Diversity and Bioactivity of Marine Bacteria Associated with the Sponges Candidaspongia flabellata and Rhopaloeides odorabile from the Great Barrier Reef in Australia
Abstract
1. Introduction
2. Materials and Methods
2.1. Sponge Collection and Isolation of Bacteria
2.2. Molecular Identification of the Bacterial Isolates
2.2.1. DNA Extraction and 16S rRNA Gene Sequencing
2.2.2. Sequence Alignment and Phylogenetic Analysis
2.3. Antimicrobial Assay
2.4. Amplification of Type 1 Polyketide Synthase and Non-Ribosomal Peptide Synthetase Genes
3. Results
3.1. Molecular Identification of the Isolates
3.1.1. Phylogenetic Analysis Based on 16S rRNA Gene Sequencing
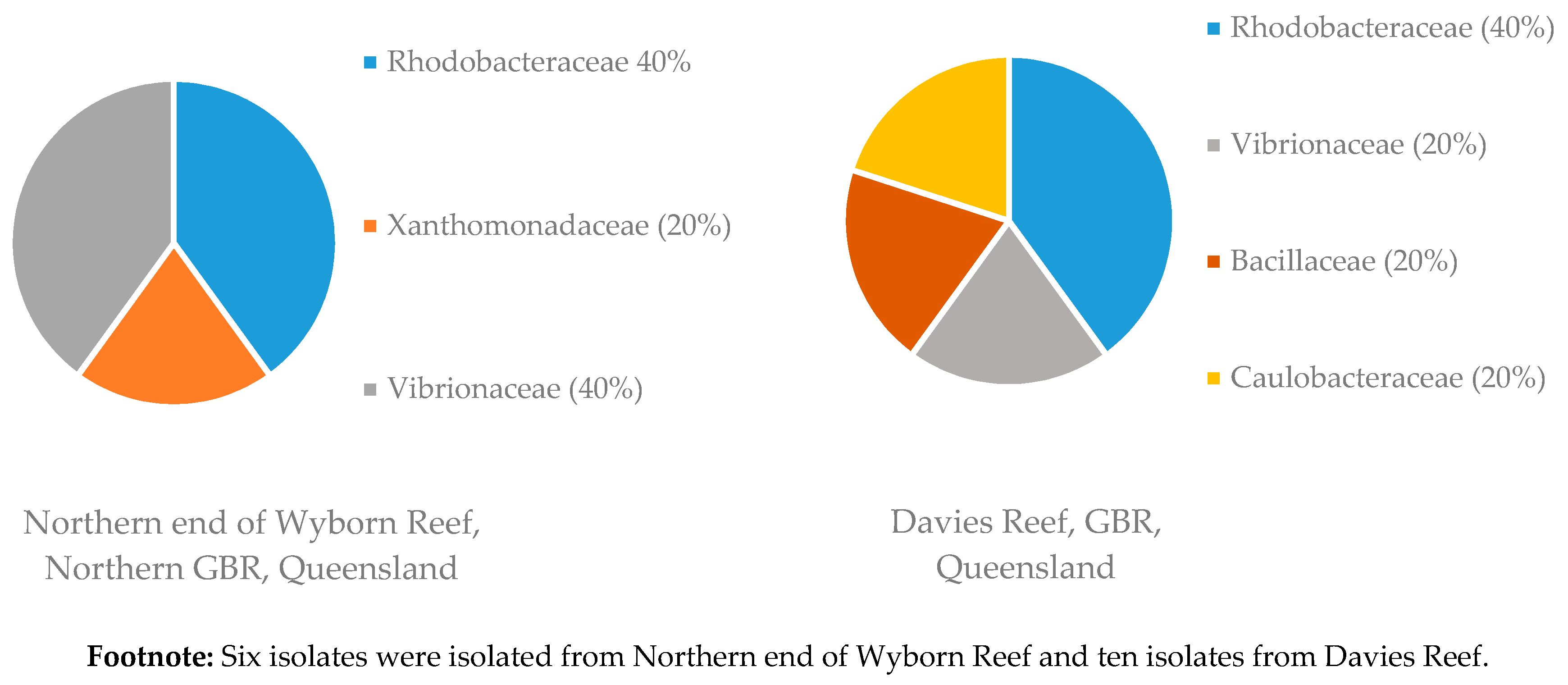
3.1.2. Distribution of Isolates in Relation to Their Sponge Hosts and Sponge Collection Locations
3.2. Antimicrobial Screening
3.3. Amplification of PKS-I and NRPS Genes
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
| Isolate Code | Closest Relative Strain | Sponge Sample Number | Purification Temperature (°C) | Days Grown | GenBank Accession Number | Latitude | Longitude | Location of Sponge Samples at the Great Barrier Reef (GBR), Queensland, Australia |
|---|---|---|---|---|---|---|---|---|
| 50161 | Vibrio harveyi strain NBRC 15634 | 16595 | 22 | 14 | KX418475 | −10.823 | 142.743 | NORTHERN END OF WYBORN REEF, NORTHERN GBR |
| 50162 | Photobacterium rosenbergii strain CC1 | 16595 | 22 | 14 | KX418476 | −10.823 | 142.743 | NORTHERN END OF WYBORN REEF, NORTHERN GBR |
| 50163 | Stenotrophomonas maltophilia strain ATCC 19861 | 16595 | 22 | 14 | KX418463 | −10.823 | 142.743 | NORTHERN END OF WYBORN REEF, NORTHERN GBR |
| 50164 | Vibrio campbellii strain ATCC 25920 | 16595 | 22 | 14 | KX418477 | −10.823 | 142.743 | NORTHERN END OF WYBORN REEF, NORTHERN GBR |
| 50165 | Vibrio harveyi strain NBRC 15634 | 16595 | 22 | 14 | KX418478 | −10.823 | 142.743 | NORTHERN END OF WYBORN REEF, NORTHERN GBR |
| 50166 | Pseudovibrio denitrificans strain NBRC 100825 | 16595 | 22 | 14 | KX418552 | −10.823 | 142.743 | NORTHERN END OF WYBORN REEF, NORTHERN GBR |
| 50895 | Microbulbifer variabilis strain Ni-2088 | 17643 | 27 | 14 | KX418479 | −18.826 | 147.64 | DAVIES REEF |
| 50897 | Microbulbifer variabilis strain Ni-2088 | 17643 | 27 | 14 | KX418480 | −18.826 | 147.64 | DAVIES REEF |
| 50898 | Microbulbifer variabilis strain Ni-2088 | 17643 | 27 | 14 | KX418464 | −18.826 | 147.64 | DAVIES REEF |
| 50899 | Micrococcus aloeverae strain AE-6 | 17643 | 27 | 14 | KX418589 | −18.826 | 147.64 | DAVIES REEF |
| 51193 | Microbulbifer agarilyticus strain JAMB A3 | 17766 | 27 | 14 | KX418502 | −21.659 | 150.324 | DAVIES REEF |
| 53189 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 19500 | 27 | 3 | KX418515 | −18.833 | 147.617 | NE-PERCY IS., NW CORNER |
| 53190 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 19500 | 27 | 3 | KX418516 | −18.833 | 147.617 | NE-PERCY IS., NW CORNER |
| 53223 | Vibrio owensii strain DY05 | 19500 | 30 | 2 | KX418481 | −18.833 | 147.617 | DAVIES REEF, GBR |
| 53224 | Bacillus safensis strain NBRC 100820 | 19500 | 30 | 2 | KX418582 | −18.833 | 147.617 | DAVIES REEF, GBR |
| 53225 | Bacillus safensis strain NBRC 100820 | 19500 | 30 | 2 | KX418572 | −18.833 | 147.617 | DAVIES REEF, GBR |
| 53226 | Bacillus safensis strain NBRC 100820 | 19500 | 30 | 2 | KX418577 | −18.833 | 147.617 | DAVIES REEF, GBR |
| 53249 | Brevundimonas diminuta strain NBRC 12697 | 19500 | 30 | 6 | KX418517 | −18.833 | 147.617 | DAVIES REEF, GBR |
| Isolate Code | Closest Relative Strain | Sponge Sample Number | Purification Temperature (°C) | Days Grown | GenBank Accession Number | Latitude | Longitude | Location of Sponge Samples at the Great Barrier Reef (GBR), Queensland, Australia |
|---|---|---|---|---|---|---|---|---|
| 53136 | Pseudomonas aeruginosa PAO1 strain PAO1 | 19496 | 27 | 10 | KX418482 | −18.820 | 147.63 | DAVIES REEF, GBR |
| 53137 | Sphingopyxis alaskensis strain RB2256 | 19496 | 27 | 6 | KX418562 | −18.820 | 147.63 | DAVIES REEF, GBR |
| 53178 | Ruegeria arenilitoris strain G-M8 | 19497 | 27 | 5 | KX418522 | −18.846 | 147.63 | DAVIES REEF, GBR |
| 53179 | Ruegeria arenilitoris strain G-M8 | 19497 | 27 | 5 | KX418523 | −18.846 | 147.63 | DAVIES REEF, GBR |
| 53651 | Micrococcus aloeverae strain AE-6 | 21250 | 26 | 19 | KX418483 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53654 | Pseudomonas aeruginosa PAO1 strain PAO1 | 21250 | 26 | 19 | KX418585 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53656 | Micrococcus aloeverae strain AE-6 | 21250 | 26 | 19 | KX418588 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53657 | Pseudomonas azotoformans strain NBRC 12693 | 21250 | 26 | 19 | KX418490 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 58101 | Ruegeria arenilitoris strain G-M8 | 21556 | 26 | 21 | KX418524 | −18.838 | 147.642 | TRUNK REEF, GBR |
| 58102 | Ruegeria arenilitoris strain G-M8 | 21556 | 26 | 21 | KX418555 | −18.332 | 146.829 | TRUNK REEF, GBR |
| 58103 | Ruegeria arenilitoris strain G-M8 | 21556 | 26 | 21 | KX418508 | −18.332 | 146.829 | TRUNK REEF, GBR |
| 58104 | Pseudomonas azotoformans strain NBRC 12693 | 21556 | 26 | 21 | KX418491 | −18.332 | 146.829 | TRUNK REEF, GBR |
| 58105 | Ruegeria arenilitoris strain G-M8 | 21556 | 26 | 21 | KX418509 | −18.332 | 146.829 | TRUNK REEF, GBR |
| 58107 | Pseudomonas azotoformans strain NBRC 12693 | 21556 | 26 | 21 | KX418492 | −18.332 | 146.829 | TRUNK REEF, GBR |
| 58111 | Pseudomonas azotoformans strain NBRC 12693 | 21556 | 26 | 21 | KX418493 | −18.332 | 146.829 | TRUNK REEF, GBR |
| 58115 | Cobetia amphilecti strain 46-2 | 21556 | 26 | 21 | KX418494 | −18.332 | 146.829 | TRUNK REEF, GBR |
| 58116 | Pseudomonas azotoformans strain NBRC 12693 | 21556 | 26 | 21 | KX418495 | −18.332 | 146.829 | TRUNK REEF, GBR |
| 58264 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21717 | 26 | 5 | KX418510 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58265 | Pseudomonas azotoformans strain NBRC 12693 | 21717 | 26 | 5 | KX418465 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58266 | Pseudomonas azotoformans strain NBRC 12693 | 21717 | 26 | 5 | KX418496 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58271 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21717 | 26 | 5 | KX418511 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58273 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21717 | 26 | 10 | KX418565 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58274 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21717 | 26 | 10 | KX418512 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58275 | Bacillus aquimaris strain TF-12 | 21717 | 26 | 5 | KX418579 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58276 | Ruegeria arenilitoris strain G-M8 | 21717 | 26 | 5 | KX418513 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58277 | Ruegeria arenilitoris strain G-M8 | 21717 | 26 | 5 | KX418514 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58278 | Ruegeria arenilitoris strain G-M8 | 21717 | 26 | 5 | KX418557 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58279 | Pseudomonas azotoformans strain NBRC 12693 | 21717 | 26 | 5 | KX418497 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58280 | Ruegeria arenilitoris strain G-M8 | 21722 | 26 | 5 | KX418518 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58281 | Pseudomonas azotoformans strain NBRC 12693 | 21722 | 26 | 5 | KX418466 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58282 | Pseudomonas azotoformans strain NBRC 12693 | 21722 | 26 | 5 | KX418467 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58283 | Bacillus cereus ATCC 14579 | 21722 | 26 | 5 | KX418567 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58285 | Pseudomonas azotoformans strain NBRC 12693 | 21722 | 26 | 5 | KX418468 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58286 | Pseudomonas azotoformans strain NBRC 12693 | 21722 | 26 | 5 | KX418469 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58287 | Bacillus aquimaris strain TF-12 | 21722 | 26 | 5 | KX418568 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58289 | Ruegeria arenilitoris strain G-M8 | 21722 | 26 | 5 | KX418525 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58290 | Ruegeria arenilitoris strain G-M8 | 21722 | 26 | 5 | KX418526 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58291 | Ruegeria arenilitoris strain G-M8 | 21722 | 26 | 5 | KX418566 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58292 | Ruegeria arenilitoris strain G-M8 | 21722 | 26 | 5 | KX418519 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58293 | Vibrio owensii strain DY05 | 21717 | 26 | 5 | KX418505 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58294 | Marinobacter vinifirmus strain FB1 | 21722 | 26 | 5 | KX418470 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58295 | Vibrio chagasii strain LMG 21353 | 21722 | 26 | 5 | KX418498 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58296 | Marinobacter vinifirmus strain FB1 | 21722 | 26 | 5 | KX418471 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58297 | Shewanella corallii strain fav-2-10-05 | 21722 | 26 | 5 | KX418503 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58298 | Ruegeria arenilitoris strain G-M8 | 21722 | 26 | 5 | KX418560 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58299 | Ruegeria arenilitoris strain G-M8 | 21722 | 26 | 5 | KX418527 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58300 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21722 | 26 | 5 | KX418528 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 53193 | Bacillus horikoshii strain DSM 8719 | 19496 | 27 | 4 | KX418578 | −18.820 | 147.630 | DAVIES REEF, GBR |
| 53208 | Kytococcus sedentarius strain DSM 20547 | 19496 | 27 | 4 | KX418583 | −18.820 | 147.630 | DAVIES REEF, GBR |
| 53209 | Kocuria rhizophilia strain TA68 | 19496 | 27 | 4 | KX418587 | −18.820 | 147.630 | DAVIES REEF, GBR |
| 53211 | Pseudoalteromonas piscicida strain NBRC 103038 | 19496 | 27 | 4 | KX418484 | −18.820 | 147.630 | DAVIES REEF, GBR |
| 58301 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21722 | 26 | 5 | KX418529 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58302 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21722 | 26 | 5 | KX418530 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58303 | Ruegeria atlantica strain NBRC 15792 | 21722 | 26 | 5 | KX418531 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58304 | Bacillus kochii strain WCC 4582 | 21722 | 26 | 5 | KX418569 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58305 | Ruegeria arenilitoris strain G-M8 | 21722 | 26 | 5 | KX418532 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58306 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21722 | 26 | 5 | KX418533 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58307 | Ruegeria arenilitoris strain G-M8 | 21722 | 26 | 5 | KX418564 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58308 | Ruegeria arenilitoris strain G-M8 | 21722 | 26 | 5 | KX418534 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58310 | Vibrio chagasii strain LMG 21353 | 21722 | 26 | 5 | KX418507 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58312 | Bacillus kochii strain WCC 4582 | 21722 | 26 | 5 | KX418574 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58313 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21722 | 26 | 5 | KX418535 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58315 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21722 | 26 | 5 | KX418536 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58316 | Ruegeria arenilitoris strain G-M8 | 21722 | 26 | 5 | KX418537 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58317 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21727 | 26 | 5 | KX418558 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58318 | Ruegeria arenilitoris strain G-M8 | 21727 | 26 | 5 | KX418538 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58319 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21727 | 26 | 5 | KX418539 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58320 | Microbulbifer variabilis strain Ni-2088 | 21727 | 26 | 5 | KX418501 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58321 | Staphylococcus warneri SG1 strain SG1 | 21727 | 26 | 5 | KX418580 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58322 | Thalassospira permensis strain SMB34 | 21727 | 26 | 5 | KX418554 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58323 | Ruegeria arenilitoris strain G-M8 | 21727 | 26 | 5 | KX418556 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58324 | Pseudomonas pachastrellae strain KMM 330 | 21727 | 26 | 5 | KX418499 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58325 | Pseudoalteromonas shioyasakiensis strain SE3 | 21727 | 26 | 5 | KX418500 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58326 | Bacillus nealsonii strain DSM 15077 | 21727 | 26 | 5 | KX418581 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58327 | Ruegeria arenilitoris strain G-M8 | 21727 | 26 | 5 | KX418540 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58328 | Ruegeria arenilitoris strain G-M8 | 21727 | 26 | 5 | KX418541 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58330 | Aquimarina spongiae strain A6 | 21727 | 26 | 5 | KX418462 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58332 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21727 | 26 | 5 | KX418520 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58333 | Ruegeria atlantica strain NBRC 15792 | 21727 | 26 | 5 | KX418542 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58334 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21727 | 26 | 5 | KX418543 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 58335 | Ruegeria arenilitoris strain G-M8 | 21727 | 26 | 5 | KX418544 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 53244 | Vibrio campbellii strain ATCC 25920 | 19496 | 30 | 2 | KX418485 | −18.82 | 147.63 | DAVIES REEF, GBR |
| 53611 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21241 | 26 | 18 | KX418545 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53613 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21241 | 26 | 18 | KX418546 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53614 | Bacillus oceanisediminis strain H2 | 21241 | 26 | 18 | KX418576 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53616 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21241 | 26 | 18 | KX418547 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53620 | Ruegeria arenilitoris strain G-M8 | 21241 | 26 | 18 | KX418548 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53622 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | 21241 | 26 | 18 | KX418549 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53623 | Vibrio fortis strain CAIM 629 | 21241 | 26 | 18 | KX418504 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53624 | Stenotrophomonas maltophilia R551-3 strain R551-3 | 21241 | 26 | 18 | KX418472 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53625 | Ruegeria arenilitoris strain G-M8 | 21241 | 26 | 18 | KX418561 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53634 | Sanguibacter inulinus strain ST50 | 21241 | 26 | 19 | KX418584 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53635 | Pseudomonas geniculate strain ATCC 19374 | 21241 | 26 | 19 | KX418506 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53636 | Pseudoalteromonas piscicida strain NBRC 103038 | 21250 | 26 | 19 | KX418486 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53640 | Pseudoalteromonas piscicida strain NBRC 103038 | 21250 | 26 | 19 | KX418487 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53641 | Ruegeria arenilitoris strain G-M8 | 21250 | 26 | 19 | KX418550 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53650 | Micrococcus yunnanensis strain YIM 65004 | 21250 | 26 | 19 | KX418586 | −18.838 | 147.642 | DAVIES REEF, GBR |
| 53240 | Bacillus safensis strain NBRC 100820 | 19496 | 30 | 2 | KX418571 | −18.820 | 147.630 | DAVIES REEF, GBR |
| 53237 | Psychrobacter celer strain SW-238 | 19496 | 30 | 2 | KX418473 | −18.820 | 147.630 | DAVIES REEF, GBR |
| 53239 | Psychrobacter celer strain SW-238 | 19496 | 30 | 2 | KX418474 | −18.820 | 147.630 | DAVIES REEF, GBR |
| 59159 | Bacillus kochii strain WCC 4582 | 22821 | 26 | 14 | KX418575 | −18.732 | 147.519 | CENTIPEDE REEF : BACK BOMMIE, GBR |
| 59161 | Vibrio alginolyticus strain ATCC 17749 | 22821 | 26 | 14 | KX418488 | −18.732 | 147.519 | CENTIPEDE REEF : BACK BOMMIE, GBR |
| 58824 | Photobacterium rosenbergii strain CC1 | 22821 | 28 | 15 | KX418489 | −18.732 | 147.519 | CENTIPEDE REEF : BACK BOMMIE, GBR |
| 58284 | Ruegeria arenilitoris strain G-M8 | 21722 | 26 | 5 | KX418551 | −18.833 | 147.626 | DAVIES REEF, GBR |
| 53260 | Brevundimonas diminuta strain NBRC 12697 | 19496 | 30 | 5 | KX418521 | −18.820 | 147.630 | DAVIES REEF, GBR |
References
- Hughes, C.C.; Fenical, W. Antibacterials from the sea. Chem. Eur. J. 2010, 16, 12512–12525. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Datta, D.; Talapatra, S.; Swarnakar, S. Bioactive compounds from marine invertebrates for potential medicines-an overview. Int. Lett. Nat. Sci. 2015, 7, 42–61. [Google Scholar] [CrossRef]
- Mehbub, M.F.; Lei, J.; Franco, C.; Zhang, W. Marine sponge derived natural products between 2001 and 2010: Trends and opportunities for discovery of bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, P.; Balasubramaniam, M.; Dam Roy, S.; Sarma, K.; Hairun, R.; Sunder, J. Characterization of the antibacterial activity of bacteria associated with stylissa sp, a marine sponge. Adv. Anim. Vet. Sci. 2014, 2, 20–25. [Google Scholar] [CrossRef]
- Burja, A.M.; Hill, R.T. Microbial symbionts of the Australian Great Barrier Reef sponge, candidaspongia flabellata. Hydrology 2001, 461, 41–47. [Google Scholar]
- Graça, A.P.; Viana, F.; Bondoso, J.; Correia, M.I.; Gomes, L.; Humanes, M.; Reis, A.; Xavier, J.R.; Gaspar, H.; Lage, O.M. The antimicrobial activity of heterotrophic bacteria isolated from the marine sponge erylus deficiens (astrophorida, geodiidae). Front. Microbiol. 2015, 6, 389. [Google Scholar] [PubMed]
- Sagar, S.; Kaur, M.; Minneman, K.P. Antiviral lead compounds from marine sponges. Mar. Drugs 2010, 8, 2619–2638. [Google Scholar] [CrossRef] [PubMed]
- Atikana, A.; Naim, M.A.; Sipkema, D. Detection of keto synthase (ks) gene domain in sponges and bacterial sponges. Ann. Bogor. 2013, 17, 27–33. [Google Scholar]
- Aguila-Ramírez, R.N.; Hernández-Guerrero, C.J.; González-Acosta, B.; Id-Daoud, G.; Hewitt, S.; Pope, J.; Hellio, C. Antifouling activity of symbiotic bacteria from sponge aplysina gerardogreeni. Int. Biodeter. Biodegr. 2014, 90, 64–70. [Google Scholar] [CrossRef]
- Thomas, T.; Moitinho-Silva, L.; Lurgi, M.; Bjork, J.R.; Easson, C.; Astudillo-Garcia, C.; Olson, J.B.; Erwin, P.M.; Lopez-Legentil, S.; Luter, H.; et al. Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 2016, 7, 11870. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, U.; Usher, K.M.; Taylor, M.W. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 2006, 55, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.J. The functional roles of marine sponges. Estuar. Coast. Shelf Sci. 2008, 79, 341–353. [Google Scholar] [CrossRef]
- Van Soest, R.W.M.; Boury-Esnault, N.; Vacelet, J.; Dohrmann, M.; Erpenbeck, D.; de Voogd, N.J.; Santodomingo, N.; Vanhoorne, B.; Kelly, M.; Hooper, J.N.A. Global diversity of sponges (porifera). PLoS ONE 2012, 7, e35105. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, W.; Feeney, R.J. Contributions to the study of marine products. XXXII. The nucleosides of sponges. I. J. Org. Chem. 1951, 16, 981–987. [Google Scholar] [CrossRef]
- Kennedy, J.; Baker, P.; Piper, C.; Cotter, P.; Walsh, M.; Mooij, M.; Bourke, M.; Rea, M.; O’Connor, P.; Ross, R.P.; et al. Isolation and analysis of bacteria with antimicrobial activities from the marine sponge haliclona simulans collected from irish waters. Mar. Biotechnol. 2009, 11, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Anand, T.P.; Bhat, A.W.; Shouche, Y.S.; Roy, U.; Siddharth, J.; Sarma, S.P. Antimicrobial activity of marine bacteria associated with sponges from the waters off the coast of south east india. Microbiol. Res. 2006, 161, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Webster, N.S.; Luter, H.M.; Soo, R.M.; Botte, E.S.; Simister, R.L.; Abdo, D.; Whalan, S. Same, same but different: Symbiotic bacterial associations in gbr sponges. Front. Microbiol. 2012, 3, 444. [Google Scholar] [CrossRef] [PubMed]
- Santos, O.C.S.; Pontes, P.V.M.L.; Santos, J.F.M.; Muricy, G.; Giambiagi-deMarval, M.; Laport, M.S. Isolation, characterization and phylogeny of sponge-associated bacteria with antimicrobial activities from brazil. Res. Microbiol. 2010, 161, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.R.A.; Kavlekar, D.P.; LokaBharathi, P.A. Marine drugs from sponge-microbe association—A review. Mar. Drugs 2010, 8, 1417–1468. [Google Scholar] [CrossRef] [PubMed]
- Amoutzias, D.G.; Chaliotis, A.; Mossialos, D. Discovery strategies of bioactive compounds synthesized by nonribosomal peptide synthetases and type-i polyketide synthases derived from marine microbiomes. Mar. Drugs 2016, 14, 80. [Google Scholar] [CrossRef] [PubMed]
- Borchert, E.; Jackson, S.A.; O’Gara, F.; Dobson, A.D.W. Diversity of natural product biosynthetic genes in the microbiome of the deep sea sponges inflatella pellicula, poecillastra compressa, and stelletta normani. Front. Microbiol. 2016, 7, 1027. [Google Scholar] [CrossRef] [PubMed]
- Santos, O.C.S.; Soares, A.R.; Machado, F.L.S.; Romanos, M.T.V.; Muricy, G.; Giambiagi-deMarval, M.; Laport, M.S. Investigation of biotechnological potential of sponge-associated bacteria collected in brazilian coast. Lett. Appl. Microbiol. 2015, 60, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.J. Engineering polyketide synthases and nonribosomal peptide synthetases. Curr. Opin. Struct. Biol. 2013, 23, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Hochmuth, T.; Piel, J. Polyketide synthases of bacterial symbionts in sponges—Evolution-based applications in natural products research. Phytochemistry 2009, 70, 1841–1849. [Google Scholar] [CrossRef] [PubMed]
- Müller, W.E.G.; Grebenjuk, V.A.; Le Pennec, G.; Schröder, H.-C.; Brümmer, F.; Hentschel, U.; Müller, I.M.; Breter, H.-J. Sustainable production of bioactive compounds by sponges—cell culture and gene cluster approach: A review. Mar. Biotechnol. 2004, 6, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Carmely, S.; Kashman, Y. Structure of swinholide-a, a new macrolide from the marine sponge theonella swinhoei. Tetrahedron. Lett. 1985, 26, 511–514. [Google Scholar] [CrossRef]
- Indraningrat, A.A.G.; Smidt, H.; Sipkema, D. Bioprospecting sponge-associated microbes for antimicrobial compounds. Mar. Drugs 2016, 14, 87. [Google Scholar] [CrossRef] [PubMed]
- Schneemann, I.; Kajahn, I.; Ohlendorf, B.; Zinecker, H.; Erhard, A.; Nagel, K.; Wiese, J.; Imhoff, J.F. Mayamycin, a cytotoxic polyketide from astreptomycesstrain isolated from the marine spongehalichondria panicea. J. Nat. Prod. 2010, 73, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Hirata, Y.; Uemura, D. Halichondrins-antitumor polyether macrolides from a marine sponge. Pure Appl. Chem. 1986, 58, 701–710. [Google Scholar] [CrossRef]
- Bai, R.L.; Paull, K.D.; Herald, C.L.; Malspeis, L.; Pettit, G.R.; Hamel, E. Halichondrin b and homohalichondrin b, marine natural products binding in the vinca domain of tubulin. Discovery of tubulin-based mechanism of action by analysis of differential cytotoxicity data. J. Biol. Chem. 1991, 266, 15882–15889. [Google Scholar] [PubMed]
- Webster, N.S.; Wilson, K.J.; Blackall, L.L.; Hill, R.T. Phylogenetic diversity of bacteria associated with the marine sponge rhopaloeides odorabile. Appl. Environ. Microbiol. 2001, 67, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Graça, A.P.; Bondoso, J.; Gaspar, H.; Xavier, J.R.; Monteiro, M.C.; de la Cruz, M.; Oves-Costales, D.; Vicente, F.; Lage, O.M. Antimicrobial activity of heterotrophic bacterial communities from the marine sponge erylus discophorus (astrophorida, geodiidae). PLoS ONE 2013, 8, e78992. [Google Scholar] [CrossRef] [PubMed]
- Webster, N.S.; Hill, R.T. The culturable microbial community of the Great Barrier Reef sponge rhopaloeides odorabile is dominated by an α-proteobacterium. Mar. Biol. 2001, 138, 843–851. [Google Scholar] [CrossRef]
- Wheeler, D.; Bhagwat, M. Blast quickstart: Example-driven web-based blast tutorial. In Comparative Genomics: Volumes 1 and 2; Bergman, N., Ed.; Humana Press: Totowa, NJ, USA, 2008. [Google Scholar]
- Ludwig, W.; Strunk, O.; Westram, R.; Richter, L.; Meier, H.; Yadhukumar; Buchner, A.; Lai, T.; Steppi, S.; Jobb, G.; et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004, 32, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Elfalah, H.W.A.; Ahmad, A.; Usup, G. Anti-microbial properties of secondary metabolites of marine gordonia tearrae extract. J. Agric. Sci. 2013, 5, 94. [Google Scholar] [CrossRef]
- Devi, P.; Wahidullah, S.; Rodrigues, C.; Souza, L.D. The sponge-associated bacterium bacillus licheniformis sab1: A source of antimicrobial compounds. Mar. Drugs 2010, 8, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, A.; Prabhakar, P.; Vijayalakshmi, M.; Venkateswarlu, Y. Purification and biological evaluation of the metabolites produced by Streptomyces sp. Tk-vl_333. Res. Microbiol. 2010, 161, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Gohar, Y.M.; El-Naggar, M.M.; Soliman, M.K.; Barakat, K.M. Characterization of marine burkholderia cepacia antibacterial agents. J. Nat. Prod. 2010, 3, 86–94. [Google Scholar]
- Dashti, Y.; Grkovic, T.; Abdelmohsen, U.R.; Hentschel, U.; Quinn, R.J. Production of induced secondary metabolites by a co-culture of sponge-associated actinomycetes, Actinokineospora sp. Eg49 and Nocardiopsis sp. Rv163. Mar. Drugs 2014, 12, 3046–3059. [Google Scholar] [CrossRef] [PubMed]
- McFarland, J. The nephelometer: An instrument for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. J. Am. Med. Assoc. 1907, 49, 1176–1178. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [PubMed]
- Palomo, S.; González, I.; de la Cruz, M.; Martín, J.; Tormo, J.R.; Anderson, M.; Hill, R.T.; Vicente, F.; Reyes, F.; Genilloud, O. Sponge-derived kocuria and micrococcus spp. As sources of the new thiazolyl peptide antibiotic kocurin. Mar. Drugs 2013, 11, 1071–1086. [Google Scholar] [CrossRef] [PubMed]
- Neilan, B.A.; Dittmann, E.; Rouhiainen, L.; Bass, R.A.; Schaub, V.; Sivonen, K.; Borner, T. Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J. Bacteriol. 1999, 181, 4089–4097. [Google Scholar] [PubMed]
- Kim, T.K.; Garson, M.J.; Fuerst, J.A. Marine actinomycetes related to the ‘salinospora’ group from the Great Barrier Reef sponge pseudoceratina clavata. Environ. Microbiol. 2005, 7, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Yang, C.; Horn, H.; Hajjar, D.; Ravasi, T.; Hentschel, U. Actinomycetes from red sea sponges: Sources for chemical and phylogenetic diversity. Mar. Drugs 2014, 12, 2771–2789. [Google Scholar] [CrossRef] [PubMed]
- Ayuso-Sacido, A.; Genilloud, O. New pcr primers for the screening of nrps and pks-i systems in actinomycetes: Detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 2005, 49, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Yoon, B.J.; You, H.S.; Lee, D.H.; Oh, D.C. Aquimarina spongiae sp. Nov., isolated from marine sponge halichondria oshoro. Int. J. Syst. Evol. Microbiol. 2011, 61, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Bondarev, V.; Richter, M.; Romano, S.; Piel, J.; Schwedt, A.; Schulz-Vogt, H.N. The genus pseudovibrio contains metabolically versatile bacteria adapted for symbiosis. Environ. Microbiol. 2013, 15, 2095–2113. [Google Scholar] [CrossRef] [PubMed]
- Pospisil, S.; Benada, O.; Kofronova, O.; Petricek, M.; Janda, L.; Havlicek, V. Kytococcus sedentarius (formerly micrococcus sedentarius) and dermacoccus nishinomiyaensis (formerly micrococcus nishinomiyaensis) produce monensins, typical streptomyces cinnamonensis metabolites. Can. J. Microbiol. 1998, 44, 1007–1011. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Yan, X.; Han, X.; Chen, H.; Lin, W.; Lee, F.S.; Wang, X. Identification of norharman as the cytotoxic compound produced by the sponge (Hymeniacidon perleve)-associated marine bacterium pseudoalteromonas piscicida and its apoptotic effect on cancer cells. Biotechnol. Appl. Biochem. 2006, 44, 135–142. [Google Scholar] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277. [Google Scholar]
- Cita, Y.P.; Suhermanto, A.; Radjasa, O.K.; Sudharmono, P. Antibacterial activity of marine bacteria isolated from sponge xestospongia testudinaria from sorong, papua. Asian. Pac. J. Trop. Med. 2017, 7, 450–454. [Google Scholar] [CrossRef]
- Zobell, C.E. Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J. Mar. Sci. 1941, 4, 42–75. [Google Scholar]
- Wei, H.; Lin, Z.; Li, D.; Gu, Q.; Zhu, T. OSMAC (one strain many compounds) approach in the research of microbial metabolites--a review. Acta Microbiol. Sin. 2010, 50, 701–709. [Google Scholar]
- Marmann, A.; Aly, A.H.; Lin, W.; Wang, B.; Proksch, P. Co-cultivation—A powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar. Drugs 2014, 12, 1043–1065. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, U.; Schmid, M.; Wagner, M.; Fieseler, L.; Gernert, C.; Hacker, J. Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the mediterranean sponges aplysina aerophoba and aplysina cavernicola. FEMS Microbiol. Ecol. 2001, 35, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.S.; Krishna, E.R.; Sujatha, P.; Kumar, B.V. Screening and isolation of associated bioactive microorganisms from fasciospongia cavernosa from of visakhapatnam coast, bay of bengal. J. Chem. 2012, 9, 2166–2176. [Google Scholar]
- Crowley, S.P.; O’Gara, F.; O’Sullivan, O.; Cotter, P.D.; Dobson, A.D.W. Marine pseudovibrio sp. As a novel source of antimicrobials. Mar. Drugs 2014, 12, 5916–5929. [Google Scholar] [CrossRef] [PubMed]
- Michel-Briand, Y.; Baysse, C. The pyocins of pseudomonas aeruginosa. Biochimie 2002, 84, 499–510. [Google Scholar] [CrossRef]
- Zheng, L.; Chen, H.; Han, X.; Lin, W.; Yan, X. Antimicrobial screening and active compound isolation from marine bacterium nj6-3-1 associated with the sponge hymeniacidon perleve. World J. Microbiol. Biotechnol. 2005, 21, 201–206. [Google Scholar] [CrossRef]
- Offret, C.; Desriac, F.; Le Chevalier, P.; Mounier, J.; Jégou, C.; Fleury, Y. Spotlight on antimicrobial metabolites from the marine bacteria pseudoalteromonas: Chemodiversity and ecological significance. Mar. Drugs 2016, 14, 129. [Google Scholar] [CrossRef] [PubMed]
- Satheesh, S.; Ba-akdah, M.A.; Al-Sofyani, A.A. Natural antifouling compound production by microbes associated with marine macroorganisms—A review. Electron. J. Biotechn. 2016, 21, 26–35. [Google Scholar] [CrossRef]
- Skariyachan, S.; G Rao, A.; Patil, M.R.; Saikia, B.; Bharadwaj Kn, V.; Rao Gs, J. Antimicrobial potential of metabolites extracted from bacterial symbionts associated with marine sponges in coastal area of gulf of mannar biosphere, India. Lett. Appl. Microbiol. 2014, 58, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Flemer, B.; Kennedy, J.; Margassery, L.M.; Morrissey, J.P.; O’Gara, F.; Dobson, A.D.W. Diversity and antimicrobial activities of microbes from two irish marine sponges, Suberites carnosus and Leucosolenia sp. Lett. Appl. Microbiol. 2012, 112, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Cuvelier, M.L.; Blake, E.; Mulheron, R.; McCarthy, P.J.; Blackwelder, P.; Thurber, R.L.V.; Lopez, J.V. Two distinct microbial communities revealed in the sponge cinachyrella. Front. Microbiol. 2014, 5, 581. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, C.C.P.; Costa, R.; Araujo, F.V.; Hajdu, E.; Peixoto, R.; Lins, U.; Rosado, A.S.; Van Elsas, J.D. Diversity of bacteria in the marine sponge aplysina fulva in brazilian coastal waters. Appl. Environ. Microbiol. 2009, 75, 3331–3343. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; He, L.; Miao, X. Cultivable bacterial community from south china sea sponge as revealed by dgge fingerprinting and 16S rDNA phylogenetic analysis. Curr. Microbiol. 2007, 55, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Turque, A.S.; Cardoso, A.M.; Silveira, C.B.; Vieira, R.P.; Freitas, F.A.D.; Albano, R.M.; Gonzalez, A.M.; Paranhos, R.; Muricy, G.; Martins, O.B. Bacterial communities of the marine sponges hymeniacidon heliophila and polymastia janeirensis and their environment in rio de janeiro, brazil. Mar. Biol. 2008, 155, 135. [Google Scholar] [CrossRef]
- Kim, T.K.; Fuerst, J.A. Diversity of polyketide synthase genes from bacteria associated with the marine sponge pseudoceratina clavata: Culture-dependent and culture-independent approaches. Environ. Microbiol. 2006, 8, 1460–1470. [Google Scholar] [CrossRef] [PubMed]
- Boddy, C.N. Bioinformatics tools for genome mining of polyketide and non-ribosomal peptides. J. Ind. Microbiol. Biot. 2014, 41, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Reen, F.J.; Romano, S.; Dobson, A.D.W.; O’Gara, F. The sound of silence: Activating silent biosynthetic gene clusters in marine microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar] [CrossRef] [PubMed]
- Ochi, K.; Hosaka, T. New strategies for drug discovery: Activation of silent or weakly expressed microbial gene clusters. Appl. Microbiol. Biotechnol. 2013, 97, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Giddings, L.-A.; Newman, D.J. Bioactive Compounds from Extremophiles: Genomic Studies, Biosynthetic Gene Clusters, and New Dereplication Methods; Briefs in Microbiology; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–124. [Google Scholar]
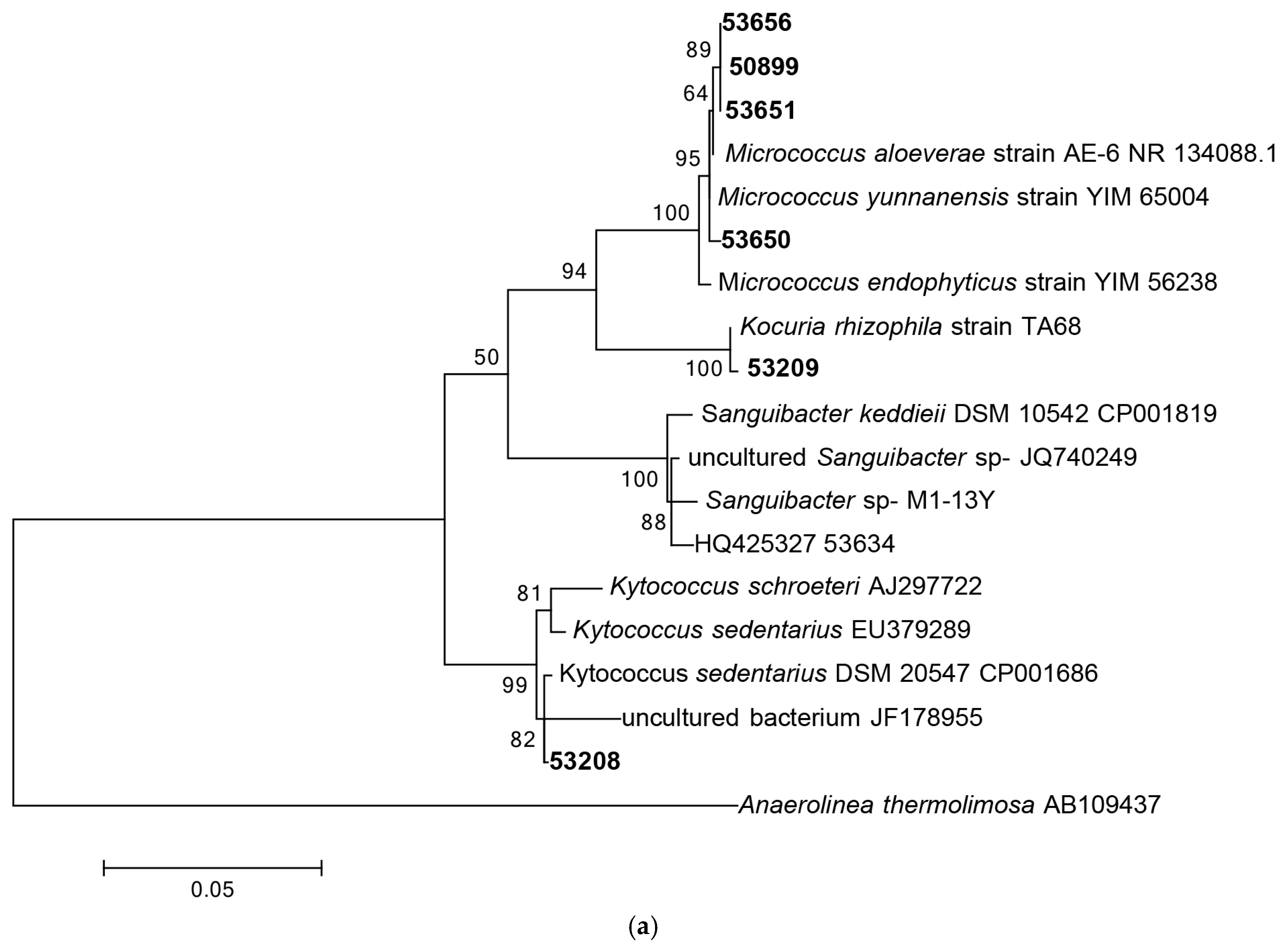

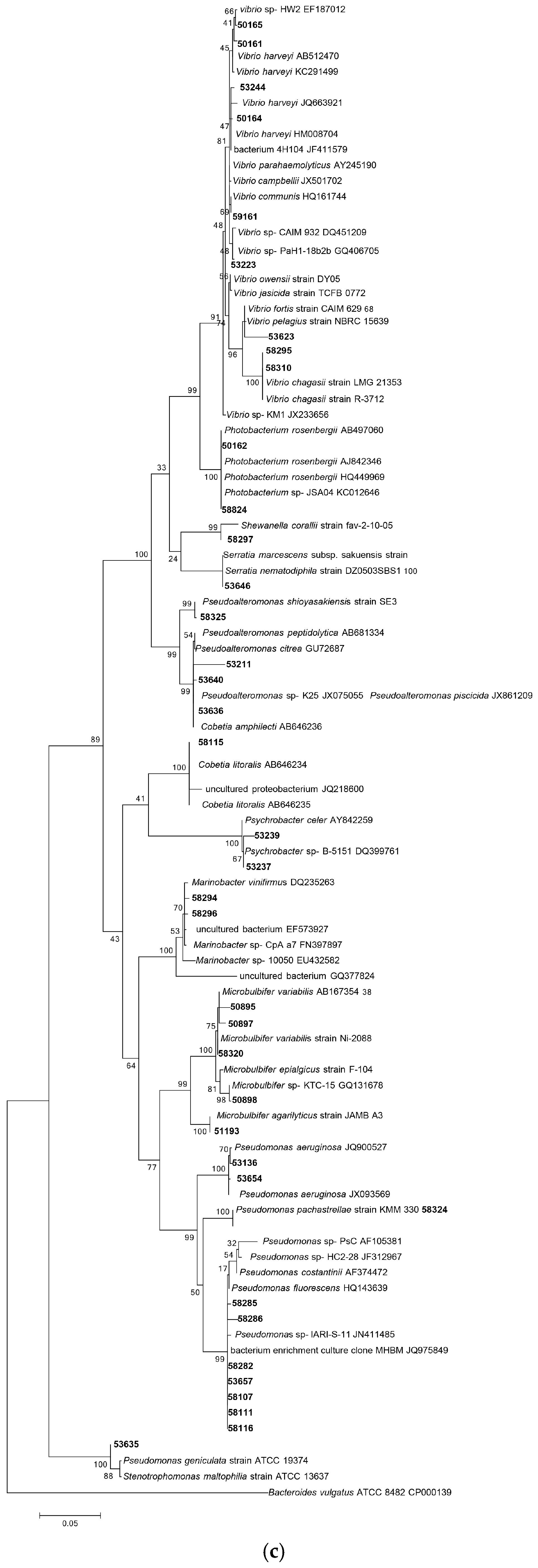
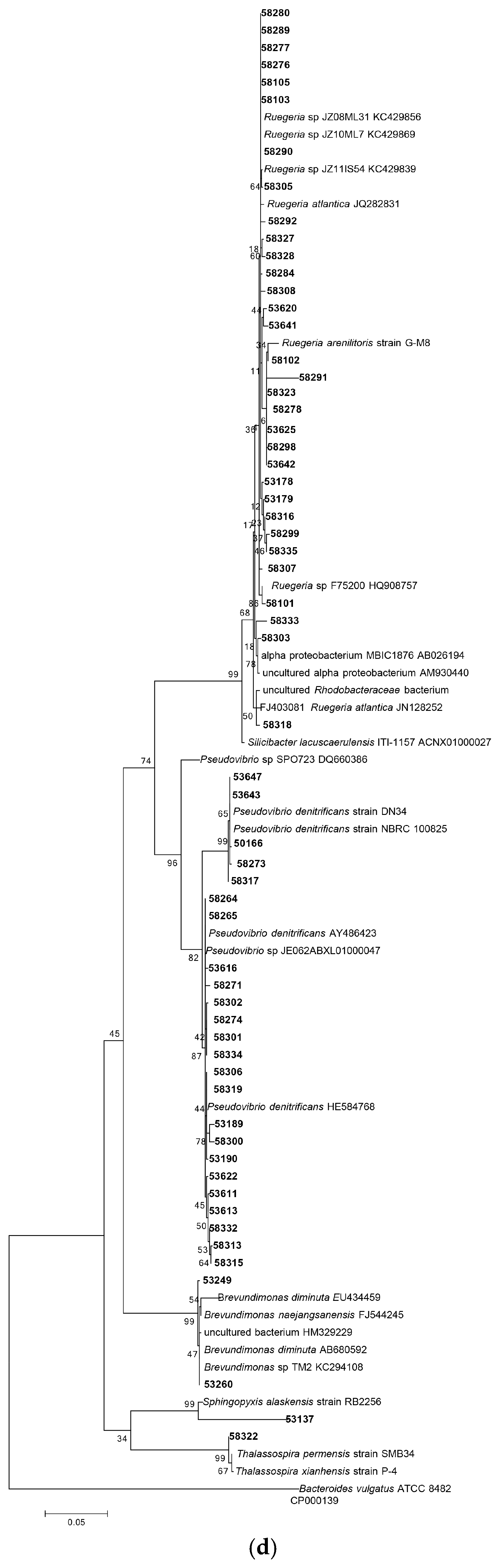
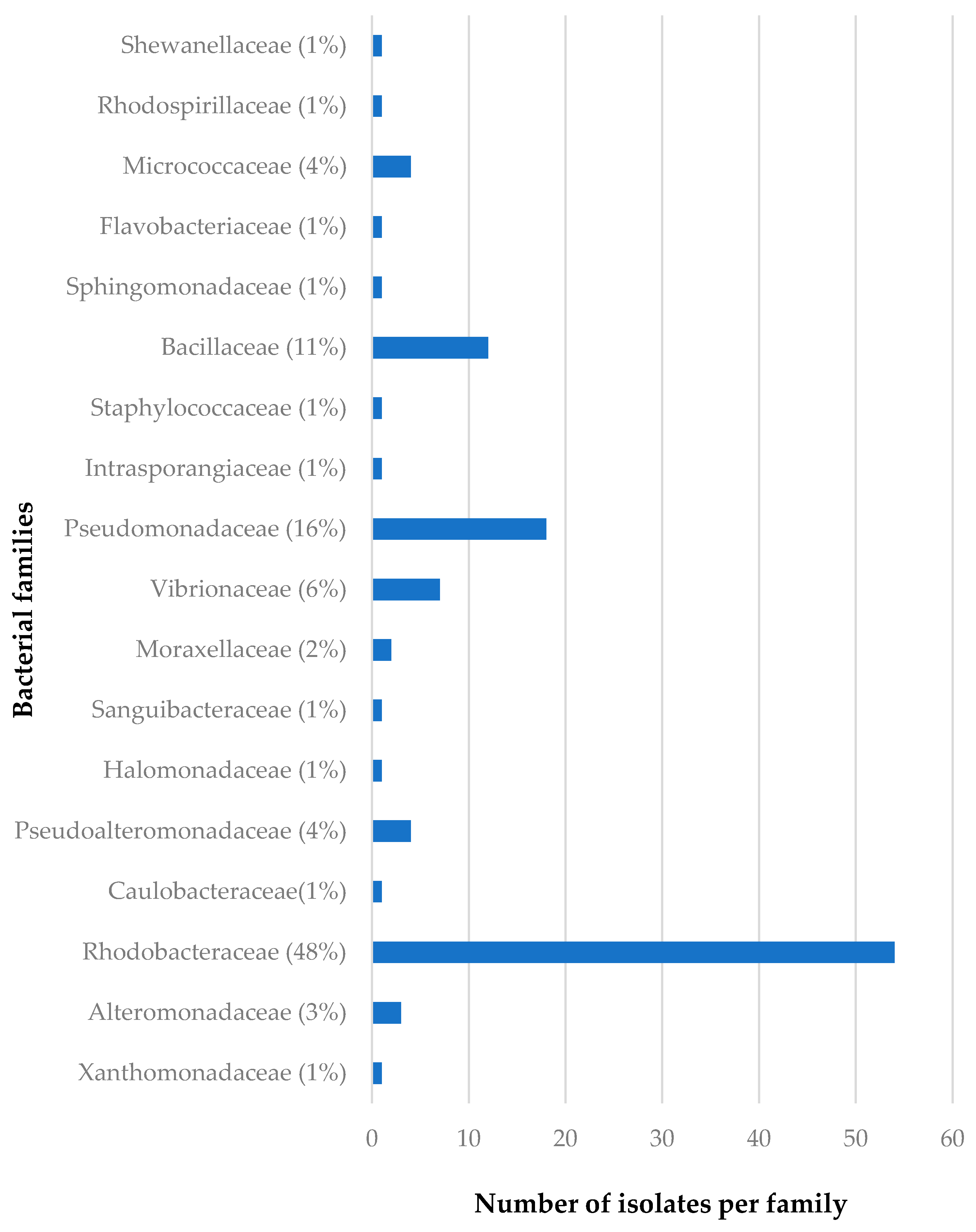
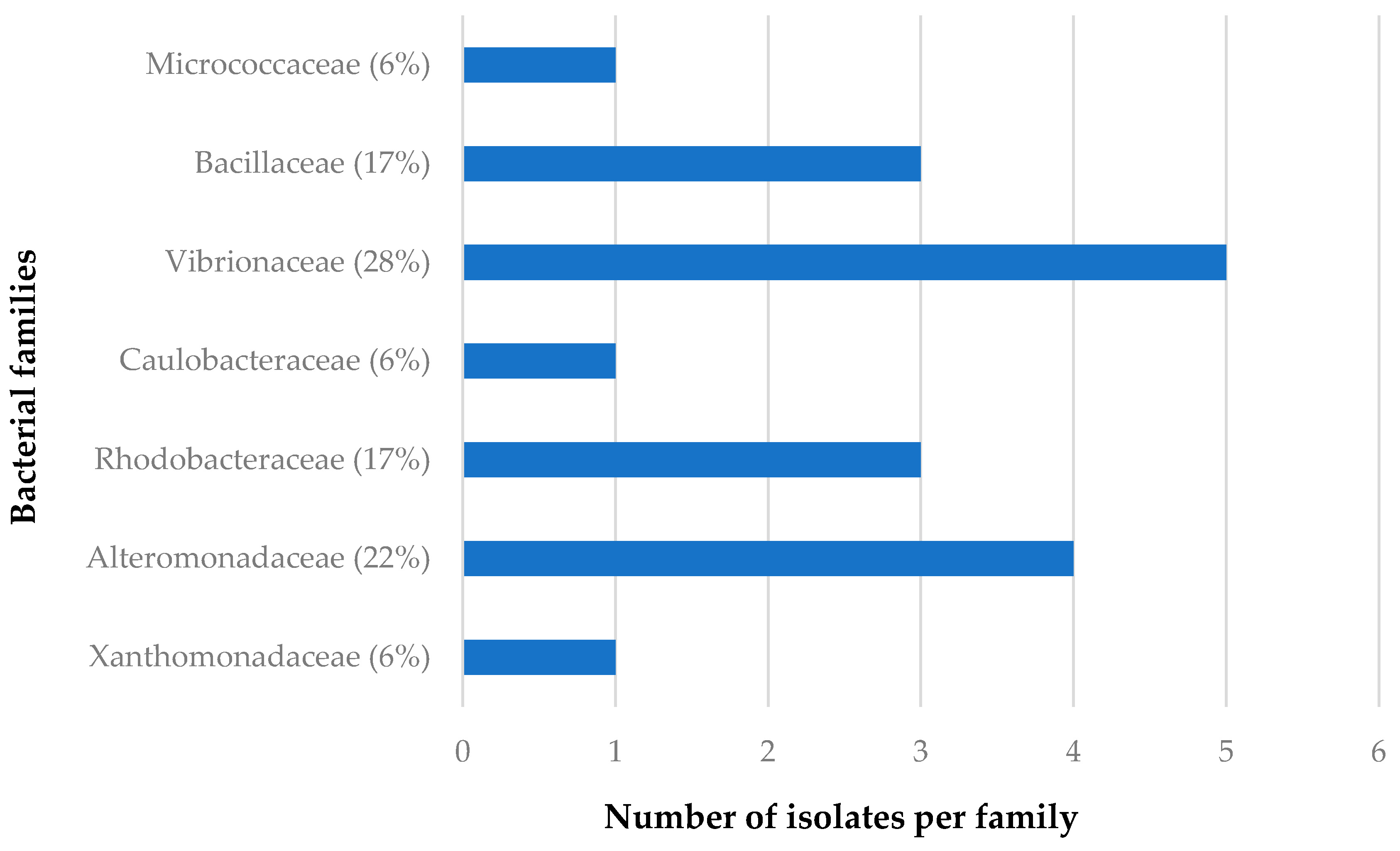
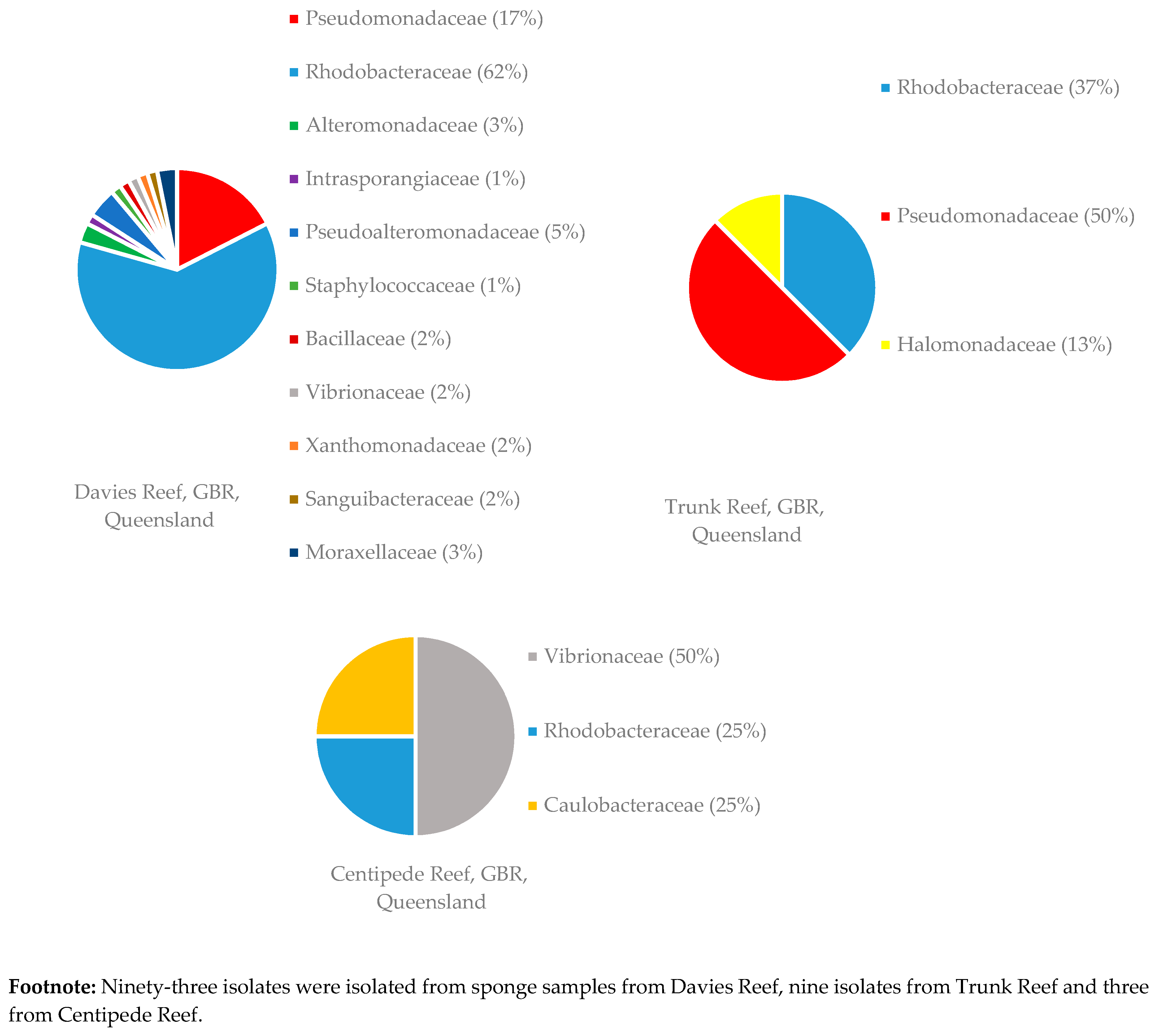

| Isolate Code | Similarity % | Closest Relative Strain | Biological Activity Spectrum | Gene Type |
|---|---|---|---|---|
| 50161 | 99.4 | Vibrio harveyi strain NBRC 15634 | N | ND |
| 50162 | 99.5 | Photobacterium rosenbergii strain CC1 | A. niger (ATCC 16888) | ND |
| 50163 | 99.6 | Stenotrophomonas maltophilia strain ATCC 19861 | N | ND |
| 50164 | 99.7 | Vibrio campbellii strain ATCC 25920 | K. pneumoniae (ATCC BAA-1705) | ND |
| 50165 | 99.4 | Vibrio harveyi strain NBRC 15634 | K. pneumoniae (ATCC BAA-1705) | ND |
| 50166 | 99.0 | Pseudovibrio denitrificans strain NBRC 100825 | K. pneumoniae (ATCC BAA-1705) | PKS-I |
| 50895 | 99.3 | Microbulbifer variabilis strain Ni-2088 | A. niger (ATCC 16888) | ND |
| 50897 | 99.6 | Microbulbifer variabilis strain Ni-2088 | N | ND |
| 50898 | 99.1 | Microbulbifer variabilis strain Ni-2088 | N | ND |
| 50899 | 99.0 | Micrococcus aloeverae strain AE-6 | A. niger (ATCC 16888) | ND |
| 51193 | 99.0 | Microbulbifer agarilyticus strain JAMB A3 | E. coli (ATCC 13706) | PKS-I |
| 53189 | 99.4 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | K. pneumoniae (ATCC BAA-1705) and S. aureus (ATCC 29247) | PKS-I |
| 53190 | 99.8 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | K. pneumoniae (ATCC BAA-1705), E. coli (ATCC 13706) and S. aureus (ATCC 29247) | PKS-I |
| 53223 | 99.6 | Vibrio owensii strain DY05 | E. coli (ATCC BAA-196) | ND |
| 53224 | 99.9 | Bacillus safensis strain NBRC 100820 | E. coli (ATCC BAA-196) | ND |
| 53225 | 99.0 | Bacillus safensis strain NBRC 100820 | N | ND |
| 53226 | 99.0 | Bacillus safensis strain NBRC 100820 | A. niger (ATCC 16888) | ND |
| 53249 | 99.8 | Brevundimonas diminuta strain NBRC 12697 | A. niger (ATCC 16888) | ND |
| Isolate Code | Similarity % | Closest Relative Strain | Biological Activity Spectrum | Gene Type |
|---|---|---|---|---|
| 53136 | 99.7 | Pseudomonas aeruginosa PAO1 strain PAO1 | E. coli (ATCC 13706), S. aureus (ATCC 29247), E. coli (ATCC 25922) | ND |
| 53137 | 92.0 | Sphingopyxis alaskensis strain RB2256 | N | ND |
| 53178 | 99.0 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 53179 | 98.9 | Ruegeria arenilitoris strain G-M8 | A. niger (ATCC 16888) | PKS-I |
| 53650 | 99.0 | Micrococcus yunnanensis strain YIM 65004 | N | ND |
| 53651 | 99.0 | Micrococcus aloeverae strain AE-6 | N | ND |
| 53654 | 99.7 | Pseudomonas aeruginosa PAO1 strain PAO1 | E. coli (ATCC 13706), E. faecalis (ATCC 51575), S. aureus (ATCC 29247), B. subtilis (ATCC 19659), C. albicans (ATCC 10231) | PKS-I |
| 53656 | 99.0 | Micrococcus aloeverae strain AE-6 | N | ND |
| 53657 | 99.6 | Pseudomonas azotoformans strain NBRC 12693 | E. coli (ATCC 13706), S. aureus (ATCC 29247), A. niger (ATCC 16888) | NRPS |
| 58101 | 98.3 | Ruegeria arenilitoris strain G-M8 | K. pneumoniae (ATCC BAA-1705) | PKS-I and NRPS |
| 58102 | 99.0 | Ruegeria arenilitoris strain G-M8 | K. pneumoniae (ATCC BAA-1705) | NRPS |
| 58103 | 99.3 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58104 | 99.9 | Pseudomonas azotoformans strain NBRC 12693 | N | ND |
| 58105 | 99.2 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58107 | 99.8 | Pseudomonas azotoformans strain NBRC 12693 | N | ND |
| 58111 | 99.9 | Pseudomonas azotoformans strain NBRC 12693 | K. pneumoniae (ATCC BAA-1705) | ND |
| 58115 | 100 | Cobetia amphilecti strain 46-2 | K. pneumoniae (ATCC BAA-1705) | PKS-I and NRPS |
| 58116 | 99.9 | Pseudomonas azotoformans strain NBRC 12693 | K. pneumoniae (ATCC BAA-1705), A. niger (ATCC 16888) | PKS-I |
| 58264 | 100 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | K. pneumoniae (ATCC BAA-1705), C. albicans (ATCC 10231), A. niger ATCC 16888) | NRPS |
| 58265 | 99.1 | Pseudomonas azotoformans strain NBRC 12693 | K. pneumoniae (ATCC BAA-1705) | ND |
| 58266 | 99.6 | Pseudomonas azotoformans strain NBRC 12693 | K. pneumoniae (ATCC BAA-1705) | ND |
| 58271 | 99.5 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | A. niger (ATCC 16888) | PKS-I |
| 58273 | 99.0 | Pseudovibrio denitrificans strain NBRC 100825 | K. pneumoniae (ATCC BAA-1705) | ND |
| 58274 | 100 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | N | ND |
| 58275 | 99.0 | Bacillus aquimaris strain TF-12 | A. niger (ATCC 16888) | ND |
| 58276 | 99.4 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58277 | 99.1 | Ruegeria arenilitoris strain G-M8 | A. niger (ATCC 16888) | PKS-I |
| 58278 | 99.0 | Ruegeria arenilitoris strain G-M8 | A. niger (ATCC 16888) | PKS-I |
| 58279 | 99.9 | Pseudomonas azotoformans strain NBRC 12693 | N | ND |
| 58280 | 99.1 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58281 | 99.6 | Pseudomonas azotoformans strain NBRC 12693 | A. niger (ATCC 16888) | PKS-I and NRPS |
| 58282 | 99.8 | Pseudomonas azotoformans strain NBRC 12693 | A. niger (ATCC 16888) | ND |
| 58283 | 99.0 | Bacillus cereus ATCC 14579 | K. pneumoniae (ATCC BAA-1705), A. niger (ATCC 16888) | ND |
| 58285 | 99.6 | Pseudomonas azotoformans strain NBRC 12693 | N | ND |
| 58286 | 99.3 | Pseudomonas azotoformans strain NBRC 12693 | A. niger (ATCC 16888) | NRPS |
| 58287 | 99.0 | Bacillus aquimaris strain TF-12 | K. pneumoniae (ATCC BAA-1705), A. niger (ATCC 16888) | PKS-I |
| 58289 | 99.1 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58290 | 99.2 | Ruegeria arenilitoris strain G-M8 | A. niger (ATCC 16888) | PKS-I |
| 58291 | 96.0 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58292 | 98.9 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58293 | 99.0 | Vibrio owensii strain DY05 | K. pneumoniae (ATCC BAA-1705) | ND |
| 58294 | 99.6 | Marinobacter vinifirmus strain FB1 | N | ND |
| 58295 | 99.0 | Vibrio chagasii strain LMG 21353 | A. niger (ATCC 16888) | ND |
| 58296 | 99.5 | Marinobacter vinifirmus strain FB1 | A. niger (ATCC 16888) | PKS-I |
| 58297 | 98.0 | Shewanella corallii strain fav-2-10-05 | A. niger (ATCC 16888) | ND |
| 58298 | 99.0 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58299 | 98.9 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58300 | 99.5 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | A. niger (ATCC 16888) | ND |
| 53193 | 99.0 | Bacillus horikoshii strain DSM 8719 | A. niger (ATCC 16888) | ND |
| 53208 | 99.6 | Kytococcus sedentarius strain DSM 20547 | K. pneumoniae (ATCC BAA-1705), A. niger (ATCC 16888) | ND |
| 53209 | 99.0 | Kocuria rhizophilia strain TA68 | K. pneumoniae (ATCC BAA-1705), E. coli (ATCC 13706), C. albicans (ATCC 10231) | NRPS |
| 53211 | 98.1 | Pseudoalteromonas piscicida strain NBRC 103038 | N | ND |
| 58301 | 99.7 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | K. pneumoniae (ATCC BAA-1705) | ND |
| 58302 | 99.9 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | N | ND |
| 58303 | 99.2 | Ruegeria atlantica strain NBRC 15792 | N | ND |
| 58304 | 99.0 | Bacillus kochii strain WCC 4582 | S. aureus (ATCC 29247) | ND |
| 58305 | 99.3 | Ruegeria arenilitoris strain G-M8 | A. niger (ATCC 16888) | PKS-I |
| 58306 | 99.8 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | A. niger (ATCC 16888) | ND |
| 58307 | 98.5 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58308 | 99.1 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58310 | 99.0 | Vibrio chagasii strain LMG 21353 | N | ND |
| 58312 | 99.0 | Bacillus kochii strain WCC 4582 | A. niger (ATCC 16888) | ND |
| 58313 | 99.8 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | N | ND |
| 58315 | 99.8 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | N | ND |
| 58316 | 98.8 | Ruegeria arenilitoris strain G-M8 | A. niger (ATCC 16888) | PKS-I |
| 58317 | 99.0 | Pseudovibrio denitrificans strain NBRC 100825 | N | ND |
| 58318 | 98.4 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58319 | 99.8 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | A. niger (ATCC 16888) | ND |
| 58320 | 99.0 | Microbulbifer variabilis strain Ni-2088 | A. niger (ATCC 16888) | ND |
| 58321 | 100 | Staphylococcus warneri SG1 strain SG1 | N | ND |
| 58322 | 99.0 | Thalassospira permensis strain SMB34 | N | ND |
| 58323 | 99.0 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58324 | 99.0 | Pseudomonas pachastrellae strain KMM 330 | N | ND |
| 58325 | 99.0 | Pseudoalteromonas shioyasakiensis strain SE3 | A. niger (ATCC 16888) | PKS-I |
| 58326 | 99.4 | Bacillus nealsonii strain DSM 15077 | N | ND |
| 58327 | 99.1 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58328 | 98.8 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 58330 | 99.0 | Aquimarina spongiae strain A6 | N | ND |
| 58332 | 99.7 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | N | ND |
| 58333 | 98.1 | Ruegeria atlantica strain NBRC 15792 | N | ND |
| 58334 | 99.9 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | A. niger (ATCC 16888) | PKS-I |
| 58335 | 98.7 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 53244 | 99.7 | Vibrio campbellii strain ATCC 25920 | N | ND |
| 53611 | 99.7 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | A. niger (ATCC 16888) | ND |
| 53613 | 99.7 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | N | ND |
| 53614 | 96.0 | Bacillus oceanisediminis strain H2 | N | ND |
| 53616 | 99.6 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | N | ND |
| 53620 | 99.0 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 53622 | 99.7 | Pseudovibrio sp. FO-BEG1 strain FO-BEG1 | N | ND |
| 53623 | 98.0 | Vibrio fortis strain CAIM 629 | A. niger (ATCC 16888) | PKS-I |
| 53624 | 98.8 | Stenotrophomonas maltophilia R551-3 strain R551-3 | A. niger (ATCC 16888) | PKS-I |
| 53625 | 99.0 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 53634 | 98.0 | Sanguibacter inulinus strain ST50 | A. niger (ATCC 16888), C. albicans (ATCC 10231) | NRPS |
| 53635 | 98.0 | Pseudomonas geniculate strain ATCC 19374 | N | ND |
| 53636 | 100 | Pseudoalteromonas piscicida strain NBRC 103038 | A. niger (ATCC 16888) | ND |
| 53640 | 99.9 | Pseudoalteromonas piscicida strain NBRC 103038 | N | ND |
| 53641 | 98.9 | Ruegeria arenilitoris strain G-M8 | N | ND |
| 53240 | 99.0 | Bacillus safensis strain NBRC 100820 | N | ND |
| 53237 | 99.3 | Psychrobacter celer strain SW-238 | N | ND |
| 53239 | 98.9 | Psychrobacter celer strain SW-238 | N | ND |
| 59159 | 99.0 | Bacillus kochii strain WCC 4582 | A. niger (ATCC 16888) | ND |
| 59161 | 99.7 | Vibrio alginolyticus strain ATCC 17749 | N | ND |
| 58824 | 99.6 | Photobacterium rosenbergii strain CC1 | N | ND |
| 58284 | 99.2 | Ruegeria arenilitoris strain G-M8 | A. niger (ATCC 16888) | ND |
| 53260 | 99.9 | Brevundimonas diminuta strain NBRC 12697 | N | ND |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brinkmann, C.M.; Kearns, P.S.; Evans-Illidge, E.; Kurtbӧke, D.İ. Diversity and Bioactivity of Marine Bacteria Associated with the Sponges Candidaspongia flabellata and Rhopaloeides odorabile from the Great Barrier Reef in Australia. Diversity 2017, 9, 39. https://doi.org/10.3390/d9030039
Brinkmann CM, Kearns PS, Evans-Illidge E, Kurtbӧke Dİ. Diversity and Bioactivity of Marine Bacteria Associated with the Sponges Candidaspongia flabellata and Rhopaloeides odorabile from the Great Barrier Reef in Australia. Diversity. 2017; 9(3):39. https://doi.org/10.3390/d9030039
Chicago/Turabian StyleBrinkmann, Candice M., Philip S. Kearns, Elizabeth Evans-Illidge, and D. İpek Kurtbӧke. 2017. "Diversity and Bioactivity of Marine Bacteria Associated with the Sponges Candidaspongia flabellata and Rhopaloeides odorabile from the Great Barrier Reef in Australia" Diversity 9, no. 3: 39. https://doi.org/10.3390/d9030039
APA StyleBrinkmann, C. M., Kearns, P. S., Evans-Illidge, E., & Kurtbӧke, D. İ. (2017). Diversity and Bioactivity of Marine Bacteria Associated with the Sponges Candidaspongia flabellata and Rhopaloeides odorabile from the Great Barrier Reef in Australia. Diversity, 9(3), 39. https://doi.org/10.3390/d9030039






