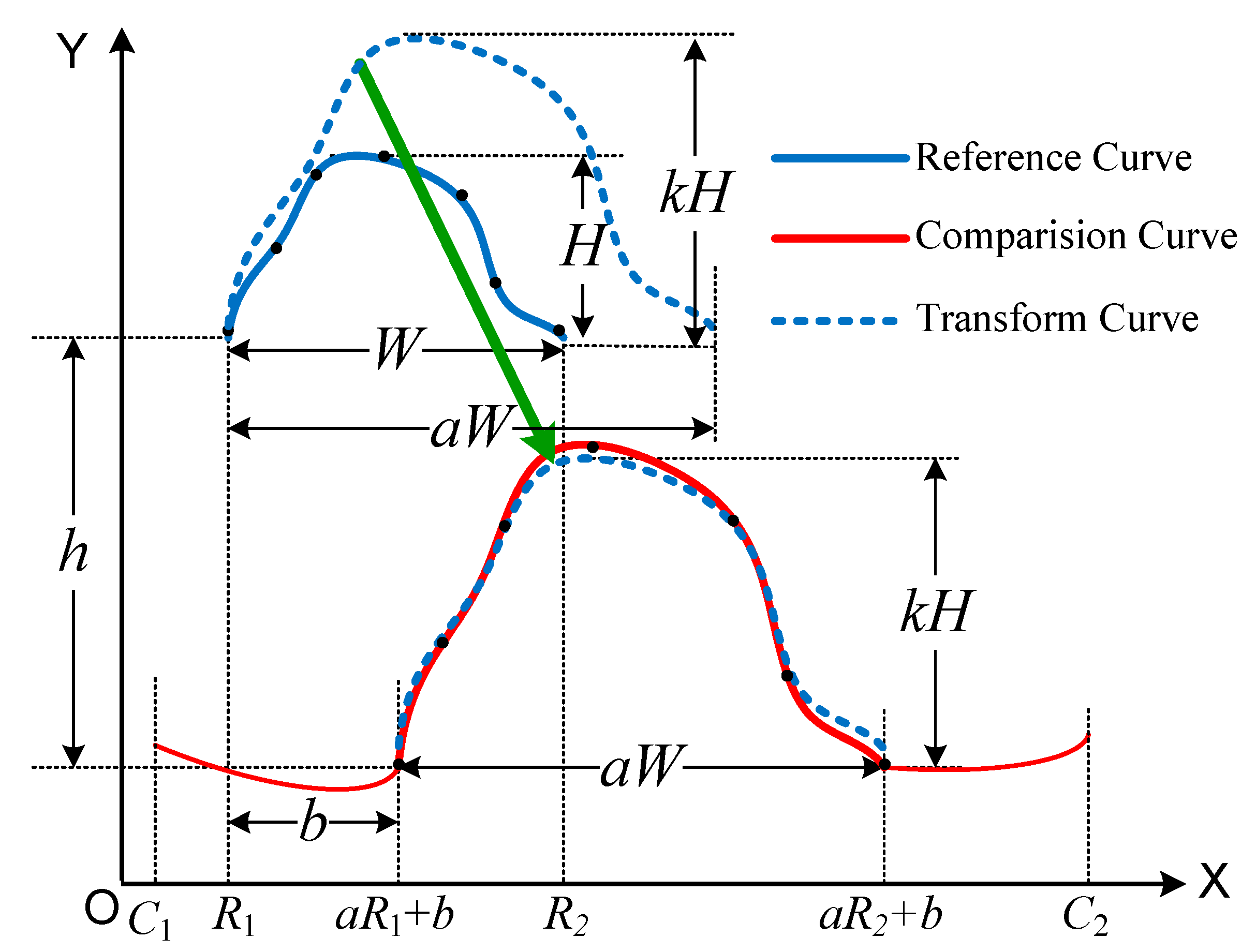
Definition 3. Given functions f1(x) defined on [R1, R2] and f2(x) defined on [C1, C2], and make transform, then the minimum distance of all the matching distances between f’1(x) and f2(x) with different transformation T is as follows:which is called the curve similarity distance of f2(x) to f1(x) under the transformation T, where f1(x) is called the reference function or the reference curve, f2(x) is called the comparison function or the comparison curve, f’1(x) is called the transform function or the transform curve, T is called a function similarity transformation or a curve similarity transformation (CST), and dis(a,b,k,h) is called the distance of the two curves under the curve similarity transformation T. Obviously, the distance between the two curves is different under different similarity transformations T, and the curve similarity distance (CSD) is the distance after the optimal matching of the reference curve for the comparison curve. After the curve similarity transformation, the curve similarity distance is denoted by min{dis(a,b,k,h)}.
Once the optimal matching of the curves is obtained, the corresponding curve similarity distance can be obtained, as shown in
Figure 2.
From
Figure 2,
a,
k are translational transformations in the horizontal and vertical directions, respectively, and
b,
h are scaling transformations in the horizontal and vertical directions, respectively.
Therefore, the similarity of curves based on curve similarity transformation is defined as follows:
Definition 4. Given a reference curve f1(x) and a comparison curve f2(x), for a given threshold ε, if d(f1,f2) ≤ ε, then the curve f2(x) is called similar to curve f1(x), and vice versa.
2.1.3. Discrete Definition
In engineering applications, the expression of the curve is therefore difficult to obtain, and can only be represented by an implicit function. By sampling, a continuous curve can be represented by a set of discrete ordered points. In this way, it is a common problem to measure the similarity of the two curves, which is to determine the similarity between two discrete ordered sets. As a result, a discrete curve similarity definition is needed. Similarly, the definition of discrete curve similarity transformation is given by reference to planar image transformation.
Definition 5. Given one discrete curve FA = {(x1, y1), (x2, y2)…(xm, ym)}, andis the curve similarity transformation matrix, where the meanings of a, b, k, h are the same as those described in Formula (1), corresponding to the translation and scale transformation of the horizontal and translation directions, respectively.
is called the similarity transformation curve of FA, where: Without any doubt, the transformation matrix T can be more complicated, such as rotation, mirroring and shearing transformation, and various combinations thereof. Moreover, the definition of discrete curve similarity distance under the condition of curve similarity transformation can be given.
Definition 6. Given a discrete curve FA = {(x1, y1), (x2, y2) … (xm, ym)} as a reference curve, another curve FB = {(x’1, y’1), (x’2, y’2) … (x’n, y’n)} is considered as a comparison curve. F’A = T • FA is transformed by a similarity transformation matrix T from FA. Among all similarity transformation matrices T, the minimum distance of all the matching distances between F’A and FB is as follows:which is called the curve similarity distance of FB to FA under the transformation T. The meanings of a, b, k, h are the same as those described in Formula (1) and Figure 2, corresponding to the translation and scale transformation of the horizontal and translation directions, respectively. Meanwhile, t is the starting point position of the optimal matching of the reference curve on the comparison curve.
Likewise, the definition of discrete curve similarity is as follows:
Definition 7. Given a discrete reference curve FA and a comparison curve FB, for a given threshold ε, if D(FA, FB) ≤ ε, then the curve FB is called similar to curve FA, and vice versa.
Finally, calculating the similarity distance of the comparison curve to the reference curve is equivalent to calculating the optimal matching of the reference curve in the sense of the average distance on the comparison curve after the transformation. Therefore, performing a similarity measure between two curves requires two steps, one is to calculate the optimal matching, and the other is to perform threshold discrimination.
In the classic curve matching algorithm DTW, once a curve of two curves is translated or stretched, the distance measurement between them will be modified. In the curve similarity measure process, the curve similarity distance calculation depends on a transformation matrix. The characteristics of the transformation matrix can well prevent the deformation process of the translation, stretching and rotation of the curve. However, the transformation matrix is sometimes difficult to solve directly.
Accordingly, an evolutionary computation (EC) [
32] algorithm has been introduced to obtain the transformation matrix and the optimal matching interval between two curves by minimizing the matching distance. The specific search algorithm is introduced later.
2.1.4. Matching Calculation
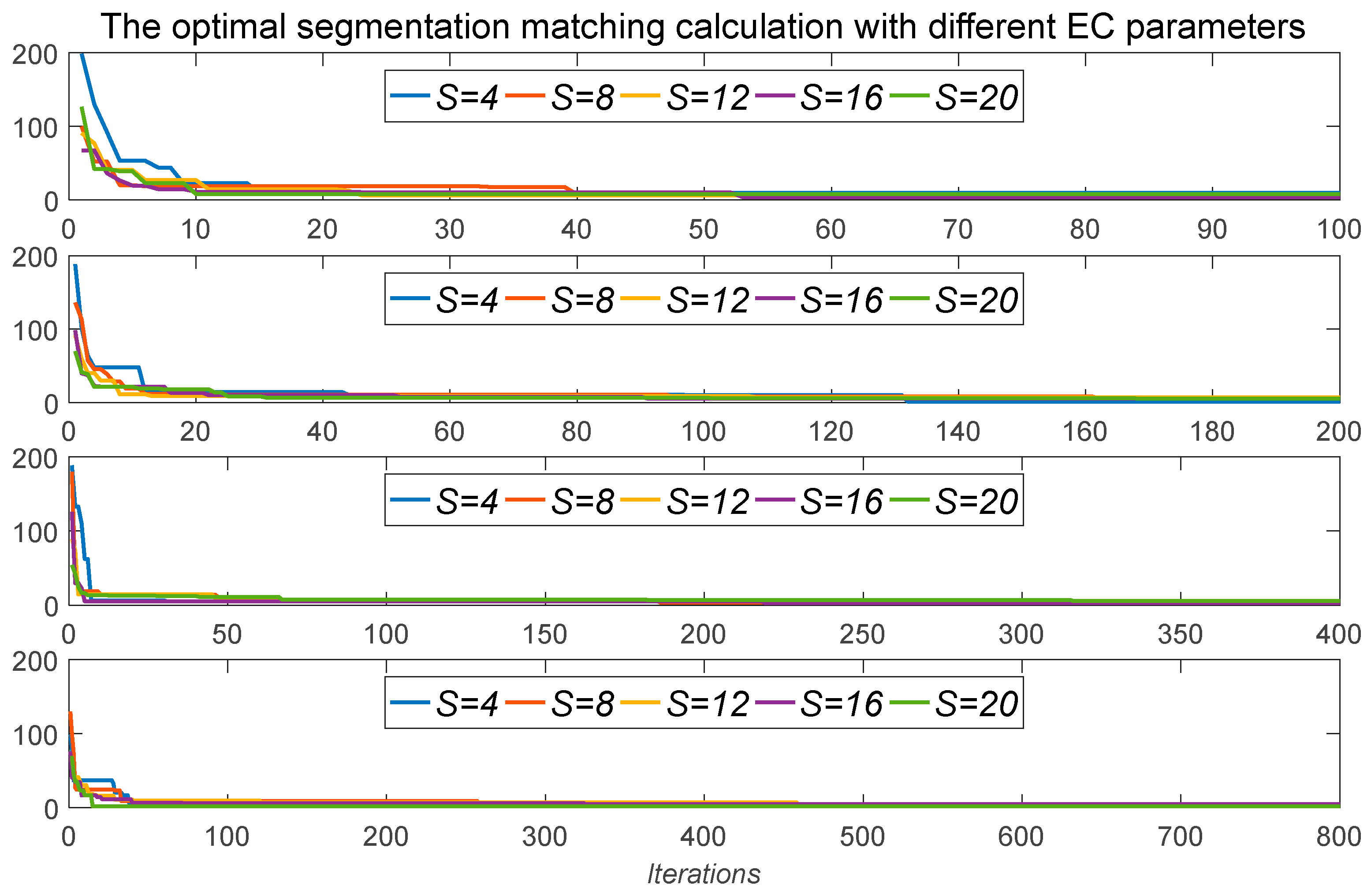
It can be observed from the definition that the curve similarity distance is the optimal matching of the reference curve with the comparison curve under different similarity transformations. Therefore, the matching distance between two curves under different CSTs can be obtained by random search algorithm. EC is a very effective algorithm for intelligent random search, where the similarity transformation matrix T is the parameter space of random search.
The whole idea of EC is to generate S random populations POP = {POP(0), POP(1), …, POP(S − 1)}, each of which corresponds to a set of parameters of CST T and a fitness value fitness = D(FA, FB) from Formula (6), where the fitness is smaller, the population is better. At the same time in each iteration search, each individual will generate new descendants near itself, and the best individuals can be chosen to enter the next iteration search.
The above process is repeated until the iterative search reaches the maximum number of iterations, at which point the optimal individual parameters and fitness will be regarded as CST and CSD, respectively.
At the same time, the parameters (t, a, b, k, h) should meet certain constraint conditions and adopt real coding for the signature curve matching. The boundary condition of the following system is 0 ≤ t < 2n, 0.90 ≤ a ≤ 1.10, 0.90 ≤ k ≤ 1.10, −100 ≤ b < 100, and −100 ≤ h ≤ 100, where a and k are elastic scales in the horizontal and vertical directions, and b and h are translations.
The Algorithm 1 process is as follows:
| Algorithm 1. Optimal matching calculation by EC |
| i = 0 |
| Forj = 0: S − 1 Do |
| //random generation of parameters t, a, b, k, h |
| initial a population POPi(j) = (t, a, b, k, h) |
| calculate fitness of each member in POPi(j) |
| End For |
| Whilei < Iterations Do |
| i = i + 1 |
| //sorting and classification |
| order POPi−1 by fitness in ascending and divide them into 4 levels |
| //acceleration search |
| For j = 0:S − 1 Do |
| If POPi−1(j) is at level kind |
| //where, kind = 1, 2, 3, 4, indicating the classification level of each population |
| //random generation of new parameters t, a, b, k, h in the neighborhood |
| //i.e., a = rand(POPi−1(j).a, kind) |
| generate new kind+1 subpopulations nPOP from POPi−1(j) |
| calculate fitness of each member in nPOP |
| //sorting subpopulations |
| order nPOP by fitness in ascending |
| //select best subpopulation nPOP as POPi−1(j + S) |
| POPi−1(j + S) = nPOP(0) |
| End If |
| End For |
| //global selection |
| order POPi−1 by fitness in ascending, where there are 2S populations |
| select the top S from POPi−1 as POPi |
| End While |
When generating new kind + 1 subpopulations from one population, each subpopulation has different parameter generation range at different level. Generally speaking, the fitness is smaller, the parameter generation range of each subpopulation is narrower.
Suppose that the
j-th population
POP(
j) is at the
kind-th level, on which parameter generation range is
GRkind = kind • GRmax/4,
kind + 1 subpopulations
nPOP can be randomly generated, here:
For example, for parameter a, GRmax = max(a) − min(a). When kind = 1 and a new random number r = −1 or 1 is generated, anew = aold + r • GRkind = aold + r • GRmax/4. At the same time, check whether anew satisfies the boundary condition.
The other parameters t, b, k, h all perform similar operations. Therefore, from the new subpopulation process generated by the parameters represented by each original population, the parameters of the generated subpopulations are determined according to the classification level of the original population. Although this parameter is also random, it varies randomly to the original population by its classification level.
2.2. Proposed System
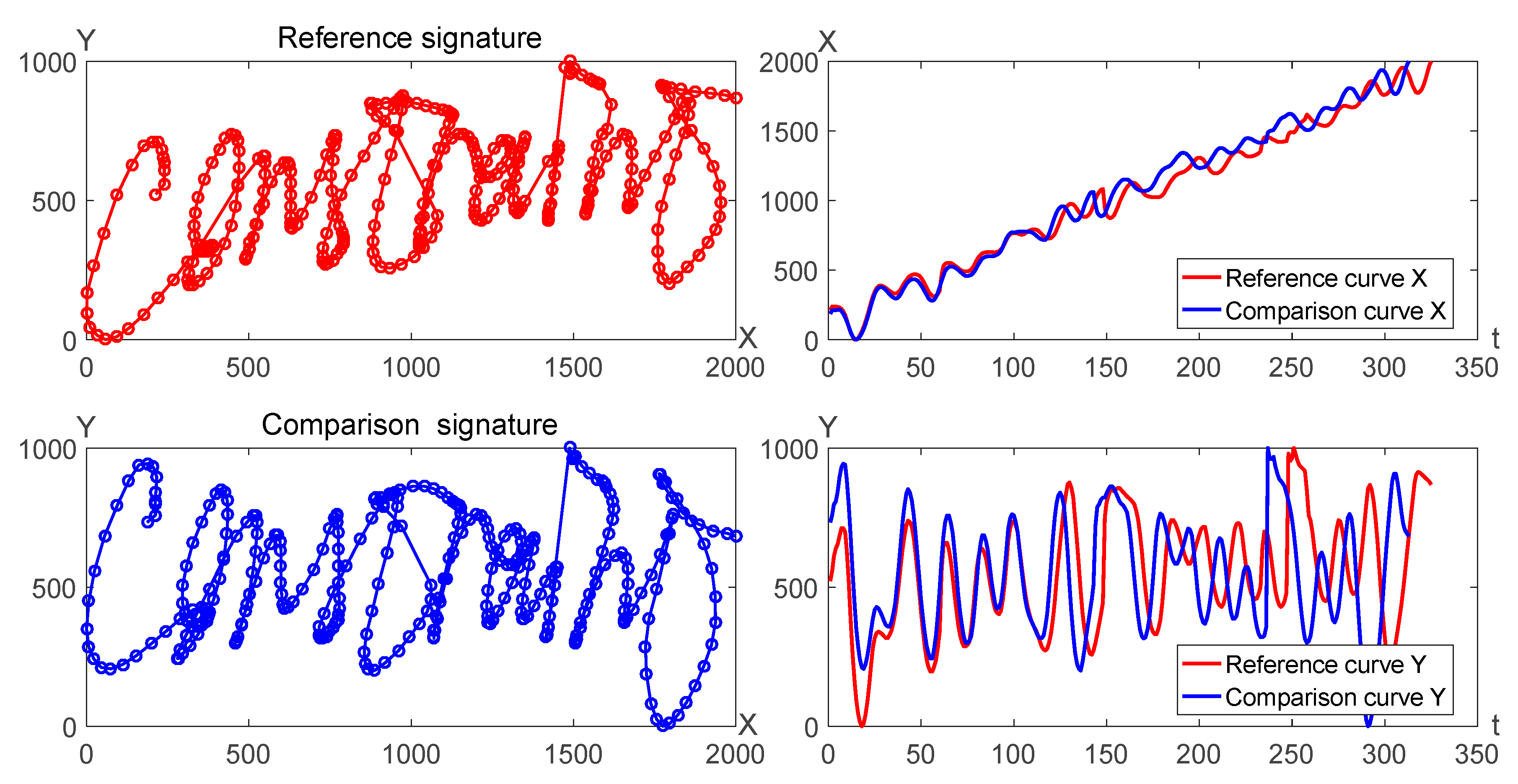
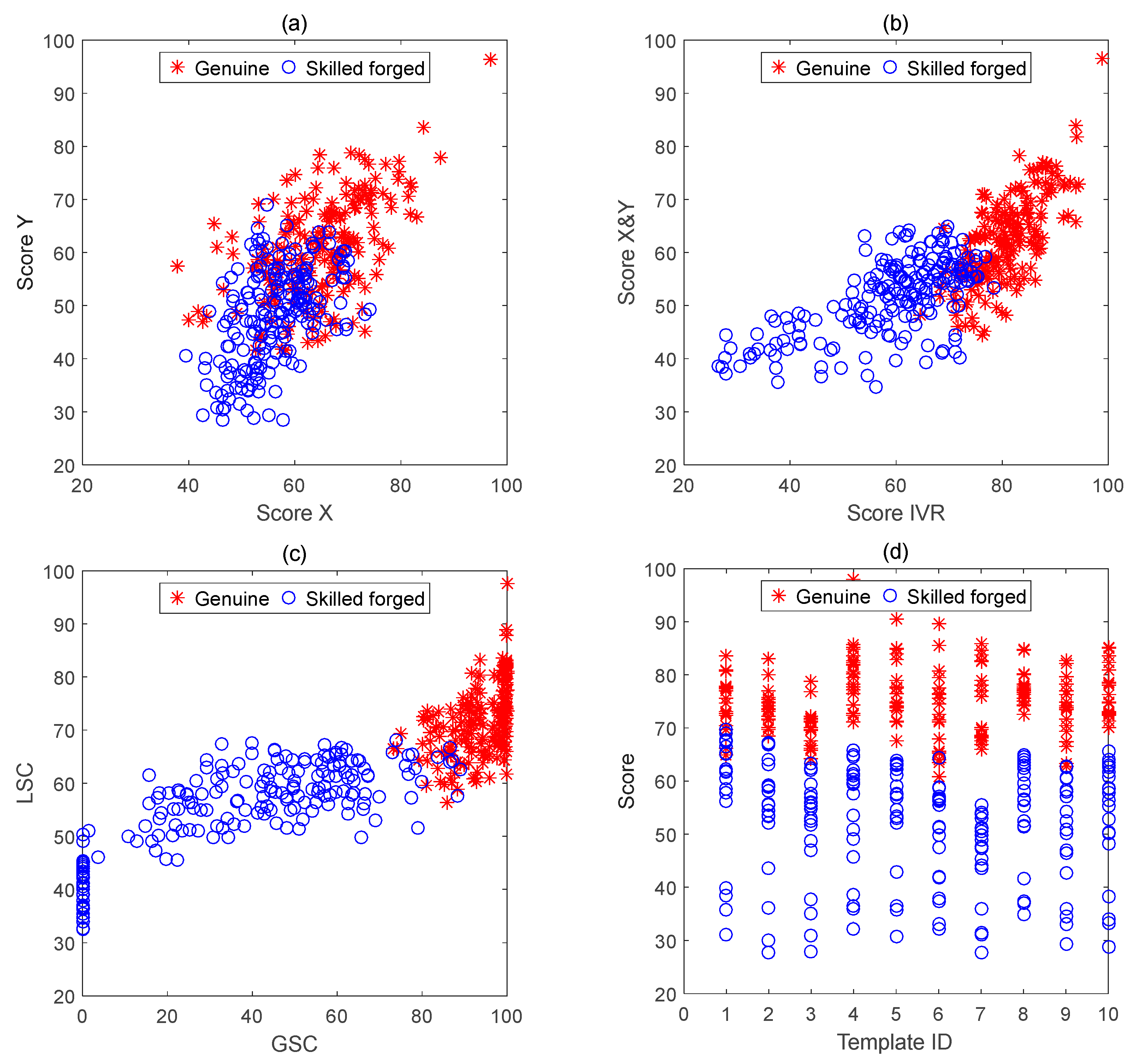
Online signature verification system can be discriminated by the similarity distance of two signature curves, where one signature can be called as the reference signature and the other can be called as the comparison signature. For instance, two signature trajectories can be considered as plane curves which can be also divided into
X curves and
Y curves, as shown in
Figure 3.
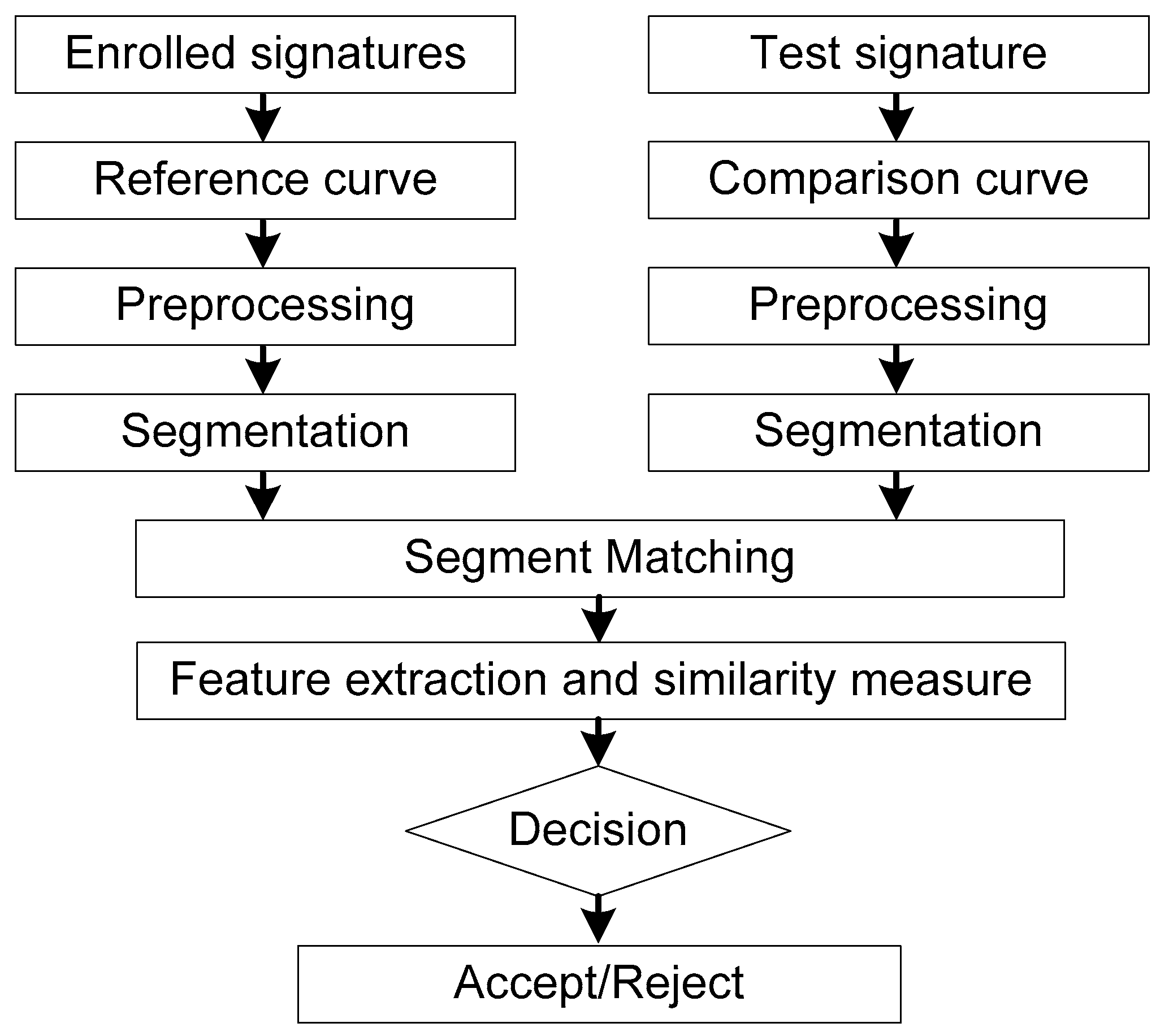
For a reference signature curve and a comparison signature curve, if the similarity distance between them is sufficiently small, it can be considered that the comparison signature is genuine one, otherwise it is a forged signature. A block diagram of the proposed system is illustrated in
Figure 4.
Given SignR = {(x1, y1), (x2, y2) … (xM, yM)} and SignC = {(x’1, y’1), (x’2, y’2) … (x’N, y’N)} as the reference curve and the comparison curve, respectively. For the calculation of the similarity distance of the signature curves, if the reference curve is calculated as a whole with the comparison signature, it will lead to greater errors. Segmentation curve matching can be used to better measure local differences between curves, which could be common in complex curve similarity measures.
2.2.1. Preprocessing
As the variations in different signatures have different dynamic ranges, min–max normalization is implemented in their
X and
Y coordinates. One signature should be reprocessed as follows:
where
x and
y are the original coordinates, and
xi and
yi are the normalized coordinates. The normalization scales of horizontal and vertical directions are different, and 2000 and 1000 are taken respectively to keep the original scale of the signature curve as much as possible.
2.2.2. Segmentation
SignR is divided into
K sections and there are
m data points of each the reference segmentation curve. Each the reference segmentation curve can be defined as
where, INT(
x) is the integral function.
Likewise for the comparison curve, each possible matching interval corresponding to the reference segmentation curve can be defined as
Here, considering the positional correlation and the deviation between the signature curves, when the reference signature length of each segmentation is m, the interval to be matched of the comparison signature is at least m in length, and is offset by n before and after the corresponding segmentation position. Equivalently, the reference curve swims within the interval to be matched on the comparison curve to obtain the optimal matching position.
2.2.3. Segmentation Matching
The process of the optimal segmentation matching is as follows:
Step 1: Take the reference signature curve as a template, and divide it into K segments according to Equation (9).
Step 2: The comparison curve should be divided into K segmentations according to Equation (10).
Step 3: For the i-th segmentation of the comparison curve, search the optimal matching with the corresponding the i-th segmentation of the reference curve by EC algorithm, and get the similarity distance di. Meanwhile, the matching distance dxi and dyi of the corresponding X, Y curves can be calculated based on the current matching result.
Step 4: Set
σxi,
σyi which are the standard deviation of
X,
Y data points in
i-th segmentation of the reference curve, compare it with the matching distance
dxi and
dyi, and calculate the similarity score
sxi and
syi of this segmentation, respectively, as in Equations (11) and (12).
Step 5: Repeat step 3 until all segmentation curves parameters are calculated.
Step 6: Calculate the average of sx, sy as the result outputs of X, Y curves’ similarity measure, and use the weighted average as the result output of the similarity measure Score of the two curves, as shown in later Formula (14).
It should be noted that when performing the optimal matching segmentation calculation, the similarity distance between the two curves can be obtained, which are considered as 2D curves. Next, the matching distances of the corresponding 1D curves X and Y can be separately calculated. Obviously, the calculated matching distances are absolute values, and if a similarity evaluation is to be performed, one threshold is needed to discriminate. For this reason, the similarity average is calculated using Gaussian functions of three different widths and the absolute distance measure is converted to a relative measure between 0 and 100. Thus, the discrimination threshold can be unified to a value between 0 and 100.
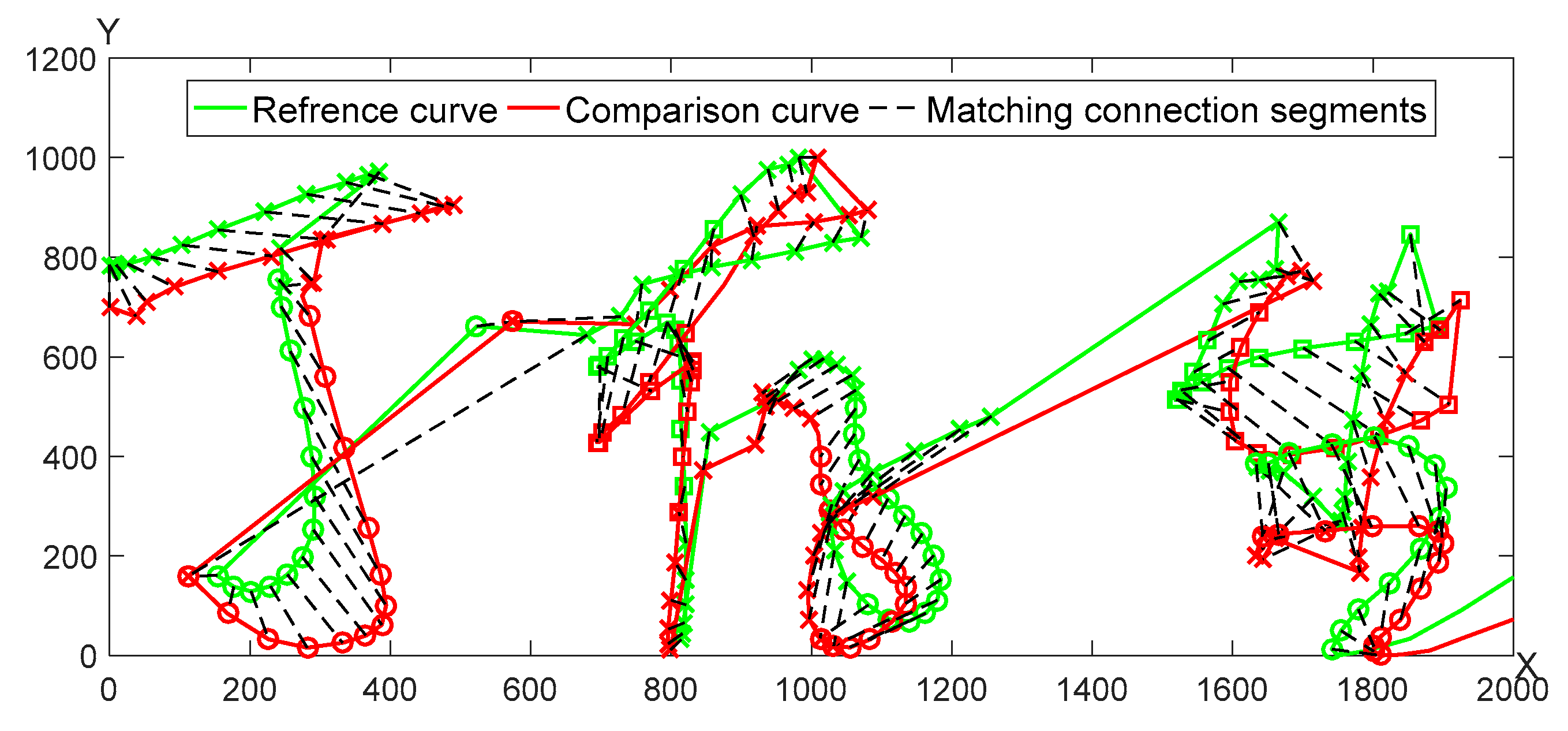
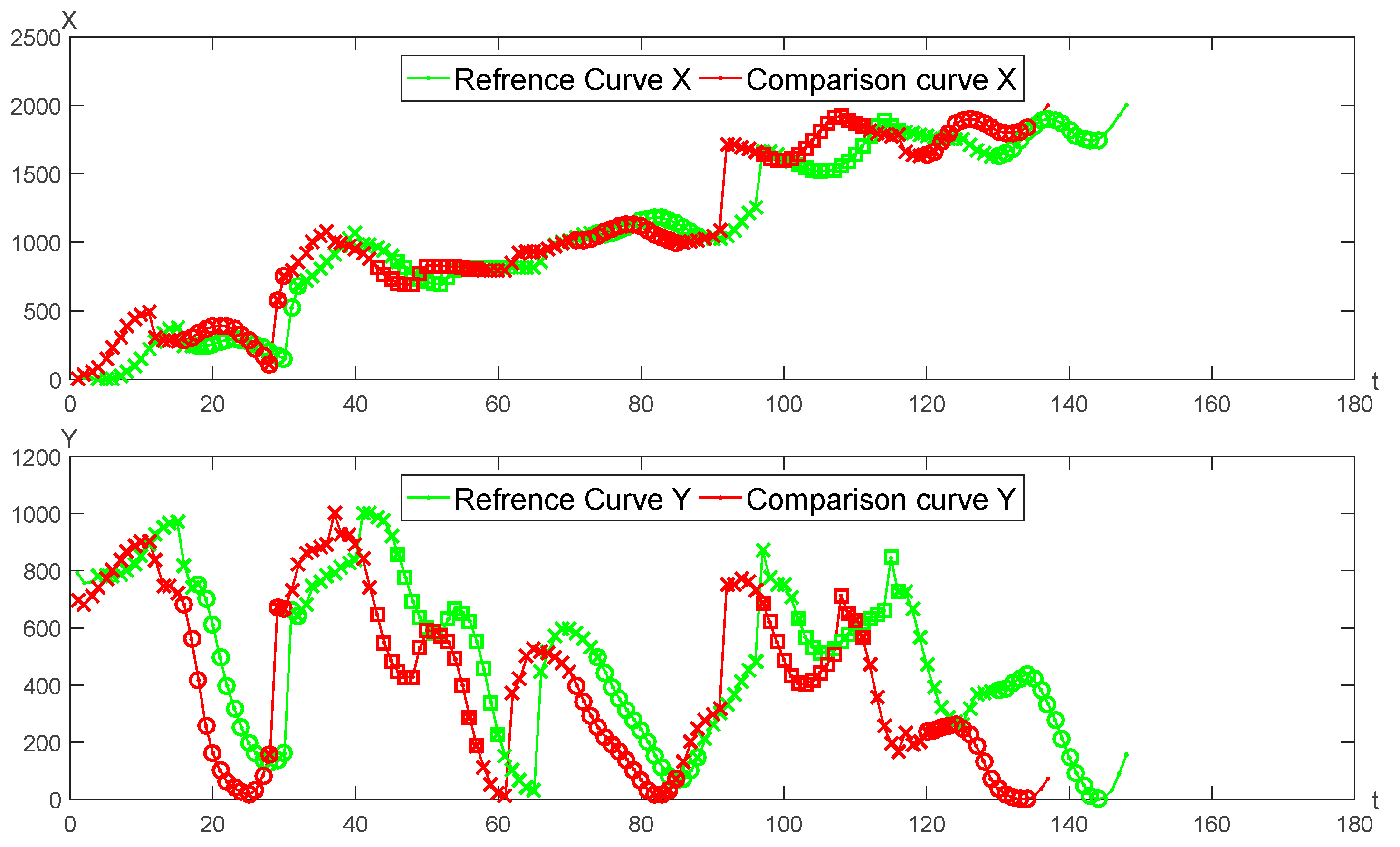
A pair of genuine signature curves is adopted for the optimal segmentation matching as seen in
Figure 5. The matching results of
X,
Y curves are shown in
Figure 6, respectively.
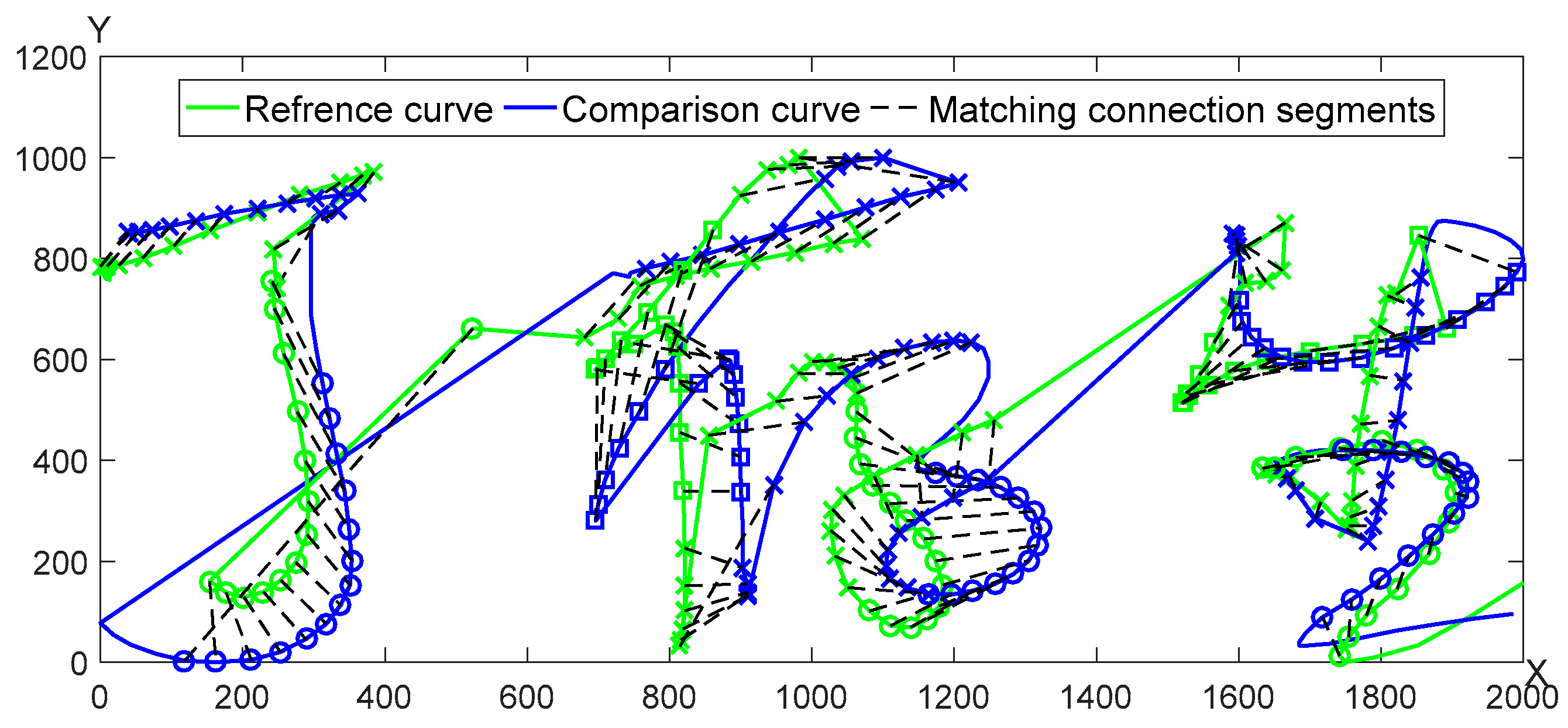
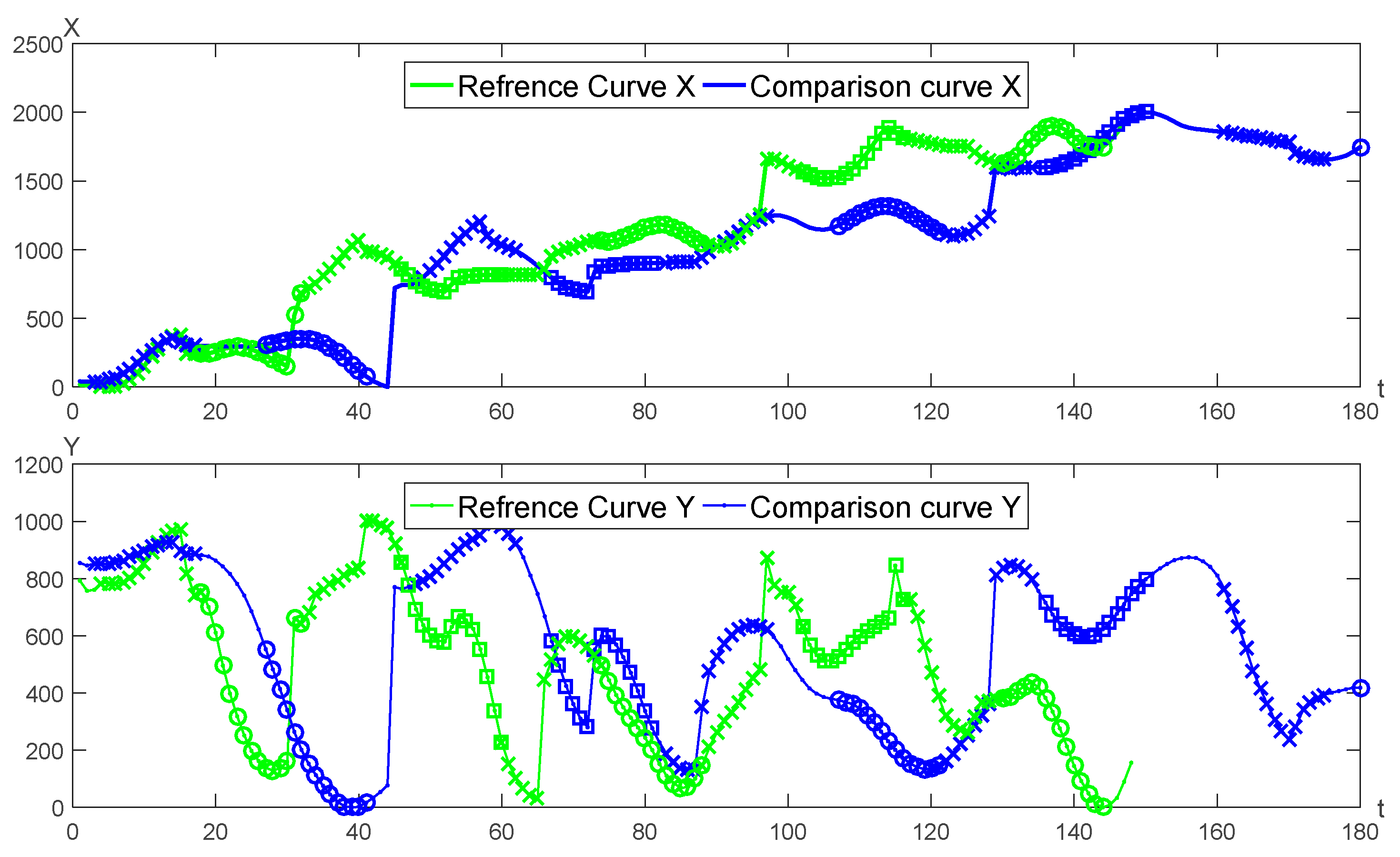
A pair of genuine and forged signature curves is adopted for the segmentation matching as seen in
Figure 7. The matching results of
X,
Y curves are shown in
Figure 8, respectively.
The optimal segmentation matching results between three curves are shown in
Table 1 and
Table 2.
It can be seen from
Table 1 and
Table 2 that for the similarity of
X,
Y curves is calculated by the same template signature and segmentations, the similarity between two genuine signatures is usually higher than that between the genuine and forged signatures. On each segmentation, the similarity calculation of the
X,
Y curves depends on the standard deviations of the respective segmentations in the template signature, as shown in Equation (11). To accurately estimate this deviation, a large number of genuine and forged signatures are needed for matching calculations and statistics. Obviously, this is difficult to obtain in practical applications. Here we use the intra-segmentation standard deviation of the signature segmentation itself and a deviation as the empirical value. In addition, the three control parameters
α,
β, and
γ are equivalent to controlling the width of the Gaussian function, and are also an empirical value. Here,
α = 0.4,
β = 1.6,
γ = 3.2 are selected. The changes in these parameters are not sensitive to the correct rate of the final evaluation results. Due to space limitations, this article will not discuss them.
2.2.4. Feature Extraction
In the
i-th matching interval [
Ri,
Ri+1], the interval velocity ratio (
IVR) of corresponding points is calculated as follows:
2.2.5. Similarity Measure
The similarity measure
Score of two signature curves is calculated as follows:
where
LSC and
GSC are local similarity score and global similarity score, respectively, while
wa and
wb are the corresponding weights.
M and
N are the lengths of the reference signature and the comparison signature, respectively.
Here:
is used to calculate score of the writing time ratio of two origin signatures.
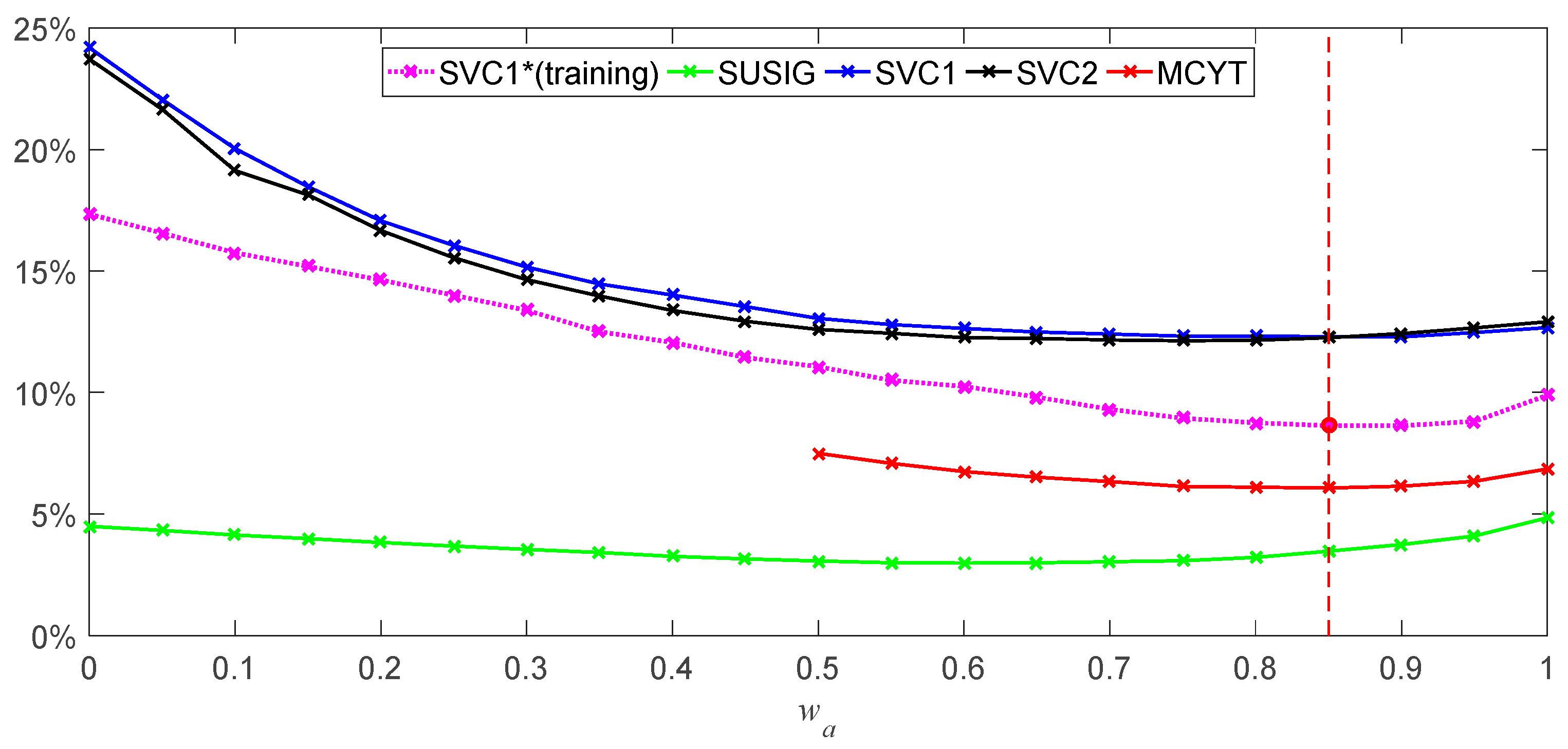
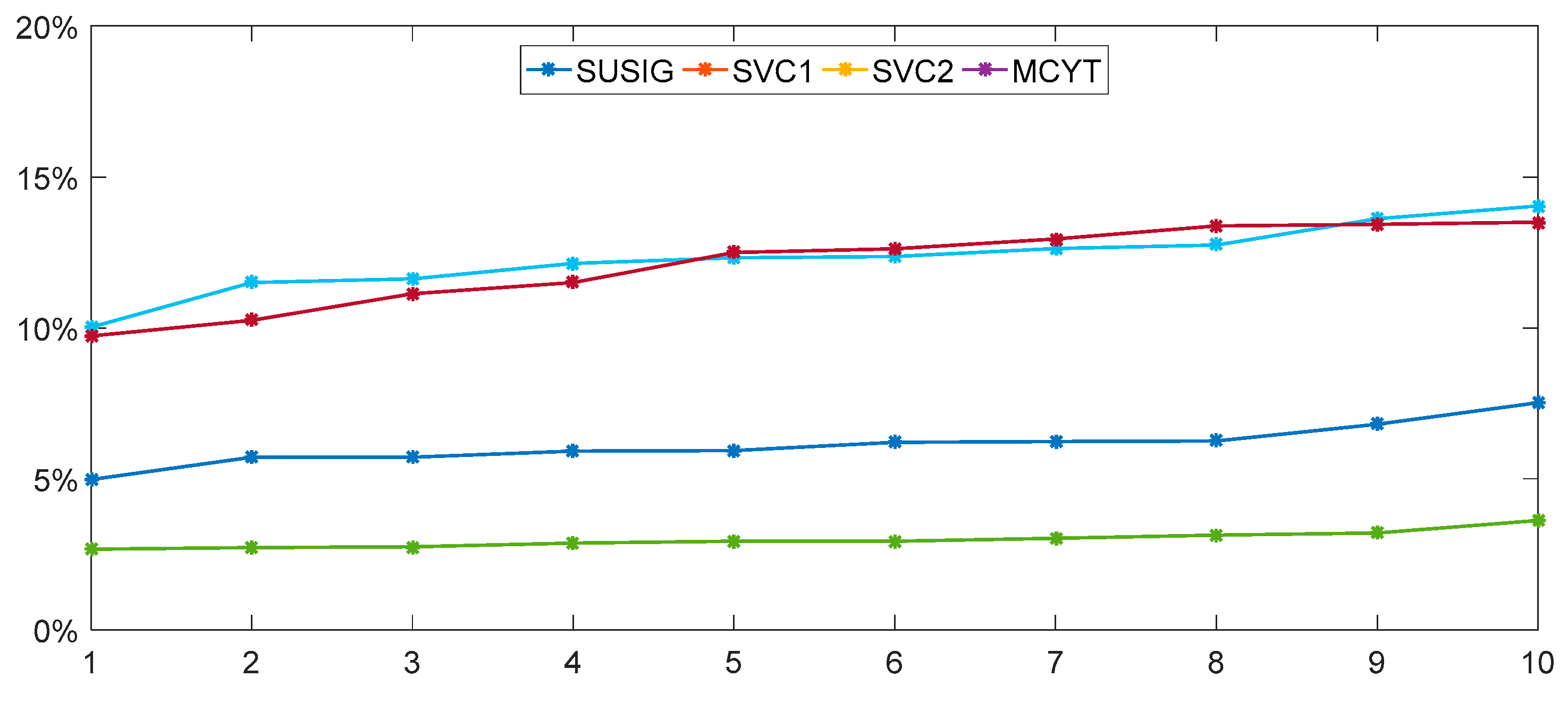
The calculation of the weight is calculated by enumeration, where
wa = 0.85 and
wb = 0.15, and the detailed process is shown in the next
Section 3.4.
It is considered that threshold ε of the signature verification system is 60 in many cases, and when Score is greater than 60, it may be distinguished into a genuine signature, and below 60 may be considered as a forged signature.
This is similar to the 100 point test. Passing more than 60 points is a pass, and below 60 is a failure. Of course, accurately determining the threshold is also a problem that needs to be studied in depth. For each user’s signature threshold determination, some other real registration signatures or even skilled forged signatures are needed. As a single template signature authentication system, only a reasonable threshold is given here, which is one of the key issues that need to be studied in the future.