Abstract
DNA barcoding is a molecular technology that allows the identification of any biological species by amplifying, sequencing and querying the information from genic and/or intergenic standardized target regions belonging to the extranuclear genomes. Although these sequences represent a small fraction of the total DNA of a cell, both chloroplast and mitochondrial barcodes chosen for identifying plant and animal species, respectively, have shown sufficient nucleotide diversity to assess the taxonomic identity of the vast majority of organisms used in agriculture. Consequently, cpDNA and mtDNA barcoding protocols are being used more and more in the food industry and food supply chains for food labeling, not only to support food safety but also to uncover food piracy in freshly commercialized and technologically processed products. Since the extranuclear genomes are present in many copies within each cell, this technology is being more easily exploited to recover information even in degraded samples or transformed materials deriving from crop varieties and livestock species. The strong standardization that characterizes protocols used worldwide for DNA barcoding makes this technology particularly suitable for routine analyses required by agencies to safeguard food safety and quality. Here we conduct a critical review of the potentials of DNA barcoding for food labeling along with the main findings in the area of food piracy, with particular reference to agrifood and livestock foodstuffs.
1. Introduction
DNA barcoding is a taxonomic method that uses a short genetic marker from a standard part of the genome of an organism’s DNA to identify it as belonging to a particular individual, breed/cultivar, or species. It represents an essential tool to vouch for quality controls of food products, to guarantee food traceability, to safeguard public health, to minimize food piracy, and to valorize local and typical agro-food production systems.
In general, DNA barcoding is based on the amplification of short DNA fragments belonging to the mitochondrial (animal foodstuffs) or chloroplast (plant foodstuffs) genomes, which are conserved at the species levels and preserved in most of the processed food products, then showing these advantages as compared to other DNA fingerprinting and genotyping approaches. DNA barcoding can be used on both fresh and raw materials for species authentication, species delimitation, and identification from different individual parts where other methods of characterization usually fail. Moreover, DNA barcoding can be applied to distinct food products and matrices deriving from single or mixed species, producing species-specific DNA sequences (i.e., barcodes). In this way this methodology could be used to discover voluntary or accidental replacements associated with foods mislabeling and commercial frauds.
Often a risk factor in identifying food mislabeling or piracy is the lack of adequate standards, and the availability of high quality repositories of reference sequences can be a critical point, so the international platform BOLD (Barcode of Life Database) supporting the collection of DNA barcodes represents a robust and accurate reference library for all living species.
New trends in agricultural practices and the recent sanitary emergencies have made producers and consumers more demanding about food authenticity, with a concurrently increasing interest for food origin, healthiness and nutraceutical properties. Modern technologies based on genomics and bioinformatics represent very efficient tools for assessing the genetic authenticity and genetic traceability of food products and beverages. DNA barcoding could eventually deal with the development or implementation of diagnostic molecular assays suitable for the genetic identification of plant foodstuffs, fresh and processed meats, and fishery derivatives by means of DNA barcoding methods. Therefore, DNA barcoding is a very robust diagnostic tool due to a number of features, including the evidence that DNA is inalterable, detectable in every cell, and resistant to heat treatments, and allows for individual, breed/cultivar or species identification. Even if results are promising, these techniques are still too expensive to be converted in routine tests, but they could be reliable molecular tools for quality controls, identity checks and discovery of fraud, and hence to prevent food piracy.
Three European Union schemes of geographical indications and traditional specialties, known as protected designation of origin (PDO), protected geographical indication (PGI), and traditional specialties guaranteed (TSG), promote and protect names of quality agricultural products and foodstuffs. Within this policy framework, food piracy can be defined as the counterfeiting of a specific food product, i.e., the practice of manufacturing, importing/exporting, distributing, selling or otherwise dealing with a food product of inferior quality and/or voluntary imitation, under a trademark that is nearly identical to or substantially indistinguishable from a registered trademark or a protected label, without any approval or oversight of the owner or local authority. Ultimately, food piracy involves a total lack of transparency concerning raw materials and the manufacturing process employed by producers, and hence does not guarantee a correct choice of the consumers and does promote speculation on profits by traders.
The phenomenon of food piracy is a well-known form of fraud in the world international trade of counterfeit and pirated products and its value is around USD 200 billion (for details, see annual report of OECD, the Organisation for Economic Co-operation and Development, 2007 [1]). This amount is larger than the national GDPs of about 150 economies of the world. The figure does not, however, include counterfeit and pirated products that are produced and consumed domestically, nor does it include non-tangible pirated digital products being distributed via the Internet. Therefore, if these items are added, the total magnitude of counterfeiting and piracy worldwide could well be several hundred billion dollars more. For these reasons food piracy—the real counterfeiting—is an illicit act punishable by law. Indeed, European food legislation is particularly strict and traceability systems, based on product labeling, have become mandatory in all European countries. However, the implementation of this system does not protect consumers against any fraud. Although barcodes are sometimes used in an effort to identify unknown species or to verify whether species should be combined or separated, the utility of DNA barcoding for these purposes is subject to debate. Moreover, the adoption of DNA barcoding would be an interesting way to check and control a number of important food chains.
The aim of the present review is to summarize the major recent advances made in the use of DNA barcoding for genetic traceability, taxonomic identification of food species and genetic authentication of food products, focusing on advantages and disadvantages of this molecular technology, and on its real and potential applications for livestock and crop plant sources in the area of commercial foodstuffs and beverages. Selected case studies related to the use of chloroplast and mitochondrial DNA barcoding to track down mislabeling and food piracy in a number of foodstuffs of plant and animal origin are critically reviewed.
Rationale on DNA Barcoding as a Molecular Assay for Species Identification
DNA barcoding involves sequencing short segments of the chloroplast or mitochondrial genome and comparing the results with orthologous reference sequences available in public database such as BOLD (www.boldsystem.org) and GenBank (www.ncbi.nlm.nih.gov/genbank). This approach is based on the analysis of the nucleotide variability existing within standard regions of the genome that are particularly informative for the identification of species. The core assumption is that DNA sequences are likely more similar to one another within species than between species as variation in the nucleotide composition is lower within one species than between different ones [2]. The system aims to detect polymorphisms of a nucleotide snippet, from ideally all organisms of the animal and plant kingdoms, so the DNA barcode sequence contains enough unique information, in terms of SNPs (single nucleotide polymorphisms) and In/Dels (insertion/deletions), shared among individuals of a species with slight variations, but specifically associated to one species with a unique haplotype. For assessing species identity and genetic traceability purposes, the DNA barcoding gap [3], intended as the difference between intra- and inter-specific divergence values, is a necessary condition because the accuracy of the method depends especially on the extent of, and separation between, intraspecific variation and interspecific differentiation of the selected markers.
The main properties of DNA barcoding include the possibility of easily associating all life cycle stages and genders, especially when the morphology, living behavior and habitat are consistently different, to identify any organism from parts or pieces and also to discriminate single species co-existing within complex matrixes containing a mixture of species.
In the last decade, DNA barcoding was proposed as a universal DNA-based tool for species identification and for authentication of processed food products. The revolution introduced by DNA barcoding is not only in the discrimination power itself, but also resides in the conjugation of distinct innovations, including the molecularization, computerization and standardization of the identification approach. Advantages of the use of DNA barcoding includes its features of being reliable and testable as long as the link between barcode sequences and reference specimens is supported, and it is also reproducible at any time and by any researcher. The experimental procedure of extracting genomic DNA and amplifying specific DNA markers is technically easy and usually does not require the destruction of the sample, which sometime needs to be safeguarded for further uses or inspections. Moreover, it allows the treatment of all kinds of biological specimens, including those non-identifiable by morphology, and it also is very fast and relatively inexpensive compared with other molecular approaches.
Mitochondrial DNA (mtDNA) was chosen as source of markers for barcoding animal species because of its features; unlike nuclear genes, mitochondrial genes belong to a haploid genome, show high copy number, lack introns, exhibit low recombination and are maternally inherited. Of the protein-coding genes present on the animal mitochondrial genome, the use of cytochrome c oxidase subunit 1 (cox1 or COI) was proposed as a standard barcode marker for animal species for two main reasons: universal primers make a 648 bp fragment at the 5′ end of this gene easy to amplify in a broad spectrum of phyla, and its nucleotide substitution rate allows not only closely related species to be distinguished but, in some taxa, also different populations, biotypes or races of the same species. Other mitochondrial genes were suggested as barcode markers, including cob, which encodes for apocytochrome b; cox2 and cox3, which encode for the cytochrome oxidase subunits 2 and 3, respectively; nad1, which encodes for NADH dehydrogenase subunit 1; and the mitochondrial 16S-rDNA gene ([4,5,6] and references therein).
Concerning chloroplast markers, establishing a standardized DNA barcoding system in plants was more challenging [7]. The proposed barcodes involved distinct combinations of seven plastid markers both from coding and non-coding regions: rpoC1 + rpoB + matK [8]; rpoC1 + matK + trnH–psbA [8]; rbcL + trnH–psbA [9]; and atpF–H + psbKI + matK [8]. The CBOL Plant Working group recommends a core barcode consisting of portions of two plastid coding regions, rbcL + matK, to be supplemented with additional markers as required. Considering, as a reference, the Arabidopsis thaliana plastid genome, the rbcL barcode consists of a 599 bp region at the 5′ end of the ribulose-bisphosphate carboxylase gene, located between positions 1–599, whereas the matK barcode includes a 841 bp region at the center of the maturase gene, located between positions 205–1046 [10]. The choice of rbcL + matK as a core barcode was based on the straightforward recovery of the rbcL region and the discriminatory power of the matK region. In fact, it is well known that a two-marker plastid barcode system enables better discrimination than single barcode markers and no other multi-marker plastid barcode permits appreciably greater species resolution than the rbcL + matK combination (CBOL Plant Working Group, 2009 [11]). In particular, matK is one of the most rapidly evolving coding sections of the plastid genome [12] and it is perhaps the closest plant analogue to the cox1 animal barcode. Unfortunately, matK can be difficult to amplify using existing primer sets, particularly in non-angiosperms. In contrast, the barcode region of rbcL is easy to amplify, sequence and align in most land plants and provides a useful backbone to the barcode dataset, despite having only modest discriminatory power [11]. Despite their high universality in terms of PCR amplification and DNA amplicon sequencing success, the analysis of these coding regions often fails due to the interspecific sharing of sequences. However, the combination of matK and rbcL with the plastid intergenic spacer region trnH−psbA increases the identification performance of DNA barcoding. This region is straightforward to amplify among land plants and also shows high variability across intergenic spacers in plants, even among closely related taxa [13,14].
2. The European Legislation on Food Safety against Food Piracy
The European Union (EU) has always paid great attention to food safety, first of all because the agro-alimentary sector on its whole is very important for European economy. The EU is the biggest producer of foodstuffs and beverages in the world with a food- and beverage-related industries production of 16% of the total EU manufacturing turnover, corresponding to more than 950 billion Euros [15].
Adulteration of foods for financial gain is prohibited. It consists in omitting any valuable constituent, substituting another component wholly or partly, or adding any substance to increase weight or bulk, worsen quality or make a product appear better than it is. Most adulterants are benign, but less expensive than some constituents of the food, and the counterfeiting products are actually food frauds, economically motivated misbranding and mislabeling, fakes based on simulation processes and imitation products. When the adulterants are toxic or allergenic, serious public health consequences may result. In this case, the food mislabeling not only robs consumers of value, but it may also endanger people who have intolerance or allergies to certain foods or their components. The most frequent incidents, based on the literature from 1980, were grouped into 11 food categories: fish and seafood, dairy products, fruit juices, oils and fats, grain products, honey and other natural sweeteners, spices and extracts, wine and other alcoholic beverages, infant formula, plant-based proteins, and other food products [16]. For processed food where the morphological characteristics of the species are removed, there is a need for inexpensive and widely available genetic testing methods.
The three most important EU documents regarding food safety are the Green Paper on the general principles of food law in the European Union (1997), the White Paper on Food Safety (2000) and the ER 178/2002, applied since the 1st of January 2005 [17,18,19,20,21].
The Green Paper on the principles of EU legislation about food products [17] is made up of six parts regarding different aspects of food safety, such as the actual legislation of member countries, the need to simplify EU legislation and, above all, the need to implement it to better protect consumers’ health, a must after the BSE outbreaks.
The White Paper on Food Safety [19], following few years later, contains strategies for updating the actual legislation, including the institution of an independent European Alimentary Authority, the risk analysis as main instrument for food safety, the application of the precaution principle, the need for controls on food products and consumer information. In addition, for the first time, it introduced the concept of traceability for feed and animal products “from farm to fork” and transparency was the leitmotiv of the entire document.
The White Paper was the basis for ER 178/2002 applied from 1 January 2005. This regulation stressed the importance of a traceability system, declaring that “the experience demonstrated how the impossibility to reconstruct the trail of a food could be a danger for the market of such product” while, a traceability system able to keep all the information regarding food production can help to proceed to its recall in case of danger without damaging the entire sector. Therefore, since 2005, the regulation has become mandatory for all member countries which must define a traceability system for the whole food sector; in addition, it permits agreement among the different member countries’ legislations in which several differences were present, leading to problems regarding the free exchange of food among them. Though ER 178/2002 is the fundamental law regarding food safety, it has been followed by several other regulations; for animal products the most important are 852/2004, 853/2004, 854/2004 and 882/2004, and all of them corroborate the importance of a traceability system and the need for authorities to control them. Not only the EU has such a strict legislation on food products traceability; indeed traceability systems based on animal identification have been implemented in several countries. In Canada, Australia and New Zealand a trace back system based on tagging was established in 2001, in Japan strict rules were established in the same year, and in Brazil and Argentina traceability systems are in use though with different depth. In the United States (USA) a trace back system was proposed even if it would not be mandatory or providing comprehensive information but still on a voluntary basis [22,23]. Moreover, in the last few years the discussion on the identification and registration of genetically modified organisms (GMOs), between the EU and the USA, contributed to an increase of the traceability requirements and transparency in food chains. Labeling of GMOs is obligatory in the USA only if the product differs essentially from the “original”, e.g., if the nutritional value differs, or if the product contains an allergen that it is not present in the original. The EU demands that all GMO-derived products, with a transgenic DNA content of 0.9% (0.1% for biological products), must be labeled as such.
3. DNA Barcoding and the Food Production Chains
In the last 15 years, researchers have done many studies on DNA barcoding reaching more than 600 scientific publications per year in the database of the ISI Web of Science. Figure 1 shows that DNA barcoding became a keyword in the scientific literature since 2000, hence it must be considered a very recent topic in the research field around the world. Applications of DNA barcoding to food safety and food piracy issues have grown in importance due to the consumers’ increasing attention to food authenticity and food safety in different products and species [24,25,26,27]. This interest seems to be due to the consumers’ lack of confidence, in particular towards food of animal origin, and it is attributable to several reasons including both food safety and socio-economic changes. Bovine spongiform encephalopathy (BSE) has certainly been the most serious food safety problem of recent years, causing a drastic reduction of beef consumption in all Europe. It was then followed by the dioxin crisis and the avian influenza in the poultry sector [28,29]. Furthermore, the incidence of food borne diseases due to microbial contamination of processed food products has increased in the last decade leading to additional food scares in the buyers [30].
Besides these “food scandals”, socio-economical reasons have also contributed to an increase in people’s interest in what they eat and in how and where it is produced. For example, it is worth mentioning that the main reasons for the negative trend of meat consumption are not only due to the negative impact of the food scandals involving meat products, but also to the new food habits of the younger generation and the progressive decline of the organoleptic meat properties [31]. At present, consumers are more aware than they were years ago of ecological and environmental matters and the demand for organic food and for products obtained in an eco-sustainable system has increased [30]; nevertheless the industrialization processes, as well as market globalization, have made it difficult for people to check on food processing methods [32].
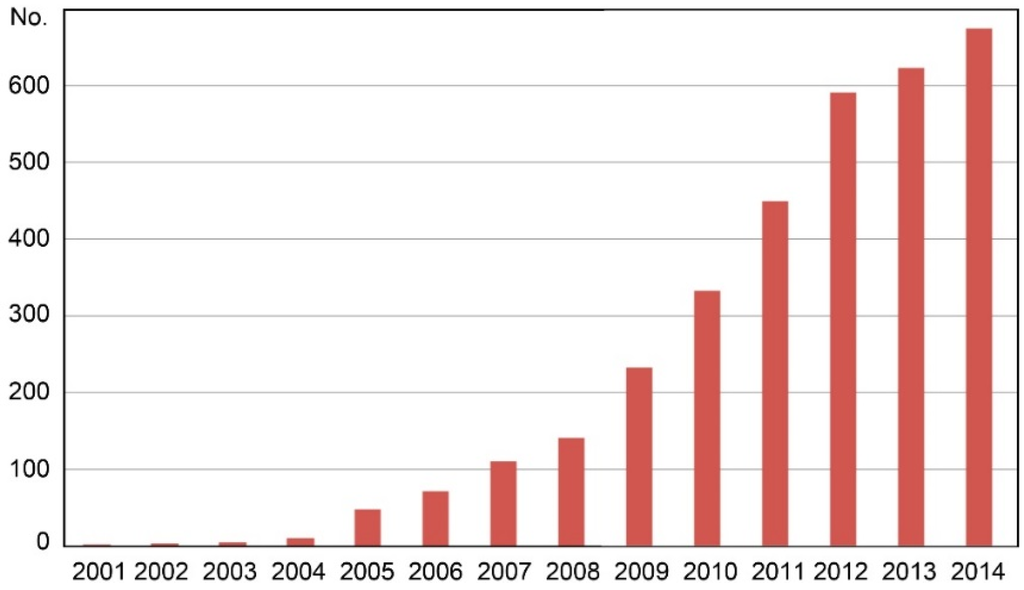
Figure 1.
Number of scientific papers found in the database of ISI Web of Science, arranged per year of publication from 2001 to 2014, using the keyword “DNA barcoding” (verified on 30 November 2015).
All these reasons have contributed to the need to find a system to trace food products. DNA barcoding might be the answer to the consumers’ demand of transparency and it is becoming synonymous with safe and high quality food. Authorities and scientists are still debating on how the perfect trace back system should work and several authors have compared, in the past, the efficacy of different traceability methods [22,33,34,35]. On the whole, a good system should be convenient, easy to use and read, durable, respecting animal and public health, and able to avoid fraud. Several determination methods have been studied, including different kinds of identification tags. At present, policymakers have implemented mandatory methods based on tags or labels, which are methods easy to use although they cannot prevent from fraud [33,34]. Debates on food safety issues and on traceability matters involve not only policymakers and scientists but also economists, as implementation of traceability systems is strongly related to cost. The realization of any kind of traceability system results in costs and benefits for both industries and consumers. In particular, on one hand, for food companies it is a tool to counterattack liability claims and to improve recall efficacy but, on the other, consumers’ willingness to pay for this service still needs to be studied.
DNA barcoding issues concern many different aspects tied not only to food safety and policymaker decisions, but also economical aspects and consumers’ decision-making behavior, so the implementation of such systems must cover all these aspects.
4. Use of DNA Barcoding and Genetic Traceability of Animal Species in Foodstuffs
DNA barcoding can be considered a molecular tool for implementation of genetic traceability that is based on the identification of both animal species and their food products through the study of extra-nuclear DNA regions. It exploits the polymorphism information content of specific DNA barcodes known to be highly variable among distinct species, inalterable during all life cycle in any animals, stable to the different treatments of processed food products, and present in every cell of all organisms.
Once the genomic DNA is extracted from the chosen matrix (e.g., animal tissue, blood, muscle, hair, sperm, faeces or even a processed food such as cheese or canned and minced meat) it is analyzed by single-locus PCR amplifications followed by direct sequencing of targeted DNA amplicons in order to obtain barcodes allowing for species identification and, in some cases, even breed discrimination. The use of this technology for the genetic traceability of animal species and their food derivatives is just an extension of the technology already in use for taxonomic and phylogenetic caseworks.
A standardized genetic traceability system applicable to species identification, in animals and their meat products, is crucial for consumers because of social, religious, health and economic implications. Nowadays animal carcasses and whole fish are rarely displayed while either fresh or frozen cuts, processed and ready to eat food are increasingly available, making species identification very difficult. For this reason, fraudulent adulteration could take place, substituting the declared meat or fish species with others of lower commercial value. Such falsification is actually very common in game meat products, resulting in a great profit due to the higher prices of these species compared to beef or pork ones. The fish industry also is involved in species counterfeits, especially in the case of canned fish such as tuna whose genus is made up of many different species characterized by different quality [36]. Moreover, the dairy sector is subject of fraud regarding milk and cheese products, concerning the species of origin; in fact the greater availability and the lower cost of cow milk rather than goat, ewe or buffalo milk lead to fraudulent substitutions in cheese manufacturing [37,38]. To understand the importance of these frauds it is worth mentioning than in Italy the addition of undeclared bovine milk to water buffalo milk for making cheese is the most frequent fraud reported by the Central Inspectorate for Repression of Frauds of the Italian Ministry of Agricultural and Forestry Policy for all foodstuffs of animal origin. In the past years, approximately 13% of cheeses tested were proven to contain undeclared non-water buffalo milk [39].
As a case study of genetic traceability, we applied mtDNA barcoding to identify the species forming a ready-to-eat coocked cod, the also-called Italian baccalà, which is largely commercialized and consumed in the Veneto region. The analysis of nucleotide sequences of different length belonging to the mitochondrial genes cox1 and cob, which encode for the cytochrome oxidase subunit I and for the apocytochrome b, respectively, allowed us to detect the Atlantic cod (Gadus morhua) along with the Argentine hake (Merluccius hubbsi) as main components (Figure 2). These two species are known to have a quite different distribution; in fact, the former is widespread from the Northwestern to the Northeastern Atlantic Ocean, whereas the latter is mainly found in the Southwestern Atlantic Ocean.
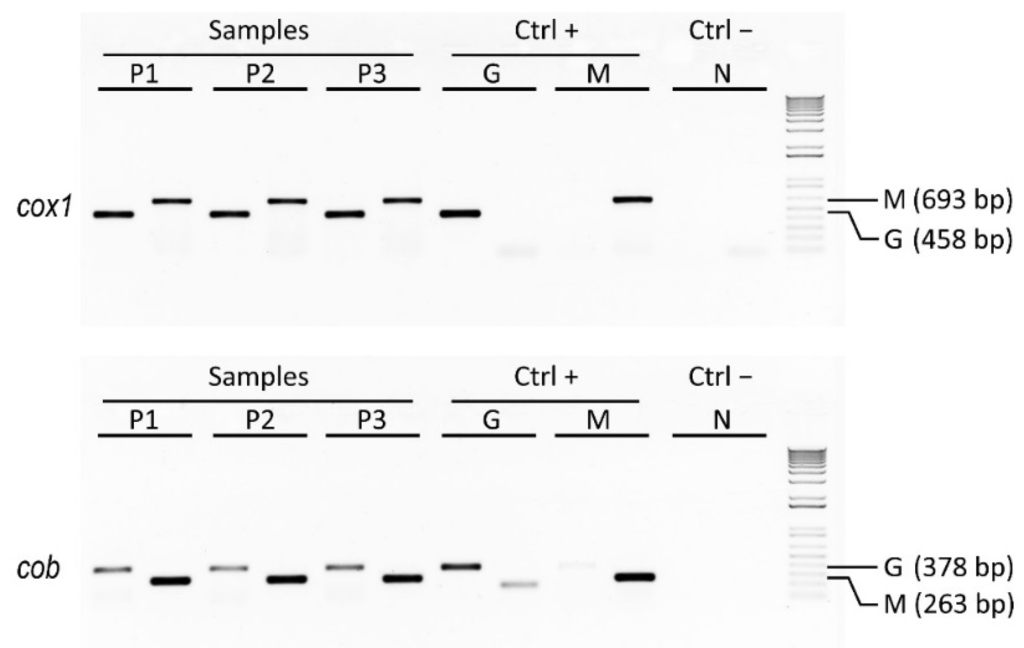
Figure 2.
Results of agarose gel electrophoresis analysis showing PCR amplicons related to three distinct samples of Italian baccalà commercial products along with their positive controls represented by pure Atlantic cod and Argentine hake species (DNA amplicons of 693 bp and 458 bp were generated using Gadus-specific, G, and Merluccius-specific, M, primers for cox1 and cob discriminant barcodes, respectively). The NCBI accessions cox1 and cob genes with 100% identity were KC015385.1 and EU492303.1 for Gadus morhus and EU074469.1 and EF362890.1 for Merluccius hubbsi, respectively (for protocol details, see Nicolè et al. [5]).
In this way we developed a two-locus approach to analyze in biological replicates discriminant barcodes of cox1 and cob genes by assaying Gadus-specific and Merluccius-specific sequences, and comparing them to reference sequences in the BOLD and GenBank online databases. This method proved to be a rapid and robust diagnostic assay that can be exploited to genetically identify different forms of cod and/or hake, and that can find utility for species authentication in order to detect mislabeling of commercially processed seafood, such as Italian baccalà.
In conclusion, most of the studies conducted on commercial samples of animal origin collected in supermarkets or butcheries uncovered several cases of counterfeits and contaminations, suggesting that controls should be more strict and analytical in order to prevent frauds and protect consumers against frauds.
5. Use of DNA Barcoding and Genetic Traceability of Plant-derived Food Products
DNA barcoding is a molecular technology suitable to greatly improve the traceability of any food from breeders or growers to dining tables. Nowadays, this approach is becaming largely exploited for genetic identification and adulteration detection in several plants species as well as their food products, also combined with high resolution melting (HRM) analysis.
DNA barcoding showed a high effectiveness in the checking of fruit-based processed products, particularly yogurts and juices. For instance, fruit juices labeled as 100% fruit represent approximately two-thirds or 10 billion liters of total EU juice sales in 2013 (European Fruit Juice Association (AIJN), 2014 [40]). This kind of juice has usually premium prices as they are more and more connected with wealthy lifestyles representing favored targets for adulteration. The adulteration of fruit juices often consists in the addition of a cheaper fruit juice to increase the production profit. DNA barcoding exploits universal well-described primers on the authentication of fresh fruit juices of the most commonly used fruit species. The use of universal primers targeting specific chloroplast genome regions is of paramount importance in the detection of unexpected adulterants. The detection of the admixture of a different fruit could be easily detected although further studies should be performed in order to evaluate the method sensitivity. Recently, it has been demonstrated the efficacy of DNA barcoding with trnL as a target sequence combined with HRM analysis for the complete fruit species discrimination (i.e., orange, mango, peach, pear and pineapple) and their quantitative evaluation in fruit juices [41].
A further application of DNA barcoding technology concerns the detection of allergenic contaminants. A similar method, described as technically rapid and highly sensitive, was recently exploited to detect the major tree nut species known for triggering adverse immune responses to individuals and quantitatively identify their allergenic components in processed foods [42]. Allergy to hazelnut is very frequent to humans with respiratory allergy to the pollen of birch, hazel and alder, causing breathlessness, so the use of hazelnuts, also in small quantities, in food industry (e.g., ice creams and biscuits) or an unintended contamination through food processing could pose a risk to consumers. The proposed method was again based on the use of universal chloroplast primers for trnL marker amplification coupled with HRM analysis for DNA barcoding tree nut species and for quantification of their allergenic components in commercial foods. Such an approach was shown to be able not only to distinguish among the different nut species, but also to reveal a ratio of 0.01% of hazelnut contamination.
DNA barcoding coupled with High Resolution Melting (Bar-HRM) analysis was exploited for genetic traceability and adulteration discovery of “Fava Santorinis” (Lathirus clymenum), a typical legume of the Mediterranean diet recognized as PDO (Protected Designation of Origin) product for its high nutritional value [43]. The four times higher cost in comparison with regular “Fava” coming from favabeans (Vicia faba) or from peas (Pisum sativum) makes this product susceptible to adulterations. The approach was based on DNA barcoding regions (trnL and rpoC) combined with HRM analysis and was proven capable not only of distinguishing among different L. clymenum legume relatives but in addition to detect a ratio of 1:100 of non-Fava Santorinis in Fava Santorinis commercial products.
A research team aimed to define a molecular system for the traceability of commercial food flavouring species based on a DNA barcoding tool [44]. Spices are a numerous group of plant species characterized by aromatic oils and secondary metabolites commonly used as flavor for cooking, essences for cosmetics, and active components in medicines. Authors investigated six major genera of cooking spices (i.e., Mentha, Ocimum, Origanum, Salvia, Thymus and Rosmarinus) belonging to the Lamiaceae family by collecting samples at different stages of the industrial supply chain and using four barcode markers (rpoB, matK, rbcL and trnH–psbA). The first result they obtained was that in almost all the cases considered, the industrial processes conducted on the commercial spices samples (e.g., crumbling, drying) did not affect the success rate of DNA extraction, amplification and sequencing therefore allowing analysis through a DNA barcoding approach. The two core-barcode markers matK and rbcL assigned species to the correct genus, but did not always reach the taxonomic classification at the species level. The non-coding trnH–psbA intergenic spacer produced species and cultivar exclusive haplotypes providing a reliable system for their identification in all six analyzed group. The building of a dedicated aromatic plants database in which all species and cultivars are described, both at morphological and molecular levels, was strongly advised to better define a more practical and standardized for spices traceability.
Olive (Olea europaea) oil is a food product particularly prone to fraudulent practices, since it commands a higher price than other vegetable oils. Olive oil is known to be one of the best vegetable fats in human diet and in recent years there has been increasing of interest in this kind of product due to its nutritional benefits and metabolic proprieties. The organoleptic characteristics of olive oil associated with its valuable effects on human health have resulted in an increasing demand in recent years, leading to frequent frauds through the mixing of lower-cost oils of plant origin, such as soya (Glycine max), canola (Brassica napus), maize (Zea mays), sunflower (Helianthus annuus) and even sesame (Sesamum indicum) oils, with olive oil. The adulteration of high-priced olive oil with low cost oils and the fraudulent labeling of oil products make the genetic traceability of vegetable oil species in the food chain very important [45].
Typical olive oil frauds include blending with lower value vegetable oil, fraudulent mislabelling on geographical origin and cultivar. The standard analysis methods for olive oil are mainly based on spectroscopy, like NMR (Nuclear Magnetic Resonance) and NIRS (Near Infra Red Spectroscopy), but these methods are based on the physical and chemical properties of edible oils, and their detection limits are not sufficient to ensure edible oil authenticity [46]. Authentication by DNA analysis is one of the most challenging issues in the area of olive oil production, especially when investigating the presence of an alien species that is not in accordance with the product label or with appropriate enforced regulations. Nevertheless, the DNA analysis in olive oil presents three major problems regarding the DNA extraction from the matrix, including DNA quantity, PCR inhibitors and storage period. The small amount of genomic DNA that can be isolated from olive oils requires amplification, but the complex olive oil matrix is known to contain substances like polysaccharides, phenolic and other compounds that are not removed with the extraction and purification steps and that act like inhibitors against the DNA polymerase activity. Concerning the storage period, the amount of time after milling is extremely important for an accurate DNA analysis. In fact, for genetic traceability purposes, the olive oil samples have to be as fresh as possible to avoid oxidation damage to DNA [47].
Like for other crop plants, the DNA marker used for the taxonomic identification of olive species is the chloroplast region belonging to the trnL gene intron. Such a DNA barcode seems to be more appropriate than other targets to detect adulteration in plant oils, considering that plastid DNA recovery is easier in oil matrices compared with nuclear DNA recovery [45]. The trnL gene intron shows a number of importart features. First of all, its evolution in land plants is well understood and it has been used in the past for reconstructing phylogenies between closely related species or for identifying plant species. Moreover, highly conserved PCR primers were designed in previous studies, exhibiting robust amplification reactions and assaying variable sites among species in terms not only of single nucleotide polymorphisms but also of insertions and/or deletions.
The DNA barcoding methodology was tested for the identification of the botanical origin of plant oils (i.e., sunflower, corn, avocado, olive, sesame, soya, almond, hazelnut, walnut, cotton and wheat), implementing an analytical method for the detection of the common adulteration of olive oil with sesame oil [48]. The differential length of DNA amplicons of the trnL gene intron enabled the specific detection of 11 plant species, except for olive and avocado, whose discrimination was only possible on the basis of SNP detection. Overall results suggested that DNA barcoding can be successfully applied to the main plant oils, including all common olive oil adulterants and oils from allergenic species, as well as mixtures of them [48].
Another key factor is the determination of geographical origin and the olive oil cultivars. The globalization of food markets and food commodities make consumers increasingly concerned about the quality of foodstuffs [45]. This represents a requirement, especially for local and typical products based on specific cultivar and/or production area. With a few exceptions (for instance, see [6,49]), chloroplast DNA barcoding must be replaced by nuclear DNA genotyping for single cultivar identification or authentication.
An interesting case study of the use of DNA barcoding is that concerning the identification of plant species in honey. Standing to the European legislation, honey “is the natural sweet substance produced by Apis mellifera bees from the nectar of plants or from secretions of living parts of plants or excretions of plant-sucking insects on the living parts of plants, which the bees collect, transform by combining with specific substances of their own, deposit, dehydrate, store and leave in honeycombs to ripen and mature” (2001/110/EC). Nectar honey can be divided into two sub-groups: mono-floral and multi-flower honey. The first type belongs to a unique plant species, whereas the second is composed by a multitude of plant species. The quality of honey is closely related to the flower composition and the climatic and environmental conditions of the production area [50].
By the 2001/110/EC, the principal labeling requirements that must be indicated are the floral origin, physicochemical proprieties, organoleptic characteristics and regional provenance. Several methods have been proposed for the determination of botanical and geographical origins of honey. The traditional approach based on the microscopic examination of pollen grains directly in the honey (e.g., melissopalynology) is time consuming since it involves laborious counting procedures, and requires specialized botanical knowledge and training. Several chemical methods were also proposed, such as aroma compounds, free amino acids or minerals and trace elements. Even if all these methods work well for identifying the geographic origins and for distinguishing honey with different botanical origins, they provide only limited information on the plant composition [51]. In fact, it is not always possibile to recognize individual species because some pollen is not well distinguishable by its morphological traits (e.g., Campulanaceae and Lamiaceae) [50].
DNA barcoding is a powerful technology for detecting frauds and mislabeling of honey. The comparison between species of the local flora with taxa detected in honey products could bring a new level of food safety for human consumption. Recently, DNA barcoding was proposed as molecular tool for honey traceability exploitable in distinct steps of the supply chain. Some reserachers demonstrated that standard barcodes, such as trnL gene intron and rbcL gene region, are suitable for identifying plant traces from different honey samples, but anable for determining the botanical composition of individual honey species [50,51]. A synergism between multiple barcode markers among those conventionally used is in fact necessary to differentiate congeneric taxa. In particular, trnH−psbA intergenic spacer was proven to be the most discriminant marker for identifying plant species in honey [50]. It is worth mentioning the potential use of DNA barcoding as a method to assess food safety as well. In fact, traces of Atropa belladonna L., a plant toxic for humans, were found in honey [50], suggesting that DNA barcoding can be used also to detect other toxic species like Rhododendorn spp. and Crotalaria spp.
As a case study, we analyzed commercial pectin samples isolated from apple (Malus domestica Borkh.) and lemon (Citrus limon L.) in order to verify their identity and purity by means of cpDNA barcoding. Conventional barcodes related to the rbcL gene region, intergenic spacer trnH−psbA and tnrL gene intron were amplified in replicated experiments using universal primers. Surprisingly none of the DNA amplicons was attributable to apple and lemon species, most likely due to the severe degradation of the genomic DNA caused by the pectin purification process. Successful amplifications were obtained only when mini-barcode regions were investigated for the universal chloroplast targets, even if their sequences failed to discriminate any species within the genera Malus and Citrus. However, it is worth mentioning that sequencing of DNA amplicons associated to the tnrL gene intron revealed environmental contaminations from cyanobacteria (Nostoc comune var. flagelliforme). Figure 3 reports the main findings from the application of DNA barcoding in a pectin sample as quality control.
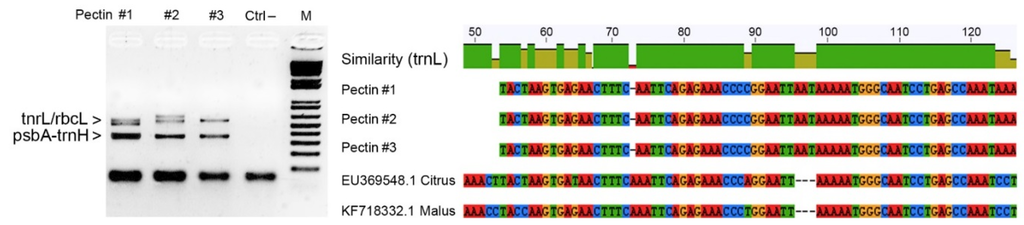
Figure 3.
Results of agarose gel electrophoresis analysis showing PCR amplicons related to three distinct samples of commercial pectins along with the negative control (DNA amplicons around 500–600 bp were generated using universal primers for rbcL barcode region and trnL gene intron, whereas DNA amlicons of about 350–400 bp refer to the intergenic spacer psbA-trnH). The NCBI accessions of Nostoc comune var. flagelliforme, scoring 99% identity with NCBI accessions are KC350486.1 and KC350471.1 (for protocol details, see Nicolè et al. [4]). Several SNPs and In/Dels were found between the aligned mini-barcode sequences recovered from pectin samples and tnrL sequences of Citrus and Malus spp.
This application shows how DNA barcoding can find utility not only for verifying the genetic identity of row materials, such as pectins, but also for assessing the purity and salubrity of food components. As the food supply chain becomes more global and complex, novel and better approaches to ensuring food safety are needed. DNA barcoding promises to meet such expectation in order to take food safety to a new level by molecular insights and understanding of the total supply chain. The synergism between the food industry and genomic laboratories should be strengthened as it could ensure the safety of our daily foodstuffs. A “microbial baseline” representing normal microbe communities would be a gold standard for food officials to understand what triggers contamination and the spread of many diseases.
6. Conclusions
DNA barcoding applied to traceability of livestock products is an essential tool to safeguard public and animal health and to endorse typical foods and their quality, and for this reason the European Union has applied since 2005 a strict legislation on labeling systems. It has been demonstrated that traceability methods based only on batch codes or papery documents are not always trustworthy, being easy to counterfeit.
In the last decade, DNA barcoding was proposed as a universal DNA-based tool for species identification [52]. The revolution introduced by this technology is not only associated to its discrimination power, but it is also related to the combination of three innovations: (i) molecularization of the identification approach, based on the use of nucleotide variation in a molecular marker as a genetic descriptor and taxonomic identifier of any form of life; (ii) computerization of the analyses by means of querying nonredundant databases of verified sequences using bioinformatic tools; (iii) standardization of the barcoding process as a whole, from sample collection to species identification through the production and interpretation of molecular data. In recent years, thanks to the use of DNA barcoding metodology, it is possible to introduce a generalization in taxonomy, allowing researchers specialized in different fields to work on shared frameworks [53].
There are many advantages in the use of DNA barcoding as it is reproducible and testable as long as the link between DNA test sequences and reference specimens is supported, and also verifiable at any time and by any researcher [4]. Moreover, the experimental procedure for extracting genomic DNA and amplifying specific DNA markers is technically easy and usually does not require the destruction of the sample, which sometimes is valuable and therefore needs to be safeguarded [6]. It allows all kinds of specimens to be treated, including those non-identifiable by morphology, and represents an universal applicable method that can be linked to any kind of biological or biodiversity information [54]. Compared with other molecular approaches, DNA barcoding as molecular protocol for genetic traceability of foodstuffs is very fast and relatively inexpensive, and the storage of DNA does not need particular attention because this molecule is very stable and any sample can be split into multiple subsamples, which can be sent to other laboratories as backups or for crosschecks. Due to all these advantages, although the use of DNA-based methods in species identification is not a new concept (e.g., use of ITS regions), DNA barcoding has asserted itself in the fields of molecular systematics and biodiversity studies, and now is becoming a challenge in the area of genetic traceability of foodstuffs.
Nowadays DNA barcoding provides an efficient molecular tool for taxonomic identifications at the species level and is contributing powerfully to animal and plant biodiversity analysis, and to genetic traceability of livestock and crop species and their food products. Since the number of DNA barcode sequences is growing more and more over the years, this information provides a unique horizontal genomics perspective with broad implications, including molecular phylogenetics and population genetics, in addition to taxonomic identifications. Theoretically, a great advantage offered by DNA barcoding is the possibility of identifying cryptic species and hence of distinguishing different related species. Such application would be feasible by identifying genetic distance limit values within which two individuals can be considered to belong to the same species, although boundaries among species seem to be somehow taxon-dependent. In fact, it has become clear that the currently used genetic distance approaches by means of DNA barcodes have great limitations, particularly when it comes to defining species boundaries.
The problem of using the barcoding gap is that lack of strong biological support could generate different type of errors. For instance, false positives could be generated if populations within one species show high rates of intraspecific differentiation, e.g., in allopatric populations with interrupted gene flow, whereas false negatives could be observed if little or no sequence variation in the barcoding region is found between different species reproductively isolated. For genetic traceability purposes, the existence of the barcoding gap could be heavily dependent of the sampling of a given species. If so, the individuals chosen to represent each taxon in the reference database should cover the vast majority of the existing diversity; otherwise an incomplete sampling could lead to a barcoding gap that does not correspond to reality.
As a consequence, accuracy is a critical issue in DNA barcoding applications for genetic traceability of food products. Hence, in order to know how well a single barcode sequence performs in delineating and identifying species, researchers must consider the extent of, and separation between, intraspecific variation and interspecific differentiation in the selected marker. This is because the more overlap there is between genetic variation within species and differentiation separating sister species, the less effective DNA barcoding becomes for taxonomic identifications [55].
Potential limitations of using mtDNA and cpDNA to infer species boundaries include hybridity and polyploidy of the genomes. While DNA barcoding using a single gene or multiple genes may be appropriate for animals, this is not so for plants due to their different life cycle characteristics, evolutionary histories, and the occurrence of hybridization and polyploidization events. Taxonomic uncertainty in dealing with hybridization phenomenon is a direct consequence of the fact that mitochondrial as well as plastid DNA is maternally inherited and any balanced or introgressed hybrid generation would have the extranuclear genome of the maternal species only. In taxonomically complex groups where species limits are often very narrowly defined, exact species identifications using a DNA barcoding approach are unlikely. One or a few markers cannot usually resolve the complexity in these groups resulting from recurrent landrace origins of taxa or where species arise through some events like recurrent interploid transition or recent hybrid speciation. Both hybridity and polyploidy can lead to shared mitochondrial and plastid haplotypes among animal and plant species, respectively.
At present molecular techniques based on DNA barcoding seem to be the most reliable and standardizable tool of authentication for food products of plant and animal origin, and researches have developed large genetic datasets and obtained significant technical improvements in the last few years. The effective applicability of DNA barcoding is not a major problem since it is a relatively simple and cheap technology, being therefore not only sustainable if these analyses are meant to be employed as routine tests in all foodstuffs, but also absolutely affordable when they are needed just as verification in particular cases (e.g., when recall of a batch is required) or authentication in specific products (e.g., any transformed or processed product). Nevertheless, any type of traceability method is associated to additional costs for food companies, but it becomes essential to analyze which part of these additional costs may be translated into benefits. Methods ensuring an efficient recall, such those based on DNA technologies, can prevent from recalling safe batches. Also, consumers’ willingness to pay for safer food should be better studied; in general consumers agree on paying extra price for food safety issues especially in developed countries. A study on consumers’ attitude towards meat products revealed that consumers’ behavior about traceability differs from country to country and it is also dependant on the perception of quality of meat [56]. For instance, the Belgian meat consumers were divided among “enthusiasts” and “pessimists” evidencing different perceptions among these groups [56]. In spite of such difference, authors highlighted that functional attributes as efficient products recall, possibility to identify an individual responsibility and a complete traceability of the meat chain may be regarded as minimum requirements of any authentication system for all consumers [35,56]. Keeping all these aspects in mind, the potential of DNA technologies appears straightforward, as recently well documented for fishery products [57,58,59].
In some cases, like for instance hybrid and polyploid complexes including fruit and crop plant species, a universal barcode marker may be insufficient for taxonomic identification due to variation in substitution rates, including occasionally low interspecific rates and high intraspecific rates. As a consequence, different DNA barcodes are likely needed for different species and their food products. This in turn will worsen the time and cost effectiveness of such methodology for identification and authentication purposes [60].
The second problem to overcome is the attainment of an agreement on markers and approaches to be utilized; in fact the scientific community is still debating distinct approaches, and common guidelines are needed. A first step has been taken by the International Society for Animal Genetics, and the Food and Agriculture Organization Standing Committee, which proposed sets of microsatellite (SSR) markers in different species for the study of animal genetic diversity and for conservation purposes. For the identification of animal species and their meat- and milk-derived products, including sausages and cheeses, it is crucial that the defition of standardized DNA markers be analyzed for genetic traceability and authentication purposes. For the determination of species in cereal flours, vegetable crops, fruit juices and seed oils, DNA barcodes provide robust information that can be exploited in many taxa.
In livestock species, among the three different levels of identification, the individual one regarding meat cut identification, appears the easiest to implement due the low number of molecular markers needed, the low costs, and to the univocal polymorphisms and simpler statistics. The main problem is eventually the organization of hair samples collection from every animal at birth. The genetic traceability of breeds and species is needed as well, but if it is true that the application of molecular methods is robust for species identification because exploits a deterministic strategy and any inference is necessary, it is also true that DNA barcoding technology is not able to ensure satisfactory levels of discrimination ability for breed determination. The same holds true in crop plant species, where the genetic traceability of cultivated varieties, such as pure lines, F1 hybrids and clones, fails when based on a DNA barcoding strategy.
In conclusion, DNA barcoding applied to genetic traceability is a useful and trustworthy tool for the identification and authentication of foodstuffs, and it is potentially the proper answer to the transformers’ and consumers’ request of quality confidence, but any DNA analysis needs more cooperation among researchers and factories involved in the food production chain and food origin inspection to be really applicable in a large scale and in order to find simpler and cheaper organizational solutions.
Acknowledgments
This research was supported by the University of Padova through special projects “Fondi ex 60%” whose principal investigators are Gianni Barcaccia, Margherita Lucchin and Martino Cassandro.
Author Contributions
All three authors conceived the study and equally contributed to the preparation and revision of the manuscript. The authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Organisation for Economic Co-operation and Development (OECD). The Economic Impact of Counterfeiting and Piracy; Executive Summary; Annual Report of the OECD: Paris, France, 2007; pp. 1–131. [Google Scholar]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef] [PubMed]
- Wiemers, M.; Fiedler, K. Does the DNA barcoding gap exist?—A case study in blue butterflies (Lepidoptera: Lycaenidae). Front. Zool. 2007, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Nicolè, S.; Erickson, D.L.; Ambrosi, D.; Bellucci, E.; Lucchin, M.; Papa, R.; Kress, W.J.; Barcaccia, G. Biodiversity studies in Phaseolus species by DNA barcoding. Genome 2011, 54, 529–545. [Google Scholar] [CrossRef] [PubMed]
- Nicolè, S.; Negrisolo, E.; Eccher, G.; Mantovani, R.; Patarnello, T.; Erickson, D.L.; Kress, W.J.; Barcaccia, G. DNA barcoding as a reliable method for the authentication of commercial seafood products. Food Technol. Biotechnol. 2012, 50, 387–389. [Google Scholar]
- Nicolè, S.; Barcaccia, G.; Erickson, D.L.; Kress, W.J.; Lucchin, M. The coding region of the UFGT gene is a source of diagnostic SNP markers that allow single-locus DNA genotyping for the assessment of cultivar identity and ancestry in grapevine (Vitis vinifera L.). BMC Res. Notes 2013, 6, 502. [Google Scholar] [CrossRef] [PubMed]
- Chase, M.W.; Salamin, N.; Wilkinson, M.; Dunwell, J.M.; Kesanakurthi, R.P.; Haidar, N.; Savolainen, V. Land plants and DNA barcodes: Short-term and long-term goals. Trends Ecol. Evol. 2005, 18, 273–273. [Google Scholar] [CrossRef] [PubMed]
- Chase, M.W.; Cowan, R.S.; Hollingsworth, P.M.; van den Berg, C.; Madrinan, S. A proposal for a standardized protocol to barcode all land plants. Taxon 2007, 56, 295–299. [Google Scholar]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef] [PubMed]
- Consortium for the Barcode of Life (CBOL). CBOL Approves matK and rbcL as the BARCODE Regions for Land Plants, Statement by the Executive Committee, Consortium for the Barcode of Life 16 November 2009. Available online: http://barcoding.si.edu/plant_working_group.html (accessed on 28 December 2015).
- Hilu, K.W.; Liang, H. The matK gene: Sequence variation and application in plant systematics. Am. J. Bot. 1997, 84, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Galimberti, A.; de Mattia, F.; Losa, A.; Bruni, I.; Federici, S.; Casiraghi, M.; Martellos, S.; Labra, M. DNA barcoding as a new tool for food traceability. Food Res. Int. 2013, 50, 55–63. [Google Scholar] [CrossRef]
- FoodDrinkEurope. Priorities for the Development of an EU Industrial Policy for Food, Competitiveness Report 2012. Available online: http://www.fooddrinkeurope.eu/uploads/press-releases_documents/FINAL_competitiveness_web.pdf (accessed on 28 December 2015).
- Everstine, K.; Spink, J.; Kennedy, S. Economically motivated adulteration (EMA) of food: Common characteristics of EMA incidents. J. Food Prot. 2013, 76, 723–735. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Green Paper on the General Principles of Food Law in the European Union; Commission of the European Communities: Brussels, Belgium, 1997; pp. 1–63. [Google Scholar]
- European Commission. Commission Regulation (EC) No. 1825/2000 of 25 August 2000 laying down detailed rules for the application of Regulation (EC) No 1760/2000 of the European Parliament and of the Council as regards the labelling of beef and beef products. Off. J. Eur. Community 2000, L216, 8–12. [Google Scholar]
- European Commission. White Paper on Food Safety; Commission of the European Communities: Brussels, Belgium, 2000; pp. 1–52. [Google Scholar]
- European Commission. Council Regulation (EC) No 178/2002 of 28 January 2002 laying down the general principles and requirements of food law, establishing the European Food Safety Authority and laying down procedures in matters of food safety. Off. J. Eur. Communities 2002, L31, 1–24. [Google Scholar]
- European Commission. Council Regulation (EC) No. 1760/2000 of 17 July 2000 establishing a system for the identification and registration of bovine animals and regarding the labeling of beef and beef products and repealing Council Regulation (EC) No 820/97. Off. J. Eur. Communities 2000, L204, 1–10. [Google Scholar]
- Marchant, J. Secure Animal Identification and Source Verification; JM Communications, Optibrand Ltd., LLC: Fort Collins, CO, USA, 2002. [Google Scholar]
- Smith, G.C.; Tatum, J.D.; Belk, K.E.; Scanga, J.A.; Grandin, T.; Sofos, J.N. Traceability from a U.S. Perspective. Meat Sci. 2005, 71, 174–193. [Google Scholar] [CrossRef] [PubMed]
- Sardina, M.T.; Tortorici, L.; Mastrangelo, S.; Di Gerlando, R.; Tolone, M.; Portolano, B. Application of microsatellite markers as potential tools for traceability of Girgentana goat breed dairy products. Food Res. Int. 2015, 74, 115–122. [Google Scholar] [CrossRef]
- Dimauro, C.; Nicoloso, L.; Cellesi, M.; Macciotta, N.P.P.; Ciani, E.; Moioli, B.; Pilla, F.; Crepaldi, P. Selection of discriminant SNP markers for breed and geographic assignment of Italian sheep. Small Rumin. Res. 2015, 128, 27–33. [Google Scholar] [CrossRef]
- Mateus, J.C.; Russo-Almeida, P.A. Traceability of 9 Portuguese cattle breeds with PDO products in the market using microsatellites. Food Control 2015, 47, 487–492. [Google Scholar] [CrossRef]
- Ng, J.; Satkoski, J.; Premasuthan, A.; Kanthaswamy, S. A nuclear DNA-based species determination and DNA quantification assay for common poultry species. J. Food Sci. Technol. 2015, 51, 4060–4065. [Google Scholar] [CrossRef] [PubMed]
- Ciampolini, R.; Leveziel, H.; Mozzanti, E.; Grohs, C.; Cianci, D. Genomic identification of an individual or its tissue. Meat Sci. 2000, 54, 35–40. [Google Scholar] [CrossRef]
- Goffaux, F.; China, B.; Dams, L.; Clinquart, A.; Daube, G. Development of a genetic traceability test in pig based on single nucleotide polymorphism detection. Forensic Sci. Int. 2005, 151, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Opara, L.; Mazaud, F. Food traceability from field to plate. Outlook Agric. 2001, 30, 239–247. [Google Scholar] [CrossRef]
- Cozzi, G.; Ragno, E. Meat production and market in Italy. Agric. Conspec. Sci. 2003, 68, 1331–7768. [Google Scholar]
- Ajmone-Marsan, P.; Milanesi, E.; Negrini, R. Breed traceability using molecular methods. In Proceedings of the 7th World Conference of the Brown Swiss Cattle Breeders, Verona, Italy, 3–7 March 2004; pp. 101–104.
- Stanford, K.; Stitt, J.; Kellar, J.A.; McAllister, T.A. Traceability in cattle and small ruminants in Canada. Rev. Sci. Tech. 2001, 20, 510–522. [Google Scholar] [PubMed]
- Barcos, L.O. Recent developments in animal identification and the traceability of animal products in international trade. Rev. Sci. Tech. 2001, 20, 640–651. [Google Scholar] [PubMed]
- Meuwissen, M.P.M.; Velthuis, A.G.J.; Hogeveen, H.; Huirne, R.B.M. Traceability and certification in meat supply chains. J. Agribus. 2003, 21, 167–181. [Google Scholar]
- Unseld, M.; Beyermann, B.; Brandt, P.; Hiesel, R. Identification of the species origin of highly processed meat products by mitochondrial DNA sequences. PCR Methods Appl. 1995, 4, 241–243. [Google Scholar] [CrossRef] [PubMed]
- Maudet, C.; Taberlet, P. Holstein’s milk detection in cheeses inferred from melanocortin receptor 1 (MC1R) gene polymorphism. J. Dairy Sci. 2002, 85, 707–715. [Google Scholar] [CrossRef]
- Maudet, C.; Luikart, G.; Taberlet, P. Genetic diversity and assignment tests among seven French cattle breeds based on microsatellite DNA analysis. J. Anim. Sci. 2002, 80, 942–950. [Google Scholar] [PubMed]
- Rea, S.; Chikuni, K.; Branciari, R.; Sangamayya, R.S.; Ranucci, D.; Avellini, P. Use of duplex polymerase chain reaction (duplex-PCR) technique to identify bovine and water buffalo milk used in making mozzarella cheese. J. Dairy Res. 2001, 68, 689–698. [Google Scholar] [CrossRef] [PubMed]
- European Fruit Juice Association (AIJN). European Fruit Juice Association 2014 Liquid Fruit Market Report. Available online: http://aijn.org/files/default/aijn2014-full.pdf (accessed on 28 December 2015).
- Faria, M.A.; Magalhães, A.; Nunes, M.E.; Oliveira, M.B.P.P. High resolution melting of trnL amplicons in fruit juices authentication. Food Control 2013, 33, 136–141. [Google Scholar] [CrossRef]
- Madesis, P.; Ganopoulos, I.; Bosmali, I.; Tsaftaris, A. Barcode High Resolution Melting analysis for forensic uses in nuts: A case study on allergenic hazelnuts (Corylus avellana). Food Res. Int. 2012, 50, 351–360. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Madesis, P.; Darzentas, N.; Argiriou, A.; Tsaftaris, A. Barcode High Resolution Melting (Bar-HRM) analysis for detection and quantification of PDO “Fava Santorinis” (Lathyrus clymenum) adulterants. Food Chem. 2012, 133, 505–512. [Google Scholar] [CrossRef] [PubMed]
- De Mattia, F.; Bruni, I.; Galimberti, A.; Cattaneo, F.; Casiraghi, M.; Labra, M. A comparative study of different DNA barcoding markers for the identification of some members of Lamiaeea. Food Res. Int. 2011, 44, 693–702. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Bazakos, C.; Madesis, P.; Kalaitzis, P.; Tsaftaris, A. Barcode DNA high-resolution melting (Bar-HMR) analysis as novel close-tubed and accurate tool for olive oil forensic use. J. Sci. Food. Agric. 2013, 93, 2281–2286. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Xu, W.; Shang, Y.; Zhu, P.; Mei, Z.; Tian, W. Development and optimization of an efficient method to detect the authenticity od edible oils. Food Control 2013, 31, 71–79. [Google Scholar] [CrossRef]
- Costa, J.; Mafra, I.; Oliveira, M.B.P.P. Advances in vegetable oil authentication by DNA-based markers. Trends Food Sci. Technol. 2012, 26, 43–55. [Google Scholar] [CrossRef]
- Spaniolas, S.; Bazakos, C.; Ntourou, T.; Bihmidine, S.; Georgousakis, A.; Kalaitzis, P. Use of lambda DNA as a marker to assess DNA stability in olive oil during storage. Eur. Food Res. Technol. 2008, 227, 175–179. [Google Scholar] [CrossRef]
- Enan, M.; Fawzi, M.; Al-Deeb, K. Amiri, DNA barcoding of Ricinus comunis from different geographical origin by using chloroplast matK and internal transcribed spacers. Am. J. Plant Sci. 2012, 3, 1304–1310. [Google Scholar] [CrossRef]
- Bruni, I.; Galimberti, A.; Caridi, L.; Scaccabarozzi, D.; de Mattia, F.; Casiraghi, M.; Labra, M. A DNA barcoding approach to identify plant species in multiflower honey. Food Chem. 2014, 170, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Valentini, A.; Miquel, C.; Taberletet, P. DNA barcoding for honey biodiversity. Diversity 2010, 2, 610–617. [Google Scholar] [CrossRef]
- Galimberti, A.; de Mattia, F.; Losa, A.; Bruni, I.; Federici, S.; Casiraghi, M.; Martellos, S.; Labra, M. DNA barcoding as a new tool for food traceability. Food Res. Int. 2013, 50, 55–63. [Google Scholar] [CrossRef]
- Casiraghi, M.; Labra, M.; Ferri, M.; Galimberti, A.; De Mattia, F. DNA barcoding: a six-question tour to improve users’ awareness about the method. Brief. Bioinform. 2010. [Google Scholar] [CrossRef] [PubMed]
- Decaëns, T.; Porco, D.; Rougerie, R. Le Barcoding ADN: Un Outil Pour Étudier la Biodiversité des Invertébrés Terrestres; Société Française d’Écologie: Cestas Cedex, France, 2013. [Google Scholar]
- Meyer, C.P.; Paulay, G. DNA barcoding: Error rates based on comprehensive sampling. PLoS Biol. 2005, 3, e422. [Google Scholar] [CrossRef] [PubMed]
- Gellynck, X.; Januszewska, R.; Verbeke, W.; Viaene, J. Firm’s costs of traceability confronted with consumer requirements. In Proceedings of the 92nd Seminar on Quality Management and Quality Assurance in Food Chains of the European Association of Agricultural Economists, Gottingen, Germany, 2–4 March 2005.
- Di Pinto, A.; Mottola, A.; Marchetti, P.; Bottaro, M.; Terio, V.; Bozzo, G.; Bonerba, E.; Ceci, E.; Tantillo, G. Packaged frozen fishery products: Species identification, mislabeling occurrence and legislative implications. Food Chem. 2015, 194, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Di Pinto, A.; Marchetti, P.; Mottola, A.; Bozzo, G.; Bonerba, E.; Ceci, E.; Bottaro, M.; Tantillo, G. Species identification in fish fillet products using DNA barcoding. Fish. Res. 2015, 170, 9–13. [Google Scholar] [CrossRef]
- Shokralla, S.; Hellberg, R.S.; Handy, S.M.; King, I.; Hajibabaei, M. A DNA mini-barcoding system for authentication of processed fish products. Sci. Rep. 2015, 5, 15894. [Google Scholar] [CrossRef] [PubMed]
- Galimberti, A.; Labra, M.; Sandionigi, A.; Bruno, A.; Mezzasalma, V.; de Mattia, F. DNA barcoding for minor crops and food traceability. Adv. Agric. 2014, 2014, 831875. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).