A Novel Pre-Processing Technique for Original Feature Matrix of Electronic Nose Based on Supervised Locality Preserving Projections
Abstract
:1. Introduction
2. Materials and Experiments
2.1. Materials and Experimental Setup
2.2. Data Collection
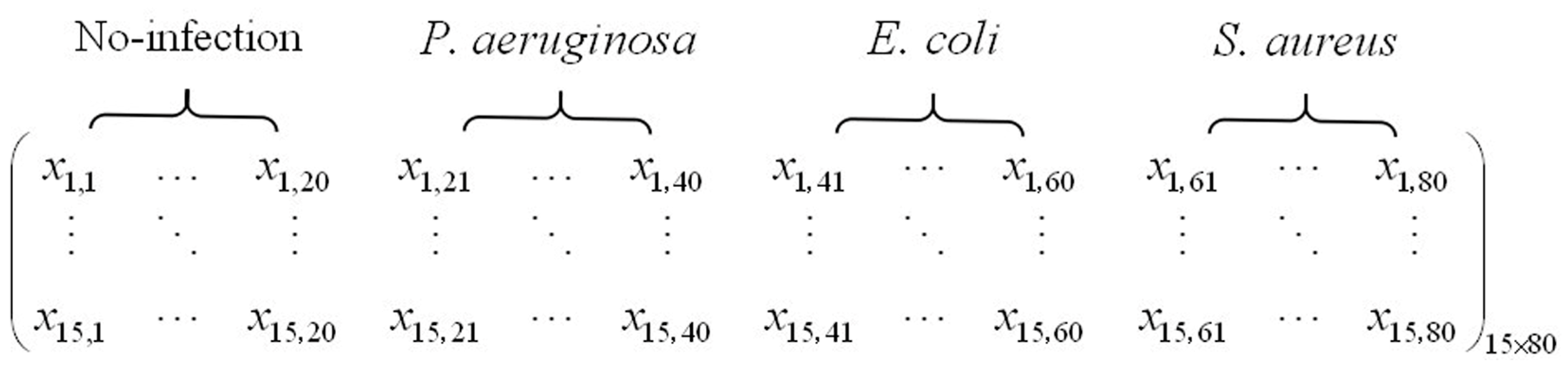
2.3. Original Feature Matrix
3. SLPP
- (1)
- Constructing the neighborhood: becomes the neighbor of only if they are from the same class and are “close”, where both and are the points of X and . Additionally, two different ways can be employed to find the neighborhood of .
- (a)
- -neighborhood: if , then can be taken as the neighbor of .
- (b)
- k-nearest-neighbors: a judgment is made on whether is among the k-nearest neighbors of .
- (2)
- Describe the relationship between and : suppose that is a variable describing the relationship between these two points, and will be “larger” if and are “closer”. There are also two different methods available to realize it.
- (a)
- simple-type: if is the neighbor of ; otherwise, .
- (b)
- heat-kernel:
- (3)
- Find the map: to make the relationship between and similar to that between and ; let Y be a “good” map to minimize the following objective function [27].under appropriate constraints, where and are the points of Y and . If and are “close” enough, then the value of will be much “larger”, and to make sure Equation (2) reaches its minimum, and must be “close” as well. In this way, Equation (2) transfers the local structure from matrix X to Y. Furthermore, because , Equation (2) can be computed aswhere . A constraint is imposed as follows [23]
4. Results and Discussion
4.1. Experimental Results
4.2. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bicego, M.; Tessari, G.; Tecchiolli, G.; Bettinelli, M. A comparative analysis of basic pattern recognition techniques for the development of small size electronic nose. Sens. Actuator B Chem. 2002, 85, 137–144. [Google Scholar] [CrossRef]
- Ciosek, P.; Wróblewski, W. The analysis of sensor array data with various pattern recognition techniques. Sens. Actuator B Chem. 2006, 114, 85–93. [Google Scholar] [CrossRef]
- Yu, J.B.; Byun, H.G.; So, M.S.; Huh, G.S. Analysis of diabetic patient’s breath with conducting polymer sensor array. Sens. Actuator B Chem. 2005, 108, 305–308. [Google Scholar] [CrossRef]
- Pavlou, A.K.; Magan, N.; McNulty, C.; Jones, J.M.; Sharp, D.; Brown, J.; Turner, A.P. Use of an electronic nose system for diagnoses of urinary tract infections. Biosens. Bioelectron. 2002, 17, 893–899. [Google Scholar] [CrossRef]
- Anh, D.T.V.; Olthuis, W.; Bergveld, P. A hydrogen peroxide sensor for exhaled breath measurement. Sens. Actuator B Chem. 2005, 111–112, 494–499. [Google Scholar] [CrossRef]
- Trihaas, J.; Nielsen, P.V. Electronic nose technology in quality assessment: monitoring the ripening process of danish blue cheese. J. Food Sci. 2005, 70, 44–49. [Google Scholar]
- Natale, C.D.; Macagnano, A.; Davide, F.; D’Amico, A.; Paolesse, R.; Boschi, T.; Faccio, M.; Ferri, G. An electronic nose for food analysis. Sens. Actuator B Chem. 1997, 44, 521–526. [Google Scholar] [CrossRef]
- Dutta, R.; Dutta, R. “Maximum probability rule” based classification of MRSA infections in hospital environment: Using electronic nose. Sens. Actuator B Chem. 2006, 120, 156–165. [Google Scholar] [CrossRef]
- Thomas, A.N.; Riazanskaia, S.; Cheung, W.; Xu, Y.; Goodacre, R.; Thomas, C.L.P.; Baguneid, M.S.; Bayat, A. Novel noninvasive identification of biomarkers by analytical profiling of chronic wounds using volatile organic compounds. Wound Repair Regen. 2010, 18, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Byun, H.; Persaud, K.C.; Pisanelli, A.M. Wound-state monitoring for burn patients using E-nose/SPME system. ETRI J. 2010, 32, 440–446. [Google Scholar] [CrossRef]
- Jia, P.; Tian, F.; He, Q.; Fan, S.; Liu, J.; Yang, S.X. Feature extraction of wound infection data for electronic nose based on a novel weighted KPCA. Sens. Actuator B Chem. 2014, 201, 555–566. [Google Scholar] [CrossRef]
- Goldberg, Y.; Zakai, A.; Kushnir, D.; Ritov, Y. Manifold learning: The price of normalization. J. Mach. Learn. Res. 2008, 9, 1909–1939. [Google Scholar]
- Roweis, S.T.; Saul, L.K. Nonlinear dimensionality reduction by locally linear embedding. Science 2000, 290, 2323–2326. [Google Scholar] [CrossRef] [PubMed]
- Tenenbaum, J.B.; Silva, V.; Langford, J.C. A global geometric framework for nonlinear dimensionality reduction. Science 2000, 290, 2319–2323. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jiang, H.; Barrio, R.; Liao, X.; Cheng, L.; Su, F. Incremental manifold learning by spectral embedding methods. Pattern Recognit. Lett. 2011, 32, 1447–1455. [Google Scholar] [CrossRef]
- Chatpatanasiri, R.; Kijsirikul, B. A unified semi-supervised dimensionality reduction framework for manifold learning. Neurocomputing 2010, 73, 1631–1640. [Google Scholar] [CrossRef]
- Shen, J.; Bu, J.; Ju, B.; Jiang, T.; Wu, H.; Li, L. Refining gaussian mixture model based on enhanced manifold learning. Neurocomputing 2012, 87, 19–25. [Google Scholar] [CrossRef]
- Han, Y.; Xu, Z.; Ma, Z.; Huang, Z. Image classification with manifold learning for out-of-sample data. Signal Process. 2012, 93, 2169–2177. [Google Scholar] [CrossRef]
- Cheon, Y.; Kim, D. Natural facial expression recognition using differential-AAM and manifold learning. Pattern Recognit. 2009, 42, 1340–1350. [Google Scholar] [CrossRef]
- Hadid, A.; Pietikäinen, M. Demographic classification from face videos using manifold learning. Neurocomputing 2012, 100, 197–205. [Google Scholar] [CrossRef]
- Lu, X.; Wang, H.; Zhou, R.; Ge, B. Autonomic failure prediction based on manifold learning for large-scale distributed systems. J. China Univ. Posts Telecommun. 2010, 17, 116–124. [Google Scholar] [CrossRef]
- Wachinger, C.; Yigitsoy, M.; Rijkhorst, E.G.; Navab, N. Manifold learning for image-based breathing gating in ultrasound and MRI. Med. Image Anal. 2012, 16, 806–818. [Google Scholar] [CrossRef] [PubMed]
- He, X. Locality preserving projections. Adv. Neural Inf. Process. Syst. 2005, 45, 186–197. [Google Scholar]
- Zhao, Z.; Zhang, L.; Zhao, M.; Hou, Z.; Zhang, C. Gabor face recognition by multi-channel classifier fusion of supervised kernel manifold learning. Neurocomputing 2012, 97, 398–404. [Google Scholar] [CrossRef]
- Zheng, Z.; Yang, F.; Tan, W.; Jia, J.; Yang, J. Gabor feature-based face recognition using supervised locality preserving projection. Signal Process. 2007, 87, 2473–2483. [Google Scholar] [CrossRef]
- Cheng, J.; Liu, Q.; Lu, H.; Chen, Y. Supervised kernel locality preserving projections for face recognition. Neurocomputing 2005, 67, 443–449. [Google Scholar] [CrossRef]
- Belkin, M.; Niyogi, P. Laplacian eigenmaps and spectral techniques for embedding and clustering. Adv. Neural Inf. Process. Syst. 2002, 14, 585–591. [Google Scholar]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Zuo, W.; Zhang, H.; Zhang, D.; Wang, K. Post-processed LDA for face and palmprint recognition: What is the rationale. Signal Process. 2010, 90, 2344–2352. [Google Scholar] [CrossRef]
- Baudat, G.; Anouar, F. Generalized discriminant analysis using a kernel approach. Neural Comput. 2000, 12, 2385–2404. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Feng, B.; Xu, W. Particle swarm optimization with particles having quantum behavior. In Proceedings of the Congress on Evolutionary Computation, Portland, OR, USA, 19–23 June 2004; pp. 325–331.
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Dasarathy, B.V. Nearest neighbor classification techniques. Stud. Fuzziness Soft Comput. 1994, 27, 181–203. [Google Scholar]




| Pathogens | Metabolites |
|---|---|
| S. aureus | Acetic acid, Aminoacetophenone, Ammonia, Ethanol, Formaldehyde, Isobutanol, Isopentyl acetate, Isopentanol, Methyl ketones, Trimethylamine, 1-Undecene, 2,5-Dimethylpyrazine isoamylamine, 2-Methylamine |
| E. coli | Acetaldehyde, Acetic acid, Aminoacetophenone, Butanediol, Decanol, Dimethyldisulfide, Dimethyltrisulfide, Dodecanol, Ethanol, Formaldehyde, Formic acid, Hydrogen sulfide, Indole, Lactic acid, Methanethiol, Methyl ketones, Octanol, Pentanols, Succinic acid, 1-Propanol |
| P. aeruginosa | Butanol, Dimethyldisulfide, Dimethyltrisulfide, Esters, Methyl ketones, Isobutanol, Isopentanol, Isopentyl acetate, Pyruvate, Sulphur compounds, Toluene, 1-Undecene, 2-Aminoacetophenone, 2-Butanone, 2-Heptanone, 2-Nonanone, 2-Undecanone |
| Sensors | Sensitive characteristic |
|---|---|
| TGS800 | Methane, Carbon monoxide, Isobutane, Hydrogen, Ethanol |
| TGS813 | Methane, Propane, Ethanol, Isobutane, Hydrogen, Carbon monoxide |
| TGS816 | Combustible gases, Methane,Propane, Butane, Carbon monoxide, Hydrogen, Ethanol, Isobutane |
| TGS822 | Organic solvent vapors, Methane, Carbon monoxide, Isobutane, n-Hexane, Benzene, Ethanol, Acetone |
| TGS825 | Hydrogen sulfide |
| TGS826 | Ammonia, Ethanol, Isobutane, Hydrogen |
| TGS2600 | Gaseous air contaminants, Methane, Carbon monoxide, Isobutane, Ethanol, Hydrogen |
| TGS2602 | VOCs, Odorous gases, Ammonia, Hydrogen sulfide, Toluene, Ethanol |
| TGS2620 | Vapors of organic solvents, combustible gases, Methane, Carbon monoxide, Isobutane, Hydrogen, Ethanol |
| WSP2111 | Benzene, Toluene, Ethanol, Hydrogen, Formaldehyde, Acetone |
| MQ135 | Ammonia, Benzene series material, Acetone, Carbon monoxide, Ethanol, Smoke |
| MQ138 | Alcohols, Aldehydes, Ketones, Aromatics |
| QS-01 | VOCs, Hydrogen, Carbon monoxide, Metane, Isobutane, Etanol, Ammonia |
| SP3S-AQ2 | VOCs, Methane, Isobutane, Carbon monoxide, Hydrogen, Ethanol |
| AQ | Carbon monoxide, Methanol, Ethanol, Isopropanol, Formaldehyde, Acetaldehyde, Sulfur dioxide, Hydrogen, Hydrogen sulfide, Phenol, Dimethyl ether, Ethylene |
| No-Infection | P. aeruginosa | E. coli | S. aureus | |
|---|---|---|---|---|
| No-infection | 1155.5567 | 1372.7781 | 1325.8864 | 1344.9724 |
| P. aeruginosa | 1372.7781 | 1461.6700 | 1488.3676 | 1499.6072 |
| E. coli | 1325.8864 | 1488.3676 | 1416.4451 | 1523.1622 |
| S. aureus | 1344.9724 | 1499.6072 | 1523.1622 | 1100.3343 |
| Methods | L | Classification Accuracy (%) | ||||
|---|---|---|---|---|---|---|
| No-Infection | P. aeruginosa | E. coli | S. aureus | Total | ||
| No-dealing | 15 | 85 | 85 | 90 | 85 | 86.25 |
| PCA | 10 | 90 | 90 | 85 | 85 | 87.5 |
| FDA | 3 | 75 | 80 | 85 | 85 | 81.25 |
| KFDA | 3 | 90 | 95 | 95 | 95 | 93.75 |
| SLPP | 7 | 100 | 95 | 100 | 100 | 98.75 |
| Methods | L | Classification Accuracy (%) | ||||
|---|---|---|---|---|---|---|
| No-Infection | P. aeruginosa | E. coli | S. aureus | Total | ||
| No-dealing | 15 | 85 | 90 | 90 | 75 | 85 |
| PCA | 10 | 90 | 80 | 90 | 85 | 86.25 |
| FDA | 3 | 75 | 80 | 70 | 95 | 80 |
| KFDA | 3 | 90 | 95 | 90 | 95 | 92.5 |
| SLPP | 7 | 100 | 95 | 90 | 100 | 96.25 |
| Methods | L | Classification Accuracy (%) | ||||
|---|---|---|---|---|---|---|
| No-Infection | P. aeruginosa | E. coli | S. aureus | Total | ||
| No-dealing | 15 | 80 | 80 | 95 | 75 | 82.5 |
| PCA | 10 | 85 | 85 | 90 | 75 | 83.75 |
| FDA | 3 | 75 | 80 | 70 | 95 | 80 |
| KFDA | 3 | 85 | 85 | 90 | 90 | 87.5 |
| SLPP | 7 | 100 | 85 | 90 | 100 | 93.75 |
| Methods | L | Classification Accuracy (%) | ||||
|---|---|---|---|---|---|---|
| No-Infection | P. aeruginosa | E. coli | S. aureus | Total | ||
| No-dealing | 15 | 85 | 80 | 80 | 85 | 82.5 |
| PCA | 11 | 90 | 85 | 75 | 85 | 83.75 |
| FDA | 3 | 85 | 80 | 75 | 85 | 81.25 |
| KFDA | 3 | 95 | 90 | 90 | 90 | 91.25 |
| SLPP | 8 | 100 | 90 | 90 | 100 | 95 |
| Methods | L | Classification Accuracy (%) | ||||
|---|---|---|---|---|---|---|
| No-Infection | P. aeruginosa | E. coli | S. aureus | Total | ||
| No-dealing | 15 | 80 | 80 | 75 | 80 | 81.25 |
| PCA | 11 | 85 | 80 | 75 | 85 | 81.25 |
| FDA | 3 | 75 | 75 | 70 | 80 | 77.5 |
| KFDA | 3 | 90 | 90 | 85 | 90 | 88.75 |
| SLPP | 8 | 100 | 90 | 85 | 100 | 93.75 |
| Methods | L | Classification Accuracy (%) | ||||
|---|---|---|---|---|---|---|
| No-Infection | P. aeruginosa | E. coli | S. aureus | Total | ||
| No-dealing | 15 | 75 | 75 | 75 | 85 | 77.5 |
| PCA | 11 | 80 | 80 | 75 | 85 | 80 |
| FDA | 3 | 75 | 75 | 70 | 85 | 76.25 |
| KFDA | 3 | 85 | 90 | 85 | 90 | 87.5 |
| SLPP | 8 | 100 | 80 | 85 | 100 | 91.25 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, P.; Huang, T.; Wang, L.; Duan, S.; Yan, J.; Wang, L. A Novel Pre-Processing Technique for Original Feature Matrix of Electronic Nose Based on Supervised Locality Preserving Projections. Sensors 2016, 16, 1019. https://doi.org/10.3390/s16071019
Jia P, Huang T, Wang L, Duan S, Yan J, Wang L. A Novel Pre-Processing Technique for Original Feature Matrix of Electronic Nose Based on Supervised Locality Preserving Projections. Sensors. 2016; 16(7):1019. https://doi.org/10.3390/s16071019
Chicago/Turabian StyleJia, Pengfei, Tailai Huang, Li Wang, Shukai Duan, Jia Yan, and Lidan Wang. 2016. "A Novel Pre-Processing Technique for Original Feature Matrix of Electronic Nose Based on Supervised Locality Preserving Projections" Sensors 16, no. 7: 1019. https://doi.org/10.3390/s16071019
APA StyleJia, P., Huang, T., Wang, L., Duan, S., Yan, J., & Wang, L. (2016). A Novel Pre-Processing Technique for Original Feature Matrix of Electronic Nose Based on Supervised Locality Preserving Projections. Sensors, 16(7), 1019. https://doi.org/10.3390/s16071019









