Aerobic Methanotrophs in Natural and Agricultural Soils of European Russia
Abstract
:1. Introduction
2. Experimental Section
2.1. Soil Sampling Sites
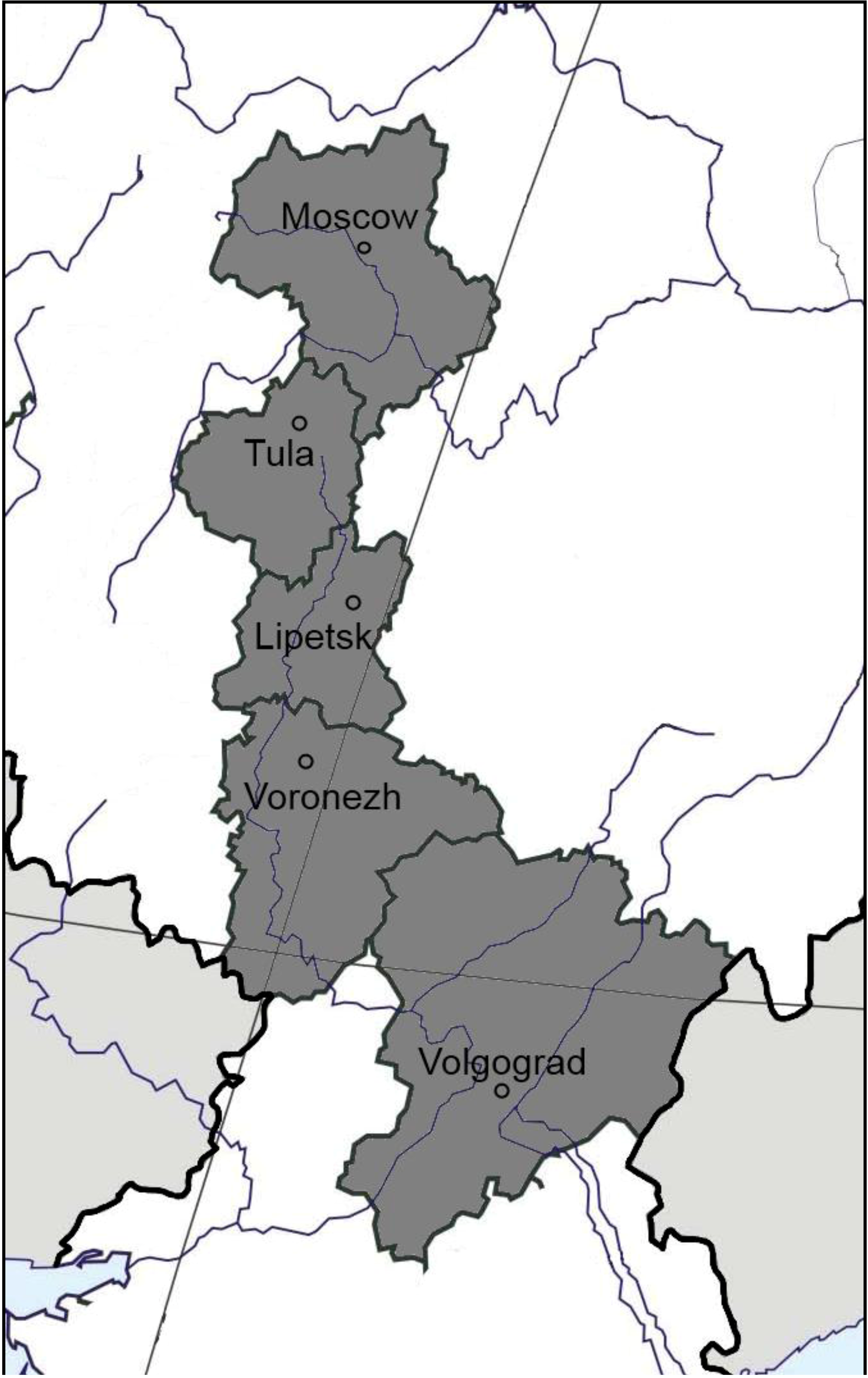
| Region, site (coordinates) | Soil type, FAO | Land management | Vegetation | C org, (%) | N tot, (%) | C:N | pH (KCl) | NO3+NH4, mg/100 g | Surface methane flux (µg CH4 m−2 h−1) |
|---|---|---|---|---|---|---|---|---|---|
| Moscow, Puschino (54.50°N, 37.37°E) | Podzoluvisol | unmanaged | Mixed forest (Pinus spp., Abies spp., Betula spp.) | 2.1 | 0.2 | 10.5 | 4.6 | 0.8 | −19.0 ± 3.4 |
| managed | Barley field (Hordeum vulgare) | 1.2 | 0.1 | 11.9 | 5.3 | 0.8 | −2.6 ± 1.2 | ||
| Tula, Schekino (54.00°N, 37.31°E) | Luvisol | unmanaged | Broadleaf forest (Tilia sp., Corylus spp., Ulmus spp.) | 1.3 | 0.1 | 10.5 | 4.2 | 2.3 | −26.0 ± 8.1 |
| managed | Wheat field (Triticum spp.) | 0.9 | 0.1 | 9.2 | 5.1 | 0.8 | −4.3 ± 1.1 | ||
| Lipetsk, Danki (53.30°N, 38.58°E) | Phaeozem | unmanaged | Broadleaf forest (Acer spp., Quercus spp., Populus spp., Tilia spp.) | 4.3 | 0.3 | 12.5 | 5.8 | 2.6 | −19.0 ± 2.0 |
| Voronezh, Bobrov (51.07°N, 40.17°E) | Solodic chernozem | unmanaged | Birch and aspen forest (Betula spp., Populus spp.) | 2.7 | 0.2 | 10.8 | 6.4 | 0.9 | +6.1 ± 0.4 |
| Voronezh, Talovaya (51.07°N, 40.43°E) | Solonetz | unmanaged | Grassland (Bromopsis spp., Vicia spp., Alopecurus spp., Phleum spp.) | 2.9 | 0.2 | 11.7 | 8.2 | 1.2 | +12.9 ± 2.0 |
| Voronezh, Talovaya (51.07°N, 40.43°E) | Chernozem | unmanaged | Grassland (Stipa spp.) | 4.3 | 0.4 | 11.5 | 6.7 | 2.0 | −7.6 ± 2.6 |
| managed | Wheat field (Triticum spp.) | 3.9 | 0.3 | 11.3 | 6.2 | 2.0 | −2.2 ± 1.2 | ||
| Volgograd, Kachalino (49.49°N, 44.32°E) | Gleyic kastanozem | unmanaged | Elm and alder forest (Ulmus spp., Alnus spp.) | 3.2 | 0.3 | 11.9 | 6.9 | 0.8 | −24.0 ± 7.0 |
| Volgograd, Ylovlya (49.47°N, 44.31°E) | Kastanozem | unmanaged | Grassland (Festuca valesiaca, Euphorbia spp., Artemisia spp.) | 1.6 | 0.1 | 10.9 | 5.5 | 0.7 | −30.0 ± 5.2 |
| managed | Wheat field (Triticum spp.) | 0.9 | 0.1 | 9.1 | 5.8 | 0.6 | −25.0 ± 7.0 |
2.2. Surface Methane Flux Measurements
2.3. Potential Methane Oxidation Rate (14CH4)
2.4. DNA Extraction and PCR Amplification
2.5. Construction of Clone Libraries
2.6. DNA Sequencing and Phylogenetic Analysis
3. Results and Discussion
3.1. CH4 Surface Flux and Potential Methane Oxidation Rate
| Soil type | Methane oxidation rate, ng CH4 g−1 day−1 | Potential methane oxidation rate, ng CH4 g−1 day−1 | |
|---|---|---|---|
| CH4-CO2 | Biomass incorporation | ||
| Podzoluvisol (natural) | 0.70 ± 0.10 | 0.51 ± 0.06 | 1.21 |
| Podzoluvisol (agricultural) | 0.28 ± 0.10 | 0.12 ± 0.08 | 0.40 |
| Luvisol (natural) | 0.78 ± 0.17 | 0.72 ± 0.06 | 1.51 |
| Luvisol (agricultural) | 0.21 ± 0.12 | 0.13 ± 0.09 | 0.34 |
| Phaeozem (natural) | 0.50 ± 0.08 | 0.04 ± 0.03 | 0.54 |
| Solodic Chernozem (natural) | 0.38 ± 0.03 | 0.14 ± 0.07 | 0.52 |
| Solonetz (natural) | 0.04 ± 0.00 | 0.00 | 0.04 |
| Chernozem (natural) | 0.27 ± 0.04 | 0.06 ± 0.03 | 0.33 |
| Chernozem (agricultural) | 0.17 ± 0.02 | 0.01 ± 0.01 | 0.18 |
| Gleyic Kastanozem (natural) | 0.07 ± 0.01 | 0.00 | 0.07 |
| Kastanozem (natural) | 0.21 ± 0.02 | 0.09 ± 0.04 | 0.30 |
| Kastanozem (agricultural) | 0.10 ± 0.03 | 0.03 ± 0.01 | 0.13 |
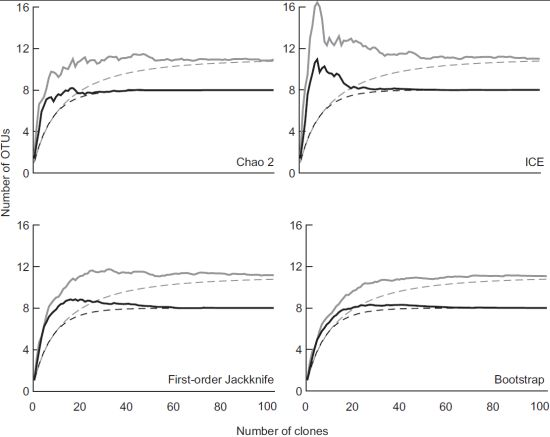
3.2. Diversity of Methanotrophs in Unmanaged and Managed Soils


4. Conclusions
Acknowledgements
Conflict of Interest
References
- Wardle, D.A.; Bardgett, R.D.; Klironomos, J.N.; Setälä, H.; van der Putten, W.H.; Wall, D.H. Ecological linkages between aboveground biota. Science 2004, 304, 1629–1633. [Google Scholar] [CrossRef]
- Dick, R.P. A review: Long-term effects of agricultural systems on soil biochemical and microbial parameters. Agric. Ecosyst. Environ. 1992, 40, 25–36. [Google Scholar] [CrossRef]
- Birkhofer, K.; Schöning, I.; Alt, F.; Herold, N.; Klarner, B.; Maraun, M.; Marhan, S.; Oelmann, Y.; Wubet, T.; Yurkov, A.; et al. General relationships between abiotic soil properties and soil biota across spatial scales and different land-use types. PLoS One 2012, 7, e43292. [Google Scholar] [CrossRef]
- Yurkov, A.M.; Kemler, M.; Begerow, D. Assessment of yeast diversity in soils under different management regimes. Fungal Ecol. 2012, 5, 24–35. [Google Scholar] [CrossRef]
- IPCC, Climate Change 2007: Synthesis Report; Cambridge University Press: Cambridge, UK, 2007.
- Mancinelli, R.L. The regulation of methane oxidation in soil. Annu. Rev. Microbiol. 1995, 49, 581–605. [Google Scholar] [CrossRef]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar]
- Stoecker, K.; Bendinger, B.; Schoning, B.; Nielsen, P.H.; Nielsen, J.L.; Baranyi, C.; Toenshoff, E.R.; Daims, H.; Wagner, M. Cohn’s Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc. Natl. Acad. Sci. USA 2006, 103, 2363–2367. [Google Scholar] [CrossRef]
- Vigliotta, G.; Nutricati, E.; Carata, E.; Tredici, S.M.; De Stefano, M.; Pontieri, P.; Massardo, D.R.; Prati, M.V.; de Bellis, L.; Alifano, P. Clonothrix fusca Roze 1896, a filamentous, sheathed, methanotrophic γ-Proteobacterium. Appl. Environ. Microbiol. 2007, 73, 3556–3565. [Google Scholar] [CrossRef]
- Pol, A.; Heijmans, K.; Harhangi, H.R.; Tedesco, D.; Jetten, M.S.; Op den Camp, H.J. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 2007, 450, 874–878. [Google Scholar] [CrossRef]
- Op den Camp, H.J.; Islam, M.T.; Stott, M.B.; Harhangi, H.R.; Hynes, A.; Schouten, S.; Jetten, M.S.M.; Birkeland, N.K.; Pol, A.; Dunfield, P.F. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 2009, 1, 293–306. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Knief, C.; Dunfield, P.F. Methylocella species are facultatively methanotrophic. J. Bacteriol. 2005, 187, 4665–4670. [Google Scholar] [CrossRef]
- Vorobev, A.V.; Baani, M.; Doronina, N.V.; Brady, A.L.; Liesack, W.; Dunfield, P.F.; Dedysh, S.N. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int. J. Syst. Evol. Microbiol. 2011, 61, 2456–2463. [Google Scholar] [CrossRef]
- McDonald, I.R.; Murrel, C.J. The particulate methane monooxygenase gene pmoA and its use as a functional gene probe for methanotrophs. FEMS Microbiol. Lett. 1997, 156, 205–210. [Google Scholar] [CrossRef]
- Dumont, M.G.; Murrell, J.C. Community-level analysis: Key genes of aerobic methane oxidation. Methods Enzymol. 2005, 397, 413–427. [Google Scholar] [CrossRef]
- Heyer, J.; Galchenko, V.F.; Dunfield, P.F. Molecular phylogeny of type II methane-oxidizing bacteria isolated from various environments. Microbiology 2002, 148, 2831–2846. [Google Scholar]
- Schnell, S.; King, G.M. Stability of methane oxidation capacity to variations in methane and nutrient concentrations. FEMS Microbiol. Ecol. 1995, 17, 285–294. [Google Scholar] [CrossRef]
- Baani, M.; Liesack, W. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc. Natl. Acad. Sci. USA 2008, 105, 10203–10208. [Google Scholar] [CrossRef]
- Semenov, V.M.; Kravchenko, I.K.; Kuznetsova, T.V.; Semenova, N.A.; Bykova, S.A.; Dulov, L.E.; Gal’chenko, V.F.; Pardini, G.; Gispert, M.; Boeckx, P.; et al. Seasonal dynamics of atmospheric methane oxidation in gray forest soils. Microbiology 2004, 73, 356–362. [Google Scholar] [CrossRef]
- Rusanov, I.; Savvichev, A.; Yusupov, S.; Pimenov, N.; Ivanov, M. Exometabolite formation during microbial oxidation of methane in marine ecosystems. Microbiology 1998, 5, 710–717. [Google Scholar]
- Hutchens, E.; Radajewski, S.; Dumont, M.G.; McDonald, I.R.; Murrell, J.C. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 2004, 6, 111–120. [Google Scholar]
- Holmes, A.J.; Roslev, P.; McDonald, I.R.; Iversen, N.; Henriksen, K.; Murrell, J.C. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl. Environ. Microbiol. 1999, 65, 3312–3318. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. 1999, 41, 95–98. [Google Scholar]
- Silvestro, D.; Michalak, I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2012, 12, 335–337. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- EstimateS: Biodiversity Estimations. Available online: http://purl.oclc.org/estimates (accessed on 6 June 2013).
- Yurkov, A.M.; Kemler, M.; Begerow, D. Species accumulation curves and incidence-based species richness estimators to appraise the diversity of cultivable yeasts from beech forest soils. PLoS One 2011, 6, e23671. [Google Scholar] [CrossRef]
- Whalen, S.C.; Reeburgh, W.S. Consumption of atmospheric methane by tundra soils. Nature 1990, 346, 160–162. [Google Scholar] [CrossRef]
- Whalen, S.C.; Reeburgh, W.S.; Barber, V.A. Oxidation of methane in boreal forest soils: A comparison of seven measures. Biogeochemistry 1992, 16, 181–211. [Google Scholar]
- Zhou, X.Q.; Wang, Y.F.; Huang, X.Z.; Hao, Y.B.; Tian, J.Q.; Wang, J.Z. Effects of grazing by sheep on the structure of methane-oxidizing bacterial community of steppe soil. Soil Biol. Biochem. 2008, 40, 258–261. [Google Scholar] [CrossRef]
- Luo, G.J.; Kiese, R.; Wolf, B.; Butterbach-Bahl, K. Effects of soil temperature and moisture on methane uptake and nitrous oxide emissions across three different ecosystem types. Biogeosciences 2013, 10, 3205–3219. [Google Scholar] [CrossRef]
- Mosier, A.; Schimel, D.; Valentine, D.; Bronson, K.; Parton, W. Methane and nitrous oxide fluxes in native, fertilized and cultivated grasslands. Nature 1991, 350, 330–332. [Google Scholar] [CrossRef]
- Maxfield, P.J.; Hornibrook, E.R.C.; Evershed, R.P. Acute impact of agriculture on high-affinity methanotrophic bacterial populations. Environ. Microbiol. 2008, 10, 1917–1924. [Google Scholar] [CrossRef]
- Dorr, N.; Glaser, B.; Kolb, S. Methanotrophic communities in Brazilian ferralsols from naturally forested, afforested, and agricultural sites. Appl. Environ. Microbiol. 2010, 76, 1307–1310. [Google Scholar] [CrossRef]
- Kravchenko, I.K.; Semenov, V.M.; Kuznetsova, T.V.; Bykova, S.A.; Dulov, L.E.; Pardini, G.; Gispert, M.; Boeckx, P.; Van Cleemput, O.; Gal’chenko, V.F. Physicochemical and biological factors affecting atmospheric methane oxidation in gray forest soils. Microbiology 2004, 74, 255–260. [Google Scholar]
- Dunfield, P.F.; Knowles, R. Kinetics of inhibition of methane oxidation by nitrate, nitrite, and ammonium in a humisol. Appl. Environ. Microbiol. 1995, 61, 3129–3135. [Google Scholar]
- Holmes, A.J.; Costello, A.; Lidstrom, M.E.; Murrell, J.C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 1995, 132, 203–208. [Google Scholar] [CrossRef]
- Bedard, C.; Knowles, R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol. Mol. Biol. Rev. 1989, 53, 68–84. [Google Scholar]
- Degelmann, D.M.; Borken, W.; Drake, H.L.; Kolb, S. Different atmospheric methane-oxidizing communities in European beech and Norway spruce soils. Appl. Environ. Microbiol. 2010, 76, 3228–3235. [Google Scholar] [CrossRef]
- McDonald, I.R.; Bodrossy, L.; Chen, Y.; Murrell, J.C. Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Environ. Microbiol. 2008, 74, 1305–1315. [Google Scholar] [CrossRef]
- Lontoh, S.; Semrau, J.D. Methane and trichloroethylene degradation by Methylosinus trichosporium OB3b expressing particulate methane monooxygenase. Appl. Environ. Microbiol. 1998, 64, 1106–1114. [Google Scholar]
- Nauer, P.A.; Dam, B.; Liesack, W.; Zeyer, J.; Schroth, M.H. Activity and diversity of methane-oxidizing bacteria in glacier forefields on siliceous and calcareous bedrock. Biogeosciences 2012, 9, 2259–2274. [Google Scholar] [CrossRef]
- Kravchenko, I.K.; Kizilova, A.K.; Bykova, S.A.; Menko, E.V.; Galchenko, V.F. Molecular analysis of high-affinity methane-oxidizing enrichment cultures isolated from a forest biocenosis and agrocenoses. Microbiology 2010, 79, 106–114. [Google Scholar] [CrossRef]
- King, G.M.; Nanba, K. Distribution of atmospheric methane oxidation and methanotrophic communities on Hawaiian volcanic deposits and soils. Microbes Environ. 2008, 23, 326–330. [Google Scholar] [CrossRef]
- Henckel, T.; Roslev, P.; Conrad, R. Effects of O2 and CH4 on presence and activity of the indigenous methanotrophic community in rice field soil. Environ. Microbiol. 2000, 2, 666–679. [Google Scholar] [CrossRef]
- Barcena, T.G.; Finster, K.W.; Yde, J.C. Spatial patterns of soil development, methane oxidation, and methanotrophic diversity along a receding glacier forefield, Southeast Greenland. Arct. Antarct. Alp. Res. 2011, 43, 178–188. [Google Scholar] [CrossRef]
- Kolb, S. The quest for atmospheric methane oxidizers in forest soils. Environ. Microbiol. Rep. 2009, 1, 336–346. [Google Scholar] [CrossRef]
- Pacheco-Oliver, M.; McDonald, I.R.; Groleau, D.; Murrell, J.C.; Miguez, C.B. Detection of methanotrophs with highly divergent pmoA genes from Arctic soils. FEMS Microbiol. Lett. 2002, 209, 313–319. [Google Scholar] [CrossRef]
- Ricke, P.; Kolb, S.; Braker, G. Application of a newly developed ARB-integrated in silico T-RFLP tool reveals the dominance of a novel pmoA cluster in a forest soil. Appl. Environ. Microbiol. 2005, 71, 1671–1673. [Google Scholar] [CrossRef]
- Uchiyama, H.; Nakajima, T.; Yagi, O.; Tabuchi, T. Aerobic degradation of trichloroethylene at high concentration by a methane-utilizing mixed culture. Agric. Biol. Chem. 1989, 53, 1019–1024. [Google Scholar] [CrossRef]
- Bussmann, I.; Rahalkar, M.; Schink, B. Cultivation of methanotrophic bacteria in opposing gradients of methane and oxygen. FEMS Microbiol. Ecol. 2006, 56, 331–344. [Google Scholar] [CrossRef]
- Smith, K.S.; Costello, A.M.; Lidstrom, M.E. Methane and trichloroethylene oxidation by an estuarine methanotroph, Methylobacter sp. strain BB5.1. Appl. Environ. Microbiol. 1997, 63, 4617–4620. [Google Scholar]
- Knief, C.; Lipski, A.; Dunfield, P.F. Diversity and activity of methanotrophic bacteria in different upland soils. Appl. Environ. Microbiol. 2003, 69, 6703–6714. [Google Scholar] [CrossRef]
- Nercessian, O.; Bienvenu, N.; Moreira, D.; Prieur, D.; Jeanthon, C. Diversity of functional genes of methanogens, methanotrophs and sulfate reducers in deep-sea hydrothermal environments. Environ. Microbiol. 2005, 7, 118–132. [Google Scholar] [CrossRef]
- Cebron, A.; Bodrossy, L.; Chen, Y.; Singer, A.C.; Thompson, I.P.; Prosser, J.I.; Murrell, J.C. Identity of active methanotrophs in landfill cover soil as revealed by DNA-stable isotope probing. FEMS Microbiol. Ecol. 2007, 62, 12–23. [Google Scholar] [CrossRef]
- Fjellbirkeland, A.; Torsvik, V.; Øvreås, L. Methanotrophic diversity in an agricultural soil as evaluated by denaturing gradient gel electrophoresis profiles of pmoA, mxaF and 16S rDNA sequences. Antonie Van Leeuwenhoek 2001, 79, 209–217. [Google Scholar] [CrossRef]
- Shrestha, M.; Abraham, W.R.; Shrestha, P.M.; Noll, M.; Conrad, R. Activity and composition of methanotrophic bacterial communities in planted rice soil studied by flux measurements, analyses of pmoA gene and stable isotope probing of phospholipid fatty acids. Environ. Microbiol. 2008, 10, 400–412. [Google Scholar] [CrossRef]
- Bodelier, P.L.E.; Roslev, P.; Henckel, T.; Frenzel, P. Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots. Nature 2000, 403, 421–424. [Google Scholar] [CrossRef]
- Noll, M.; Frenzel, P.; Conrad, R. Selective stimulation of type I methanotrophs in a rice paddy soil by urea fertilization revealed by RNA-based stable isotope probing. FEMS Microbiol. Ecol. 2008, 65, 125–132. [Google Scholar] [CrossRef]
- Dunfield, P.F. The soil methane sink. In Greenhouse Gas Sinks; Reay, D., Hewitt, C.N., Smith, K., Grace, J., Eds.; CAB International: Wallingford, UK, 2007; pp. 152–170. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kizilova, A.; Yurkov, A.; Kravchenko, I. Aerobic Methanotrophs in Natural and Agricultural Soils of European Russia. Diversity 2013, 5, 541-556. https://doi.org/10.3390/d5030541
Kizilova A, Yurkov A, Kravchenko I. Aerobic Methanotrophs in Natural and Agricultural Soils of European Russia. Diversity. 2013; 5(3):541-556. https://doi.org/10.3390/d5030541
Chicago/Turabian StyleKizilova, Anna, Andrey Yurkov, and Irina Kravchenko. 2013. "Aerobic Methanotrophs in Natural and Agricultural Soils of European Russia" Diversity 5, no. 3: 541-556. https://doi.org/10.3390/d5030541
APA StyleKizilova, A., Yurkov, A., & Kravchenko, I. (2013). Aerobic Methanotrophs in Natural and Agricultural Soils of European Russia. Diversity, 5(3), 541-556. https://doi.org/10.3390/d5030541