Assessing the Diversity and Composition of Bacterial Communities across a Wetland, Transition, Upland Gradient in Macon County Alabama
Abstract
:1. Introduction
2. Experimental Section
2.1. Sites and Sampling
2.2. Soil pH and SOC
2.3. Soil Enzyme Activity
2.4. DNA Extraction, Amplification, and Sequencing
2.5. Bioinformatics and Statistical Analysis
3. Results
3.1. Soil Properties
3.1.1. Physio- and Biochemical Properties
3.1.2. Bacterial Richness
| USS | TSS | WSS | |
|---|---|---|---|
| Soil Chemical Properties | |||
| pH | 5.21a | 4.91 | 5.03a |
| SOC | 1.57a | 0.73 | 1.67a |
| Soil Enzymatic Properties | |||
| ACP | 1.10a | 0.50 | 1.41a |
| APA | 0.09 | 0.04 | 0.24a |
| Soil Bacterial Diversity/Richness* | |||
| Shannon | 5.38 | 4.80 | 5.20 |
| Sobs | 1105 | 863 | 1040 |
| Chao1 | 3530 | 2385 | 3545 |
| ACE | 7126 | 4449 | 6960 |
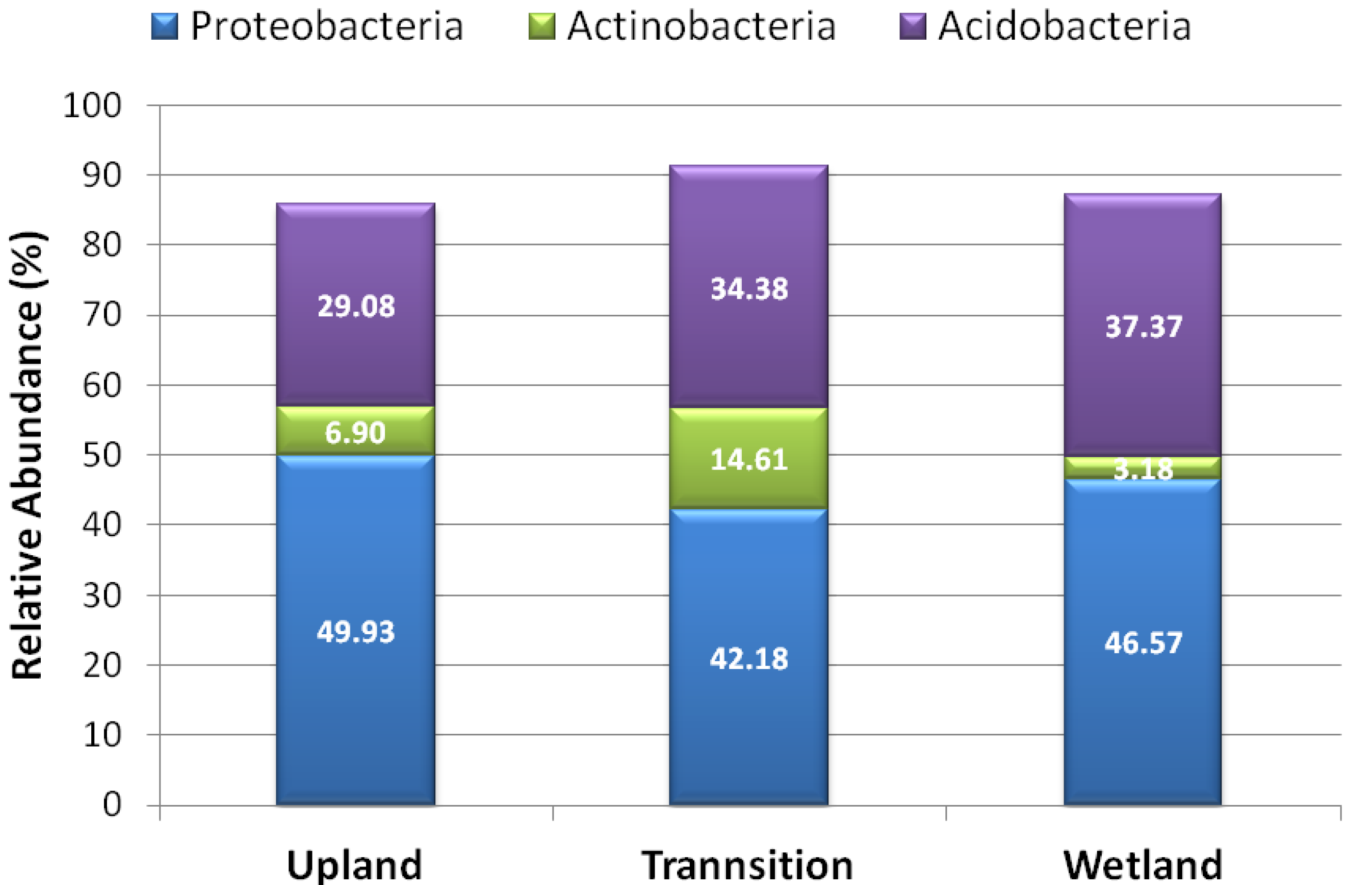
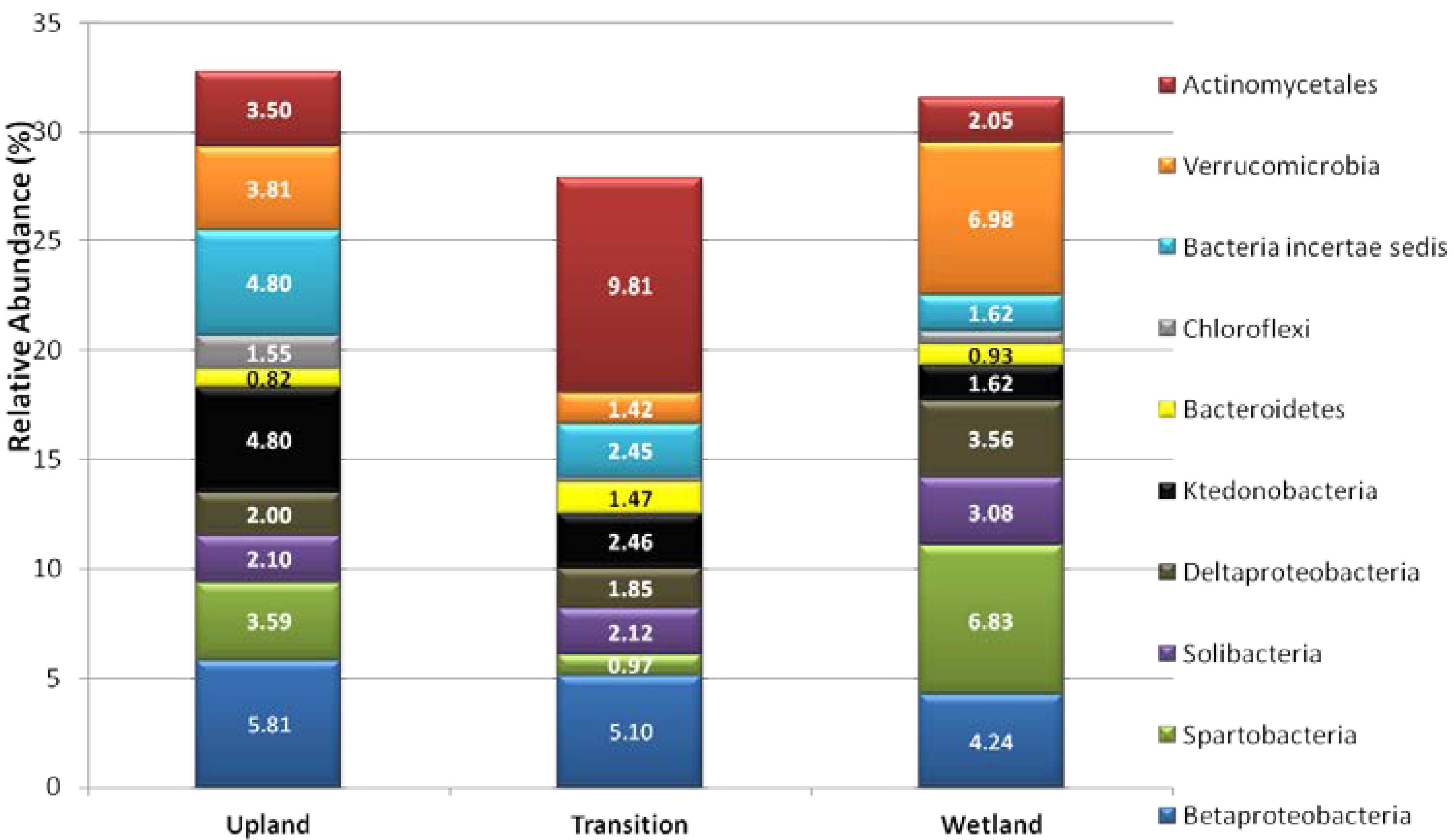
3.2. Relative Abundance of Selected Taxa
| Species | Upland | Transition | Wetland |
|---|---|---|---|
| Acidiphilium cryptum | 0.67a | 1.62b | 1.37ab |
| Acidisphaera rubrifaciens | 0.34a | 1.61b | 0.57ab |
| Acidothermus cellulolyticus | 0.34a | 0.97b | 0.32ab |
| Caldilinea sp | 1.06a | 0.09 | 0.31 |
| Candidatus Reyranella sp | 1.82 | 0.92 | 0.40 |
| Conexibacter woesei | 0.34ab | 1.45a | 0.02b |
| Cryptosporangium minutisporangium | 0.19 | 0.93a | 0.04 |
| Marinobacter excellens | 1.51ab | 0.61a | 2.17b |
| Mycobacterium holsaticum | 0.05 | 1.47a | 0.00 |
| Nonomuraea kuesteri | 0.24 | 1.04a | 0.09 |
| Rhodoplanes roseus | 3.88 | 3.75 | 1.46a |
| Syntrophus aciditrophicus | 0.41 | 0.13 | 1.30a |
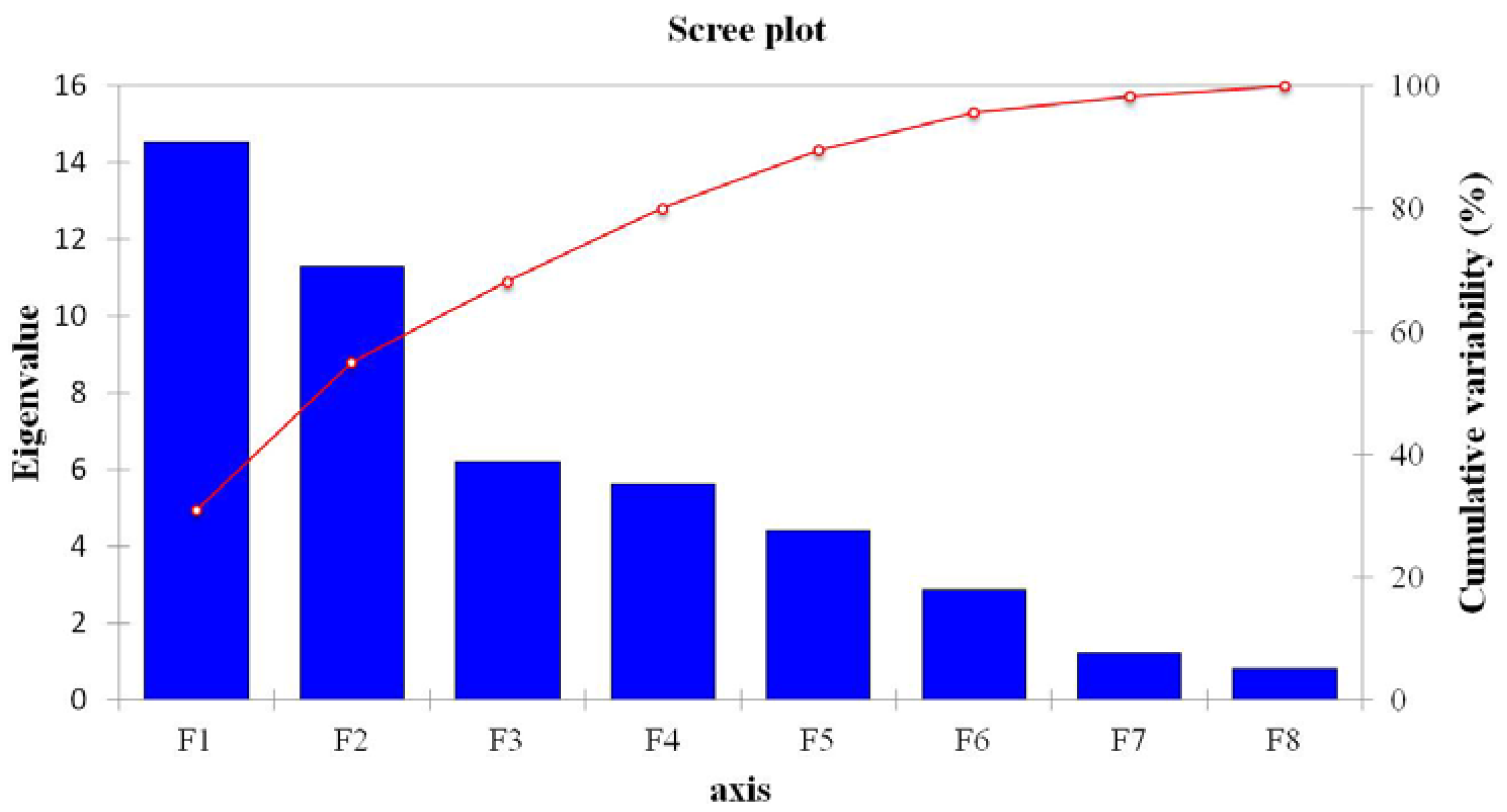
3.3. Contributions of Bacterial Taxa to Principal Components
| F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | |
|---|---|---|---|---|---|---|---|---|
| Eigenvalue | 14.555 | 11.286 | 6.220 | 5.617 | 4.414 | 2.881 | 1.236 | 0.791 |
| Variability (%) | 30.969 | 24.013 | 13.233 | 11.951 | 9.392 | 6.129 | 2.630 | 1.683 |
| Cumulative (%) | 30.969 | 54.982 | 68.215 | 80.166 | 89.558 | 95.688 | 98.317 | 100.000 |

4. Discussion
4.1. Soil Chemical, Biochemical, and Biological Properties
4.2. Bacterial Community Membership
4.3. Clustering of Samples According to Community Structure
5. Conclusions
Acknowledgments
Conflict of Interest
References
- Costanza, R.; d’Arge, R.; de Groot, R.; Farber, S.; Grasso, M.; Hannon, B.; Limburg, K.; Naeem, S.; O’Neill, R.V.; Paruelo, J.; et al. The value of the world’s ecosystem services and natural capital. Nature 1997, 387, 253–260. [Google Scholar] [CrossRef]
- Dahl, T.E. Wetlands Losses in the United States 1780’S to 1980’s; Fish and Wildlife Service: Washington, DC, USA, 1990; p. 13. [Google Scholar]
- Dahl, T.E. Status and Trends of Wetlands in the Conterminous United States 1998 to 2004; Fish and Wildlife Service: Washington, DC, USA, 2006; p. 112. [Google Scholar]
- Lilly, J.P. Soil Facts Wetland Issues. Available online: http://www.soil.ncsu.edu/publications/Soilfacts/AG-439-26/AG-439-26.pdf (accessed on 15 April 2013).
- Whittecar, G.R.; Daniels, W.L. Use of hydrogeomorphic concepts to design created wetlands in southeaster Virginia. Geomorphology 1999, 31, 355–371. [Google Scholar] [CrossRef]
- Faulkner, S.P.; Patrick, W.H., Jr.; Gambrell, R.P. Field techniques for measuring wetland soil parameters. Soil Sci. Soc. Am. J. 1989, 53, 883–890. [Google Scholar] [CrossRef]
- Stolt, M.H.; Genthner, M.H.; Daniels, W.L.; Groover, V.A.; Nagle, S.; Haering, K.C. Comparison of soil and other environmental conditions in constructed and adjacent palustrine reference wetlands. Wetlands 2000, 20, 671–683. [Google Scholar]
- Campbell, D.A.; Cole, C.A.; Brooks, R.P. A comparison of created and natural wetlands in Pennsylvania, USA. Wetl. Ecol. Manag. 2002, 10, 41–49. [Google Scholar]
- Bruland, G.L.; Richardson, C.J. Comparison of soil organic matter in created, restored and paired natural wetlands in North Carolina. Wetl. Ecol. Manag. 2006, 14, 245–251. [Google Scholar] [CrossRef]
- Acosta-Martinez, V.; Dowd, S.; Yung, S.; Allen, V. Tag encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 2008, 40, 2762–2770. [Google Scholar]
- Shange, R.S.; Ankumah, R.O.; Ibekwe, A.M.; Zabawa, R.; Dowd, S.E. Distinct soil bacterial communities revealed under a diversely managed agroecosystem. PLoS One 2012, 7, e40338. [Google Scholar]
- Dick, W.A.; Ali Tabatabai, M. Significance and Potential Uses of Soil Enzymes. In Soil Microbial Ecology: Applications in Agricultural and Environmental Management; Metting, F.B., Ed.; Marcel Dekker, Inc.: New York, NY, USA, 1992; pp. 95–127. [Google Scholar]
- Acosta-Martínez, V.; Cruz, L.; Sotomayor-Ramirez, D.; Pérez-Alegría, L. Enzyme activities as affected by soil properties and land use in a tropical watershed. Appl. Soil Ecol. 2007, 35, 35–45. [Google Scholar] [CrossRef]
- Shange, R.S.; Ankumah, R.O.; Githinji, L.; Zabawa, R. Spatial assessment of selected soil properties within an industrial poultry production site. Air Soil Water Res. 2012, 5, 59–68. [Google Scholar]
- Davidson, E.A.; Ackerman, I.L. Changes in soil carbon inventories following cultivation of previously untilled soils. Biogeochemistry 1993, 20, 161–193. [Google Scholar]
- Mann, L.K. Changes in soil carbon after cultivation. Soil Sci. 1986, 142, 279–288. [Google Scholar] [CrossRef]
- Schlesinger, W.H. Changes in Soil Carbon Storage and Associated Properties with Disturbance and Recovery. In The Changing Carbon Cycle: A Global Analysis; Trabalka, J.R., Reichle, D.E., Eds.; Springer-Verlag: Berlin, Germany, 1985; pp. 194–220. [Google Scholar]
- Post, W.M.; Mann, L.K. Changes in Soil Organic Carbon and Nitrogen as a Result of Cultivation. In Soils and the Greenhouse Effect; Bouwman, A.F., Ed.; John Wiley & Sons: New York, NY, USA, 1990; pp. 401–406. [Google Scholar]
- Mankolo, R.; Reddy, C.; Senwo, Z.; Nyakatawa, E.; Sajjala, S. Soil biochemical changes induced by poultry litter application and conservation tillage under cotton production systems. Agronomy 2012, 2, 187–198. [Google Scholar] [CrossRef]
- Song, Y.; Song, C.; Yang, G.; Miao, Y.; Wang, J.; Guo, Y. Changes in labile organic carbon fractions and soil enzyme activities after marshland reclamation and restoration in the Sanjiang Plain in Northeast China. Environ. Manag. 2012, 50, 418–426. [Google Scholar] [CrossRef]
- McArthur, J.V. Bacteria as Biomonitors. In Bioassessment and Management of North American Fresh Water Wetlands; Rader, R.B., Batzer, D.P., Wissinger, S.A., Eds.; John Wiley & Sons: New York, NY, USA, 2001; pp. 249–261. [Google Scholar]
- Van der Heijden, M.G.; Bardgett, R.D.; van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 3, 296–310. [Google Scholar] [CrossRef]
- Wright, A.L.; Reddy, K.R. Phosphorus lading effects on extracellular enzyme activity in Everglades wetland soils. Soil Sci. Soc. Am. J. 2001, 65, 588–595. [Google Scholar] [CrossRef]
- Soil Survey of Macon County Alabama. Available online: http://soildatamart.nrcs.usda.gov/Manuscripts/AL087/0/Macon.pdf (accessed on 15 April 2013).
- Tabatabai, M.A. Enzymes. In Methods of Soil Analysis, Part 2: Microbiological and Biochemical properties; Weaver, R.W., Augle, S., Bottomly, P.J., Bezdicek, D., Smith, S., Tabatabai, M.A., Wollum, A., Eds.; Soil Science Society of America: Madison, WI, USA, 1994; pp. 775–833. [Google Scholar]
- Bandick, A.K.; Dick, R.P. Field management effects on soil enzyme activities. Soil Biol. Biochem. 1999, 31, 1471–1479. [Google Scholar] [CrossRef]
- Elsgaard, L.; Anderson, G.H.; Eriksen, J. Measurement of arylsulphatase activity in agricultural soils using a simplified assay. Soil Biol. Biochem. 2002, 34, 79–82. [Google Scholar] [CrossRef]
- Dowd, S.E.; Sun, Y.; Secor, P.R.; Rhoads, D.D.; Wolcott, B.M.; James, G.A.; Wolcott, R.D. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008, 8, 43. [Google Scholar] [CrossRef]
- Dowd, S.E.; Sun, Y.; Wolcott, R.D.; Carroll, J.A. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: Bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog. Dis. 2008, 5, 459–472. [Google Scholar] [CrossRef]
- Gontcharova, V.; Youn, E.; Wolcott, R.D.; Hollister, E.B.; Gentry, T.J.; Dowd, S.E. Black Box Chimera Check (B2C2): A windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene datasets. Open Microbiol. J. 2010, 4, 47–52. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: open source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Roesch, L.F.; Fulthrope, R.R.; Riva, A.; Casella, G.; Hadwin, A.K.M.; Kent, A.D.; Ddaroub, S.H.; Camargo, F.A.; Farmerie, W.G.; Triplett, E.W. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007, 1, 283–290. [Google Scholar]
- Cole, J.R.; Chai, B.; Farris, R.J.; Wang, Q.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Bandela, A.M.; Cardenas, E.; Garrity, G.M.; Tiedje, J.M. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007, 35, D169–D172. [Google Scholar] [CrossRef]
- Chao, A.; Ma, M.C.; Yang, M.C.K. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika 1993, 80, 193–201. [Google Scholar] [CrossRef]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Clymo, R.S.; Reddaway, E.J.F. Productivity of Sphagnum (bog-moss) and peat accumulation. Hydrobiologia 1971, 12, 181–192. [Google Scholar]
- Mausbach, M.J.; Richardson, J.L. Biogeochemical processes in hydric soil formation. Curr. T. Wetl. Biogeochem. 1994, 1, 68–127. [Google Scholar]
- Pimentel, D.; Harvey, C.; Resosudarmo, P.; Sinclair, K.; Kunz, D.; McNair, M.; Crist, S.; Shpritz, L.; Fitton, L.; Sa.ouri, R.; et al. Environmental and economic costs soil erosion and conservation benefts. Science 1995, 267, 1117–1123. [Google Scholar] [CrossRef]
- Hartanto, H.; Prabhu, R.; Widayat, A.S.E.; Asdak, C. Factors affecting runoff and soil erosion: Plot-Level soil loss monitoring for assessing sustainability of forest management. For. Ecol. Manage 2003, 180, 361–374. [Google Scholar] [CrossRef]
- Gardner, T.; Acosta-Martinez, V.; Calderón, F.; Zobeck, T.; Baddock, M.; Van Pelt, R.; Senwo, Z.; Dowd, S.; Cox, S. Pyrosequencing reveals bacteria carried in different wind-eroded sediments. J. Environ. Qual 2011, 41, 744–53. [Google Scholar]
- Spain, A.M.; Krumholz, L.R.; Elshahed, M.S. Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 2009, 3, 992–1000. [Google Scholar] [CrossRef]
- Kersters, K.; de Vos, P.; Gillis, M.; Swings, J.; Vandamme, P.; Stackebrandt, E. Introduction to the Proteobacteria. In The Prokaryotes: A Handbook on the Biology of Bacteria; Dwarkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 3–37. [Google Scholar]
- Gardner, T.G.; Acosta-Martínez, V.; Senwo, Z.; Dowd, S.E. Soil rhizosphere microbial communities and enzyme activities under organic farming in Alabama. Diversity 2011, 3, 308–328. [Google Scholar]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Jangid, K.; Williams, M.A.; Franzluebbers, A.J.; Sanderlin, J.S.; Reeves, J.H.; Jenkins, M.B.; Endale, D.M.; Coleman, D.C.; Whitman, W.B. Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biol. Biochem. 2008, 40, 2843–2853. [Google Scholar] [CrossRef]
- Nacke, H.; Thurmer, A.; Wollherr, A.; Will, C.; Hodac, L.; Herold, N.; Schöning, I.; Schrumpf, M.; Daniel, R. Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS One 2011, 6, e17000. [Google Scholar] [CrossRef]
- Acosta-Martínez, V.; Dowd, S.E.; Sun, Y.; Webster, D.; Allen, V.G. Pyrosequencing analysis for characterization of soil bacterial populations as affected by an integrated livestock-cotton production system. Appl. Soil Ecol. 2010, 45, 13–25. [Google Scholar] [CrossRef]
- Empadinhas, N.; da Costa, M.S. Diversity, biological roles and biosynthetic pathways for sugar-glycerate containing compatible solutes in bacteria and Archaea. Environ. Microbiol. 2011, 13, 2056–2077. [Google Scholar] [CrossRef]
- Bergmann, G.; Bates, S.; Eilers, K.; Lauber, C.; Caporaso, J.; Walters, W.; Knight, R.; Fierer, N. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol. Biochem. 2011, 43, 1450–1455. [Google Scholar] [CrossRef]
- Singh, B.K.; Bardgett, R.D.; Smith, P.; Reay, D.S. Microorganisms and climate change: Terrestrial feedbacks and mitigation options. Nat. Rev. Microbiol. 2010, 8, 779–790. [Google Scholar] [CrossRef]
- Dedysh, S.N. Cultivating uncultured bacteria from northern wetlands: Knowledge gained and remaining gaps. Front. Microbiol. 2011, 2, 184. [Google Scholar] [CrossRef]
- Janssen, P.H. Identifying the dominant soil bacteria taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.M.; Piceno, Y.M.; DeSantis, F.Z.; Andersen, G.L.; Bakker, P.; et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Bardhan, S.; Jose, S.; Jenkins, M.A.; Webster, C.R.; Udawatta, R.P.; Stehn, S.E. Microbial community diversity and composition across a gradient of soil acidity in spruce–fir forests of the southern Appalachian Mountains. Appl. Soil Ecol. 2012, 61, 60–68. [Google Scholar] [CrossRef]
- Burke, R.A.; Molina, M.; Cox, J.E.; Osher, L.J.; Piccolo, M.C. Stable carbon isotope ratio and composition of microbial fatty acids in tropical soils. J. Environ. Qual. 2003, 32, 198–206. [Google Scholar]
- Waldrop, M.P.; Balser, T.C.; Firestone, M.K. Linking microbial community composition to function in a tropical soil. Soil Biol. Biochem. 2000, 32, 1837–1846. [Google Scholar] [CrossRef]
- Hill, P.; Kristufek, V.; Dijfkhuizen, L.; Boddy, C.; Kroetsch, D.; van Elsas, J.D. Land use intensity controls actinobacterial community structure. Microb. Ecol. 2011, 61, 286–302. [Google Scholar] [CrossRef]
- Hartman, W.H.; Richardson, C.J.; Vilgalys, R.; Bruland, G.L. Environmental and anthropogenic controls of bacterial communities in wetland soils. Proc. Natl. Acad. Sci. USA 2008, 105, 17842–17847. [Google Scholar] [CrossRef]
- Lazzarini, A.; Cavaletti, L.; Toppo, G.; Marinelli, F. Rare genera of actinomycetes as potential producers of new antibiotics. Antonie van Leeuwenhoek 2000, 78, 399–405. [Google Scholar] [CrossRef]
- Lee, J.P.; Hwang, B.Y. Diversity of antifungal actinomycetes in various vegetative soils of Korea. Can. J. Microbiol. 2002, 48, 407–417. [Google Scholar] [CrossRef]
- Bouizgarne, B.; El Hadrami, I.; Ouhdouch, Y. Novel production of isochainin by a strain of Streptomyces sp. isolated from rhizosphere soil of the indigenous Moroccan plant Argania. spinosa L. World J. Microbiol. Biotechnol. 2006, 22, 423–429. [Google Scholar] [CrossRef]
- Nurjasmi, R.; Widada, J. Diversity of actinomycetes at several forest types in Wanagama I Yogyakarta and their potency as a producer of antifungal compound. Indo J. Biotech. 2009, 14, 1196–1205. [Google Scholar]
- Fierer, N.; Schimel, J.; Holden, P. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 2003, 35, 167–176. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Shange, R.; Haugabrooks, E.; Ankumah, R.; Ibekwe, A.M.; Smith, R.C.; Dowd, S. Assessing the Diversity and Composition of Bacterial Communities across a Wetland, Transition, Upland Gradient in Macon County Alabama. Diversity 2013, 5, 461-478. https://doi.org/10.3390/d5030461
Shange R, Haugabrooks E, Ankumah R, Ibekwe AM, Smith RC, Dowd S. Assessing the Diversity and Composition of Bacterial Communities across a Wetland, Transition, Upland Gradient in Macon County Alabama. Diversity. 2013; 5(3):461-478. https://doi.org/10.3390/d5030461
Chicago/Turabian StyleShange, Raymon, Esther Haugabrooks, Ramble Ankumah, Abasiofiok M. Ibekwe, Ronald C. Smith, and Scot Dowd. 2013. "Assessing the Diversity and Composition of Bacterial Communities across a Wetland, Transition, Upland Gradient in Macon County Alabama" Diversity 5, no. 3: 461-478. https://doi.org/10.3390/d5030461
APA StyleShange, R., Haugabrooks, E., Ankumah, R., Ibekwe, A. M., Smith, R. C., & Dowd, S. (2013). Assessing the Diversity and Composition of Bacterial Communities across a Wetland, Transition, Upland Gradient in Macon County Alabama. Diversity, 5(3), 461-478. https://doi.org/10.3390/d5030461





