Assessment of Genetic Diversity in Faba Bean Based on Single Nucleotide Polymorphism
Abstract
:1. Introduction
2. Experimental Section
2.1. Plant Material and DNA Extraction
2.2. SNP Genotyping
| Genotype | Breeding History | Seed Size (g/100 sd) | Major Trait | Origin of Source Germplasm |
|---|---|---|---|---|
| Cultivars | ||||
| Aquadulce | Introduced variety, reselected in Aust | 130 | Broad bean | Spain |
| Ascot | Seln ex Fiord | 45 | Asco res | Greece |
| Cairo | Seln ex open pollinated ILB 2282 | 65 | Adaptation | Cyprus |
| Doza | Acc 157 × STW | 50 | Rust res | England, Sudan |
| Farah | Seln ex Fiesta | 65 | Asco res | Spain |
| Fiesta | Seln ex BPL 1196 B8817 | 65 | Adaptation | Spain |
| Fiord | Seln ex landrace | 45 | Adaptation | Greece |
| Icarus | Seln ex BPL 710 | 80 | Choc spot res | Ecuador |
| Manafest | Seln ex ILB 3026 | 80 | Choc spot res | Ecuador |
| Nura | Icarus × Ascot | 60 | Asco res | Ecuador, Greece |
| PBA Kareema | Seln ex Aquadulce | 140 | Broad bean | Spain |
| PBA Rana | Manafest × (Acc 611 × Manafest) (breeding line AF01006-1) | 75 | Asco res | Ecuador, Lebanon |
| PBA Warda | SP99046 × SP99081 | 60 | Rust res | Ecuador, Greece, Ethiopia |
| Divine | European variety, not cultivated in Australia | 50 | Low v/cv | Europe |
| Australian Breeding Lines | ||||
| 1269*483/6-1 | BPL 710 × Farah | 70 | Cerco res | Ecuador, Spain |
| AF03063-1 | (Icarus × Acc286) × Farah | 70 | Asco res | Spain, Ecuador, Greece |
| AF04053 | (Acc 735 × Acc 683) × Farah | 65 | Asco res | Spain, Syria, Ecuador, Turkey |
| AF05069 | (Acc 611 × Acc 722) x ((Icarus × Ascot) × Farah) | 70 | Asco res | Lebanon, UK, Spain, Ecuador, Greece |
| AF05095 | Acc 920 × Farah | 80 | Asco res | Morocco, Spain |
| IX220d/2-5 | IX38 × IX4 | 65 | BLRV res | China, Sudan, Italy |
| Germplasm Accessions | ||||
| Acc 278 | Seln ex BPL 112 | 75 | Asco res | UK |
| Acc 286 | Seln ex landrace | 110 | Early flowering | Crete |
| Acc 391#5188 | Seln ex ILB 142 | 55 | Rust res | Morocco |
| Germplasm Accessions | ||||
| Acc 611 | Seln ex BPL 472 | 80 | Asco res | Lebanon |
| Acc 624#8103 | Seln ex ILB 397 | 45 | Rust res | Tunisia |
| Acc 683 | ILB 938 × BPL 1390 | 100 | Choc spot | Ecuador, Turkey |
| Acc 737 | Seln ex ILB 3033 B 88176 | 120 | Choc spot | Ecuador |
| Acc 805#4912 | Seln ex ILB 445 | 35 | BLRV res | Yemen |
| Acc 868 | ILB 68 × Fiord | 65 | Zero tannin | UK, Greece |
| Acc 920/3 | Seln ex landrace | 90 | Asco res | Morocco |
| Acc 970 | Seln ex ILB 752 | 100 | Asco res | Sweden (Donor) |
| Acc 973 | Seln ex ILB 3025 | 55 | Rust res | Ecuador |
| Acc 1107/2 | Seln ex ILB 2582 | 100 | Cerco res | Morocco |
| Acc 1108/1 | Seln ex ILB 2583 | 80 | Cerco res | Morocco |
| Acc 1129/2 | Seln ex ILB 3362 | 90 | Salt tol | China (Zhejiang) |
| Acc 1206#4919 | Seln ex ILB 5220 | 110 | BLRV res | China (Yunnan) |
| Acc 1248/4 | Seln ex ILB 5333 | 90 | Choc spot res | Ecuador |
| Acc 1257-3 | Seln ex ILB 5353 | 80 | Choc spot res | Ecuador |
| Acc 1322/2 | L 82003 B8833 (BPL 472 × BPL 261) | 75 | Cerco res | Lebanon, Greece |
| Acc 1430/3 | Seln ex BPL 217 | 150 | Asco res | Morocco |
| Acc 1477/4 | Seln ex landrace | 70 | Asco res | Crete |
| Acc 1487/7 | Seln ex ILB 917 | 70 | Salt tol | Algeria |
| Acc 1604#6108 | Seln ex BPL 5247 | 70 | BYMV res | Afghanistan |
| Acc 1714/1 | Seln ex ILB 5561 | 140 | Choc spot res | China (Gansu) |
| Acc 1727 | Seln ex landrace | 100 | Early flowering | China (Guangxi) |
2.3. Genetic Diversity and Population Structure Analysis
3. Results and Discussion
3.1. SNP Polymorphism
3.2. Genetic Diversity Analysis

3.3. Relationships between Genetic Diversity and Breeding History
3.4. Population Structure Analysis
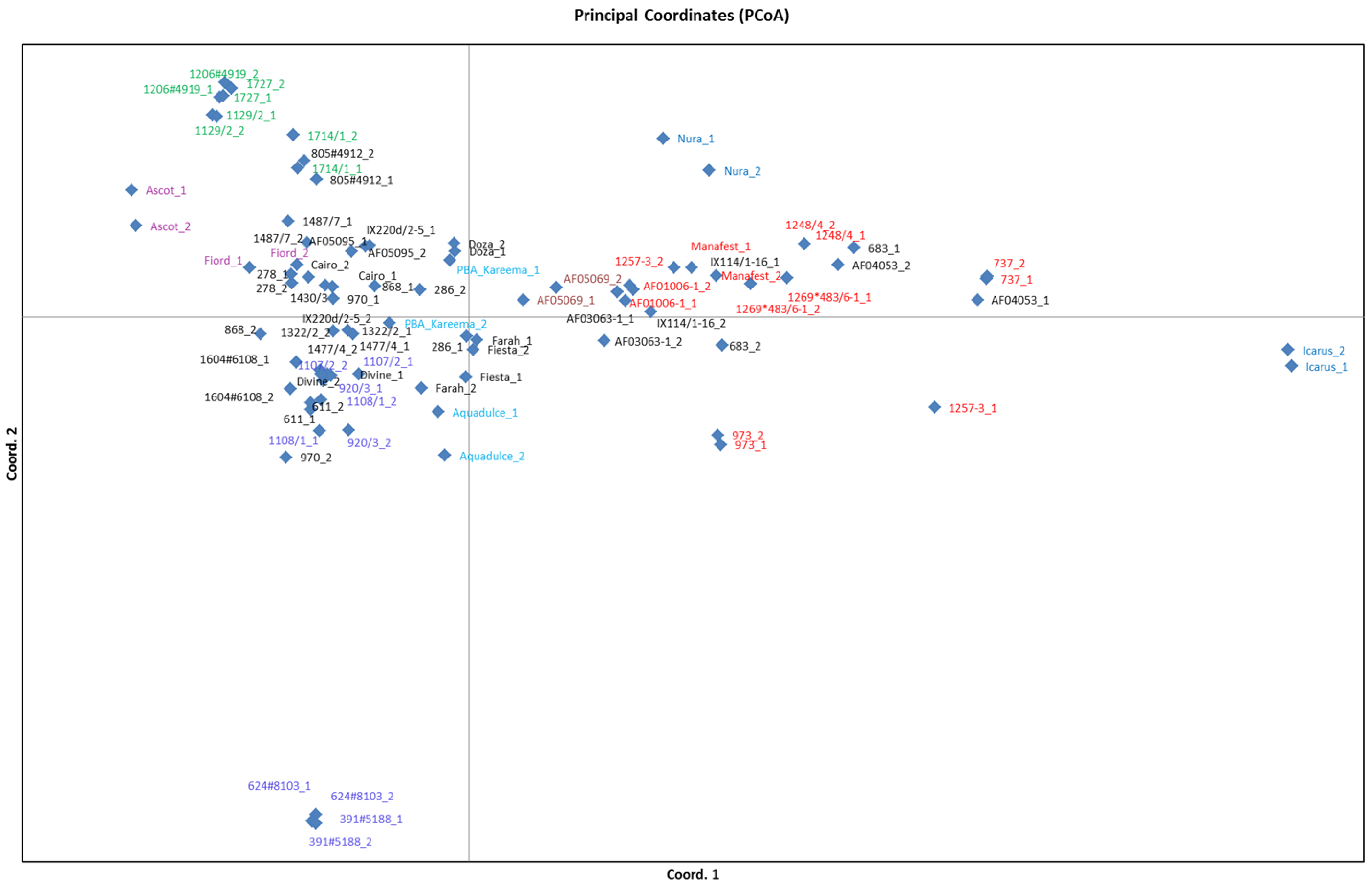
4. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
References and Notes
- FAOSTAT. Crops. Available online: http://faostat.fao.org/site/567/default.aspx#ancor/ (accessed on 17 January 2014).
- Cubero, J. On the evolution of Vicia faba L. Theor. Appl. Genet. 1974, 45, 47–51. [Google Scholar] [CrossRef]
- Alghamdi, S.; Al-Faifi, S.A.; Migdadi, H.M.; Ammar, M.H.; Siddique, K.H.M. Inter-simple sequence repeat (ISSR)-based diversity assessment among faba bean genotypes. Crop Pasture Sci. 2013, 62, 755–760. [Google Scholar]
- Ye, Y.; Lang, L.; Xia, M.; Tu, J. Faba beans in China; (in Chinese). China Agriculture Press: Beijing, China, 2003; pp. 1–15. [Google Scholar]
- Raina, S.; Ogihara, Y. Ribosomal DNA repeat unit polymorphism in 49 Vicia species. Theor. Appl. Genet. 1995, 90, 477–486. [Google Scholar]
- Bennett, M.; Smith, J.B. Nuclear DNA amounts in angiosperms. Proc. Royal Soc. B: Biol. Sci. 1982, 216, 179–199. [Google Scholar] [CrossRef]
- Johnston, J.; Bennett, M.D.; Rayburn, A.L.; Galbraith, D.W.; Price, H.J. Reference standards for determination of DNA content of plant nuclei. Am. J Bot. 1999, 86, 609–613. [Google Scholar] [CrossRef]
- Duc, G.; Bao, S.Y.; Baum, M.; Redden, B.; Sadiki, M.; Suso, M.J.; Vishniakova, M.; Zong, X.X. Diversity maintenance and use of Vicia faba L. genetic resources. Field Crops Res. 2010, 115, 270–278. [Google Scholar] [CrossRef]
- Link, W.; Dixkens, C.; Singh, M.; Schwall, M.; Melchinger, A.E. Genetic diversity in European and Mediterranean faba bean germplasm revealed by RAPD markers. Theor. Appl. Genet. 1995, 90, 27–32. [Google Scholar]
- Zeid, M.; Schon, C.C.; Link, W. Genetic diversity in recent elite faba bean lines using AFLP markers. Theor. Appl. Genet. 2003, 107, 1304–1314. [Google Scholar] [CrossRef]
- Upadhyaya, H.; Dwivedi, S.L.; Ambrose, M.; Ellis, N.; Berger, J.; Smykal, P.; Debouck, D.; Duc, G.; Dumet, D.; Flavell, A.; Sharma, S.K.; et al. Legume genetic resources: Management, diversity assessment, and utilization in crop improvement. Euphytica 2011, 180, 27–47. [Google Scholar] [CrossRef]
- Zeid, M.; Schon, C.C.; Link, W. Genetic diversity in a group of recent elite faba bean lines. Czech J. Genet. Plant Breed. 2001, 37, 34–40. [Google Scholar]
- Kwon, S.; Hu, J.; Coyne, C.J. Genetic diversity and relationship among faba bean (Vicia faba L.) germplasm entries as revealed by TRAP markers. Plant Genet. Resour. Char. Util. 2010, 8, 204–213. [Google Scholar] [CrossRef]
- Wang, H.; Zong, X.; Guan, J.; Yang, T.; Sun, X.; Ma, Y.; Redden, R. Genetic diversity and relationship of global faba bean (Vicia faba L.) germplasm revealed by ISSR markers. Theor. Appl. Genet. 2012, 124, 789–797. [Google Scholar] [CrossRef]
- Kaur, S.; Pembleton, L.; Cogan, N.; Savin, K.; Leonforte, T.; Paull, J.; Materne, M.; Forster, J. Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genom. 2012, 13, 104. [Google Scholar] [CrossRef]
- Akash, M.; Myers, G.O. The development of faba bean expressed sequence tag—simple sequence repeats (ESTSSRs) and their validity in diversity analysis. Plant Breed. 2012, 131, 522–530. [Google Scholar] [CrossRef]
- Yang, T.; Bao, S.; Ford, R.; Jia, T.; Guan, J.; He, Y.; Sun, X.; Jiang, J.; Hao, J.; Zhang, X.; et al. High-throughput novel microsatellite marker of faba bean via next generation sequencing. BMC Genom. 2012, 13, 602. [Google Scholar] [CrossRef]
- Kaur, S.; Kimber, R.B.E.; Cogan, N.O.I.; Materne, M.; Forster, J.W.; Paull, J.G. SNP discovery and high-density genetic mapping in faba bean (Vicia faba L.) permits identification of QTLs for ascochyta blight resistance. Plant Sci. 2014, 217-218, 47–55. [Google Scholar] [CrossRef]
- Rohlf, F. NTSYS-pc Numerical Taxonomy and Multivariate Analysis System, version 2.1; Stony Brook: New York, NY, USA, 2000. [Google Scholar]
- Liu, K.; Muse, S.V. PowerMarker: Integrated analysis environment for genetic marker data. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar]
- Perrier, X.; Flori, A.; Bonnot, F. Data analysis methods. In Genetic Diversity of Cultivated Tropical Plants; Hamon, P., Seguin, M., Perrier, X., Glaszmann, J.C., Eds.; Enfield Science Publishers: Montpellier, France, 2003; pp. 43–76. [Google Scholar]
- Peakell, R.; Smouse, P.E. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Cottage, A.; Gostkiewicz, K.; Thomas, J.E.; Borrows, R.; Torres, A.M.; O’Sullivan, D.M. Heterozygosity and diversity analysis using mapped SNPs in a faba bean inbreeding programme. Mol. Breed. 2012, 30, 1799–1809. [Google Scholar] [CrossRef]
- Emanuelli, F.; Lorenzi, S.; Grzeskowiak, L.; Catalano, V.; Stefanini, M.; Troggio, M.; Myles, S.; Martinez-Zapater, J.M.; Zyprian, E.; Moreira, F.M.; et al. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol. 2013, 13, 39. [Google Scholar] [CrossRef]
- Wurschum, T.; Langer, S.M.; Friedrich, C.; Longin, H.; Korzun, V.; Akhunov, E.; Ebmeyer, E.; Schachschneider, R.; Schacht, J.; Kazman, E.; et al. Population structure, genetic diversity and linkage disequilibrium in elite winter wheat assessed with SNP and SSR markers. Theor. Appl. Genet. 2013, 126, 1477–1486. [Google Scholar] [CrossRef]
- Zong, X.; Ren, J.; Guan, J.; Wang, S.; Liu, Q.; Paull, J.G.; Redden, R. Molecular variation among Chinese and global germplasm in spring faba bean areas. Plant Breeding 2010, 129, 508–513. [Google Scholar]
- Ouji, A.; Suso, M.J.; Rouaissi, M.; Abdellaoui, R.; Gazzah, M.E. Genetic diversity of nine faba bean (Vicia faba L.) populations revealed by isozyme markers. Genes Genom. 2011, 33, 31–38. [Google Scholar] [CrossRef]
- Gnanasambandam, A.; Paull, J.; Torres, A.; Kaur, S.; Leonforte, T.; Li, H.; Zong, X.; Yang, T.; Materne, M. Impact of molecular technologies on faba bean (Vicia faba L.) breeding strategies. Agronomy 2012, 2, 132–166. [Google Scholar] [CrossRef]
- Piepho, H.; Laidig, F. A Review of Methods for Cluster Analysis of Marker Data . In proceedings of the UPOV—Working Group on Biochemical & Molecular Techniques & DNA-Profiling in Particular, Cambridge, UK, 11–13 March 1997.
- Van Leur, J.A.G. New South Wales Department of Primary Industries, Tamworth, Australia. Personnal communication, 2013. [Google Scholar]
- Hanounik, S.B.; Maliha, N. Horizontal and vertical resistance in Vicia faba to chocolate spot caused by Botrytis fabae. Plant Dis. 1986, 70, 770–773. [Google Scholar] [CrossRef]
- Hanounik, S.B.; Robertson, L.D. New sources of resistance in Vicia faba to chocolate spot caused by Botrytis fabae. Plant Dis. 1988, 72, 696–698. [Google Scholar] [CrossRef]
- Bond, D.A.; Jellis, G.J.; Rowland, G.G.; Le Guen, J.; Robertson, L.D.; Khalil, S.A.; Li-Juan, L. Present status and future strategy in breeding faba beans (Vicia faba L.) for resistance to biotic and abiotic stresses. Euphytica 1994, 73, 151–166. [Google Scholar] [CrossRef]
- Zong, X.; Liu, X.; Guan, J.; Wang, S.; Liu, Q.; Paull, J.G.; Redden, R. Molecular variation among Chinese and global winter faba bean germplasm. Theor.Appl. Genet. 2009, 118, 971–978. [Google Scholar] [CrossRef]
- Kimber, R.B.E.; Davidson, J.A.; Paull, J.G. Using Genetic Diversity within Faba Bean Germplasm to Develop Resistance to Ascochyta Blight. In Proceedings of the 1st International Ascochyta Workshop on Grain Legumes, Le Tronchet, France, 2006.
- Knight, R. Ascot VF. Plant Var. J. 1997, 10, 33. [Google Scholar]
- Paull, J. Farah. Plant Var. J. 2003, 16, 64. [Google Scholar]
- Paull, J. PBA Kareema. Plant Var. J. 2010, 23, 51. [Google Scholar]
- Van de Ven, M.; Powell, W.; Ramsay, G.; Waugh, R. Restriction fragment length polymorphisms as genetic markers in Vicia. Heredity 1990, 65, 329–342. [Google Scholar] [CrossRef]
- Glaszmann, J.; Kilian, B.; Upadhyaya, H.D.; Varshney, R.K. Accessing genetic diversity for crop improvement. Curr. Opin. Plant Biol. 2010, 13, 167–173. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kaur, S.; Cogan, N.O.I.; Forster, J.W.; Paull, J.G. Assessment of Genetic Diversity in Faba Bean Based on Single Nucleotide Polymorphism. Diversity 2014, 6, 88-101. https://doi.org/10.3390/d6010088
Kaur S, Cogan NOI, Forster JW, Paull JG. Assessment of Genetic Diversity in Faba Bean Based on Single Nucleotide Polymorphism. Diversity. 2014; 6(1):88-101. https://doi.org/10.3390/d6010088
Chicago/Turabian StyleKaur, Sukhjiwan, Noel O. I. Cogan, John W. Forster, and Jeffrey G. Paull. 2014. "Assessment of Genetic Diversity in Faba Bean Based on Single Nucleotide Polymorphism" Diversity 6, no. 1: 88-101. https://doi.org/10.3390/d6010088
APA StyleKaur, S., Cogan, N. O. I., Forster, J. W., & Paull, J. G. (2014). Assessment of Genetic Diversity in Faba Bean Based on Single Nucleotide Polymorphism. Diversity, 6(1), 88-101. https://doi.org/10.3390/d6010088




