Abstract
We used near-term climate scenarios for the continental United States, to model 12 invasive plants species. We created three potential habitat suitability models for each species using maximum entropy modeling: (1) current; (2) 2020; and (3) 2035. Area under the curve values for the models ranged from 0.92 to 0.70, with 10 of the 12 being above 0.83 suggesting strong and predictable species-environment matching. Change in area between the current potential habitat and 2035 ranged from a potential habitat loss of about 217,000 km2, to a potential habitat gain of about 133,000 km2.
1. Introduction
Invasive species are a major threat to ecosystems worldwide. They play a major role in displacing native species [1,2,3] and cause deterioration of many ecosystem functions [4,5]. The spread of invasive species is the second leading threat to biodiversity following habitat destruction [5], and costs the United States up to $120 billion per year [6]. Resource managers today face the challenge of determining where an invasive species outbreak may occur, and where an invasive species will move next.
Early detection of invasive plants is of the utmost importance, especially discovering and mitigating invasions when they are small [7,8,9]. This issue becomes particularly relevant in the face of climate change [10,11]. There is a potential for the area of habitat that is suitable for any given species to shift with rapid climate change. Those populations that are on the edges of the invasion will have the potential to move with the climate change. Some areas will go from being unsuitable habitat to suitable habitat; these areas will be the leading edge of the suitable habitat. Other areas will remain stable as suitable habitat, and a final area will cease to be suitable habitat [12]. This third category of habitat does not imply that species will cease to exist in these areas where it is already established, these species may persist from the seed bank for many years in suitable micro-habitats within unsuitable areas [10,13,14]. There is also the potential for an adaptable species such as a habitat generalist to continue to adapt to new climates and not shift at all, populations of species have the opportunity to migrate, adapt, or be extirpated.
Species-environment matching models (or niche models, or species distribution models) provide a method to calculate potential suitable habitat. This method uses various algorithms to examine the habitat of a species and match that environmental space to areas where the species does not currently exist, locating areas of potential suitable habitat beyond the original data points. Hijmans and Graham [15] offer an evaluation of several niche modeling methods. Until fairly recently this type of modeling was computationally intense, and model creation could take days to weeks to complete. Increases in computing power and subsequent improvement in methods have helped develop the field of niche models from theory to practice. A review of the early methods of species distribution modeling can be found in Guisan and Zimmerman [16]. Since the turn of the century there has been an explosion in the methodology and abilities of species distribution modeling, and in the number of publications on the topic. A Web of Science search on “species distribution model*” from 1960 to 2000 produced 5,155 entries while one third of that time, 2000–2009, produced double the results, 10,296.
Species-environment matching models start from a distribution of presence points on a landscape. These data do have drawbacks; they do not provide any information on abundance or absence of the species of interest. There also may be many gaps in the data that are used. Less than 1% of any landscape can be affordably measured [17], and we must use what data we have available. These models are a starting point in an iterative process, informing managers of where potential distributions may lie and giving an idea of where to add surveys, and where to sample in the future [18]. Distribution models form the basis of an excellent first approximation map that is especially applicable for early detection programs [19].
Many techniques of climate environment matching models for invasive species do not require absence data [20]. This is useful for invasive species models because there is no guarantee that a point that is collected as an absence point is truly unsuitable habitat; it may be suitable, but the species has not yet germinated in or migrated to that location. Maximum entropy modeling (Maxent [21]) uses presence data and background data in lieu of true absence data. These models are well suited to generate maps of potential distribution and habitat suitability from current point distributions given the caveats above.
Broad scale distribution models are rapidly gaining acceptance. Morisette et al. [22] used climate envelope modeling to model current tamarisk potential habitat at a 1 km2 resolution for the continental United States using logistic regression with an Area Under the receiver operating characteristic Curve (AUC) of 0.95, but this model did not address climate change. A worldwide bullfrog model was created at a 10-minute resolution for current conditions [23]. This model had a very wide distribution, but a coarse resolution. Bradley et al. [12] created models at a 4-km2 resolution using Mahalanobis distance for five invasive plants in the United States for the year 2100, a management level geographic scale, but a very coarse time scale. We integrated aspects of these studies, by examining a management level resolution, a country level distribution, and a near term time scale.
We recognized that these maps of potential habitat suitability do not address propagule pressure, predation by natural enemies, or other biotic interactions [23]. The next step was to examine how a species might spread and where. We used the current potential suitable habitat model as a mask and developed a distance from seed source surface as a proxy for propagule pressure, a rudimentary invasibility index to address this issue.
Our objectives were to: (1) provide a strategic methodology to forecast scenarios of potential spread based on point distributions; (2) create potential distributions of invasive species with Maxent and examine the relationship of these species to their environment; and (3) consider data gaps, distance from seed source and suitable habitat (a surrogate for propagule pressure) to assess risks to invasion.
1.1. Methods
Data
We gathered point data for twelve invasive plant species in the continental United States (Table 1). These data were not exhaustive of all locations for each of these species, but were the available data from accurate sources (e.g., the Biota of North America Program, www.BONAP.org) gathered for these species. There were gaps in the data; some areas were more completely sampled than other areas. That said, points are our first and best descriptors of distributions [18]. From points we created a systematic methodology for assessing point distributions relative to environmental predictors and created models of potential suitability.

Table 1.
Species modeled. The sample size is the number of points used, after duplicates were removed. The training Area Under the Curve (AUC) measures model performance of 70% of the data. The Test AUC measures model performance on the remaining 30% of the data. The Threshold value uses the 10 percentile logistic threshold to distinguish between potentially suitable or unsuitable habitat. The area on the leading edge of the invasion is the area of potential habitat that has shifted from potentially unsuitable to potentially suitable between the current model and the 2035 model. The area on the trailing edge of the invasion is the area of habitat that has shifted from potentially suitable habitat to potentially unsuitable habitat between the current model and the 2035 model.
| Scientific name | Common name | Sample size | Training AUC | Test AUC | Threshold value | Area on Leading Edge of invasion (~km2) | Area on Trailing Edge of invasion (~km2) | Change in Area(~km2) | Habitat specialist? | Introduced after 1900? |
|---|---|---|---|---|---|---|---|---|---|---|
| Bromus tectorum | Cheat grass | 9517 | 0.84 | 0.83 | 0.43 | 131,000 | 131,000 | 0 | No | No |
| Carduus nutans | Muskthistle | 4670 | 0.88 | 0.88 | 0.39 | 372,000 | 383,000 | −11,000 | No | No |
| Celastrus orbiculatus | Oriental bittersweet | 282 | 0.84 | 0.79 | 0.24 | 103,000 | 156,000 | −53,000 | No | No |
| Centaurea stoebe | Spotted knapweed | 5899 | 0.91 | 0.90 | 0.46 | 195,000 | 245,000 | −50,000 | No | No |
| Cirsium arvense | Canada thistle | 7960 | 0.86 | 0.86 | 0.45 | 206,000 | 423,000 | −217,000 | No | No |
| Cynoglossum officinale | Houndstounge | 1884 | 0.89 | 0.88 | 0.34 | 228,000 | 219,000 | 9,000 | No | No |
| Lepidium latifolium | Perrennial pepperweed | 1015 | 0.93 | 0.91 | 0.36 | 546,000 | 451,000 | 95,000 | No | Yes |
| Linaria dalmatica | Dalmation toadflax | 1372 | 0.92 | 0.91 | 0.39 | 239,000 | 284,000 | −45,000 | No | No |
| Lonicera japonica | Japaneese honeysuckle | 2771 | 0.70 | 0.68 | 0.40 | 170,000 | 144,000 | 26,000 | No | No |
| Lythrum salicaria | Purple loosestrife | 4921 | 0.85 | 0.84 | 0.37 | 376,000 | 293,000 | 83,000 | Yes | No |
| Microstegium vimineum | Japanese stiltgrass | 321 | 0.78 | 0.68 | 0.34 | 243,000 | 110,000 | 133,000 | No | Yes |
| Pennisetum ciliare | Buffelgrass | 1876 | 0.92 | 0.91 | 0.37 | 20,000 | 20,000 | 0 | Yes | Yes |
We chose species with a broad range of current distributions, narrow to wide; recently introduced to well established; and with more than 250 known locations (Table 1). We considered two species as habitat specialists; Lythrum salicaria as it is generally confined to wetlands [24]; and Pennisetum ciliare being confined to sandy soils, that do not freeze for extended periods, with precipitation from 200 to 1200 mm per year [25]. We harvested most of the species location data from on-line sources, especially the National Institute of Invasive Species Science [26]. NIISS is a data clearinghouse on the internet that has quality control measures on the data that it ingests [27]. Each data source is listed in Appendix A.
The independent variables used for this study were 19 bioclimatic layers created using combinations of minimum and maximum temperatures and precipitation [28]. These bioclimatic layers were created as variables to capture climatic seasonality important for organisms. The climate variables for current conditions were from the DAYMET dataset for the years 1980–1997 [29]. The modeling resolution for current conditions was approximately 1-km2 pixels, the finest resolution we could find for each of the data layers over such a broad extent. The climate projection data were derived from Parameter-elevation Regressions on Independent Slopes Method (PRISM) data at a 4-m2 resolution (PRISM data available at http://www.prism.oregonstate.edu/, PRISM Group, [30]). We used predictions for 2020 and 2035 created by Jarnevich and Stohlgren [31] by extrapolating climate conditions from the PRISM data for 1895 to 2006. We chose years in the near future to be of imminent use to land managers.
Modeling Techniques
Once the data were compiled we used maximum entropy modeling (Maxent 3.2.9; [21]) to create three predictions for each species: (1) current potential habitat suitability; (2) potential habitat suitability in 2020; and (3) potential habitat suitability in 2035. Maximum entropy modeling is a machine learning method that requires only presence data. This algorithm estimated potential habitat distribution by finding the distribution of maximum entropy, or furthest from random [21]. Maxent used background data, or the environmental layers as model inputs [15]. The program removed duplicate records within a 1-km2 pixel.
We tested each species for correlations between the variables using Systat v 12 [32]. We removed variables with correlations of r ≤ −0.8 or r ≥ +0.8. The remaining variables were clipped to the counties containing data for the species, constraining the model to counties of known realized habitat [33]. These variables were used to train each model, creating a potential habitat suitability surface at approximately 1-km2 resolution for the current climate. We ran each model 25 times, withholding a different 30% of the presence locations from each model run as a test dataset for model evaluation, and averaged the results of the runs.
Then, we applied the model to the entire United States for the current climate and the climate scenarios for 2020 and 2035. For overall performance, the models were assessed using the Area Under the Receiver Operating Characteristic Curve, or AUC. This is a threshold independent indication of model performance [21]. To distinguish the threshold between suitable and unsuitable habitat for further analyses, we used the 10 percentile training presence logistic threshold as determined by Maxent (Table 1). This created an average potential suitability surface and a clamping surface for each species. The clamping surface shows the areas of the map where the model is extended beyond the climatic conditions that it was trained on, and can show areas that the model may be less reliable. We chose to mask out these locations from our analyses. Next, we used raster calculator in ArcMap [34] to calculate areas of potential habitat stability, potential habitat increase, and potential habitat decrease by comparing the current suitability map with the future suitability maps. Stable areas were defined as those with suitable habitat across all three time steps, habitat increase was defined as areas that went from unsuitable habitat in current conditions to suitable habitat in 2020 or 2035, and habitat decrease was defined as areas that went from suitable habitat in current conditions to unsuitable habitat in 2020 or 2035.
Finally, we were interested in finding a surrogate for dispersal of the species. We did not have detailed data available for dispersal mechanisms of each species, so we used distance from known seed source, or nearest data point. For this part of the analysis, we used the clamping and unsuitable habitat surfaces for each species as a mask, effectively excluding these areas from the analysis. We used the straight line distance function in Spatial Analyst of ArcMap [34] to create a surface of the distance from the nearest known presence point. This is a simple first order approximation of invasibility as defined by both suitable habitat and available propagules.
2. Results and Discussion
We had a range of available sample sizes from 282 to 9517 (Table 1), all of which represent quite a small proportion of the land in the conterminous United States. Ten of twelve species modeled exhibited excellent performance with AUC values from 0.84 to 0.93 ([35]; Table 1). Both Lonicera japonica (Japaneese honeysuckle) and Microstegium vimineum (Japanese stiltgrass) had AUC values at or below 0.70 indicating only acceptable model performance. There was little variation between training data AUC and test data AUC, suggesting repeatable and robust models (test data included 30% of the full suite of data). Each model run used a different 30% of the available data as test data, yet produced a similar result to the majority of the points modeled. The exception here, again, was M. vimineum which had a test AUC value 0.1 less than its training AUC value, possibly due to the relatively small number of data points available for the species.
The change in potentially suitable habitat area varied dramatically among species (Table 1). The largest increase in potentially suitable habitat was about 133,000 km2 for Microstegium vimineum, a species that has been introduced to the United States after 1900. The largest decrease in potential suitable habitat was about 217,000 km2 for Cirsium arvense a habitat generalist.
We achieved excellent model performance based on AUC with the data and computing power that are currently available, with the understanding that we did not capture every location of each species modeled and that the available data were not collected with probabilistic sampling. And for the locations for which we had data, this was still a resolution that may miss patchily distributed resources at the microscale, focusing mostly on large-scale climate patterns of the invaders [36]. These models were reliant not only on the quality and number of species data points available, but also on predictor layers used and the extent and resolution of the models being considered [37]. Future climate scenario models also contain their own uncertainty, especially concerning frequency of climate station data and interpolation techniques used to create a continuous surface between stations [36]. We presented first iteration models that can point managers and field crews to gaps in information and guide resource managers to suitable areas to collect more information.
This analysis defines the leading and trailing edge of invasion for these 12 species. Thuiller et al. [10] suggested looking at these leading and trailing edges, and incorporating migration into the modeling of the leading edge and persistence into the modeling on the trailing edge. Finding a way to incorporate these variables into such a large-scale model is a challenging and worthwhile task for future analysis.
The invasibility index is a simple distance function that does not include human accelerated dispersal such as commerce and trade, trucks, landscaping and disturbance facilitated invasion [38,39,40]. These are all important factors in the spread of invasive plants and excellent future projects. The models were also limited by the patchy nature of the presence data we were able to compile. Populations may exist in areas where there were data gaps, and these locations will be under-valued in the index.
Current species-environmental matching models performed well for this group of species with 10 of 12 models having AUC’s indicating excellent model performance. The two species that did not perform as well may need additional predictor layers such as canopy cover, soils, or elevation. We also recognize that we had access to large amounts of data, and the models may not perform as well with a smaller sample size (the smallest sample size was 282). However, Maxent models have been used to model rare and threatened species using even fewer than 30 data points [41]. Additionally, due to our lack of absence data, we were only able to calculate AUC as a performance metric using a semi-independent data set. Ideally, we would calculate multiple metrics and examine model performance across them all [37], while using an independent data set that may not suffer from the same biases as the dataset used to train the model.
However, given these caveats, this systematic approach has many advantages to resource managers and policy makers. The models are easily updated as information becomes available. Providing predictions of current habitat suitability, highlighting data gaps, and showing maps of clamping may entice resource managers to collect more data and amend the information that currently exists, allowing for the creation of even better models.
2.1. Climate Scenarios
Climate change in the last half-century has been highly variable in space and over time. Some areas warmed, others cooled in consistent trends for greater than 50 years in either direction. There is no reason, based on observed historical patterns, to suspect climate change in the future will be uniform in direction or intensity at local or regional scales. The standard Global Climate Model (GCM) scenarios are based on coarse-resolution global models and tend to show unidirectional trends (warming). There is no evidence of cooling for any local or regional area, despite historical observations to the contrary. We have not found any logical reason, based on observed historical patterns, to accept unidirectional projections for all areas [42]. Additionally, precipitation patterns are poorly measured, and even more-poorly modeled. Observed increases in precipitation are not uniform or unidirectional. Because of measurement and modeling errors, few scientists are confident in projections of precipitation over long periods. The uncertainty is too high to justify the use of GCMs for precipitation.
Our own analysis of climate change patterns suggests higher spatial variation in trends as shorter intervals of less than 100 years are investigated. Global Climate Models may be inappropriate for county-scale or regional-scale projections. We believe our methods to be the most appropriate for the questions we ask. We don’t know of any scientific evidence or papers that suggest that long-term GCM model results can be effectively used to predict near-term climate at local and regional scales. The climate scenario methods and approaches we have used are carefully explained in Jarnevich and Stohlgren, 2009 [31].
Forecasts are a tricky business. This does not stop the land managers and policy makers from asking scientists and experts to set priorities or give guidelines for setting priorities for rapid response and containment of invasive species. Forecasts are simply a tool to assess potential spread of a species. These forecasts are particularly important for invasive species early in the invasion process that might not have filled all of their niches. For example, the model predicts that Microstegium vimineum will have 133,000 additional km2 of potential suitable habitat by the year 2035. If land managers are able to keep a watch on these areas before the species spreads it may be possible to contain the invasion [8]. The fundamentals of early detection have not changed, but the habitat that is potentially suitable for species may be shifting with the changing climate.
Most of the species modeled in this study were considered habitat generalists. Lobo et al. [37] have claimed that Maxent models do not perform well on habitat generalists, yet these models did perform well. For the two species that had models that did not perform as well, we may need to add additional predictive layers to the model, use a different resolution, or even a different modeling technique. Not every species will respond to the same modeling format, and there are many techniques to choose from [16,43,44].
2.2. Invasibility Index
While we were not able to address biotic interaction or competition in these models, they could be incorporated by using predictor variables of the competition [45,46]. However, we did make an initial attempt at addressing propagule pressure, by devising the invasion index and looking at distance from known seed source within potential suitable habitat as a proxy for propagule pressure (Figure 1). We recognize that we do not have data for every location of the species, yet these data give a first order approximation of the potential range of invasion (or invasibility).
In addition to looking at the invasibility index, it is important to keep in mind the method of invasion for each species. Some invasive plant species spread primarily by runners and do not move very far with each season, while others spread primarily by seeds on the wind giving them farther reaching potential for spread. Plant dispersal is another worthy task of future analysis.
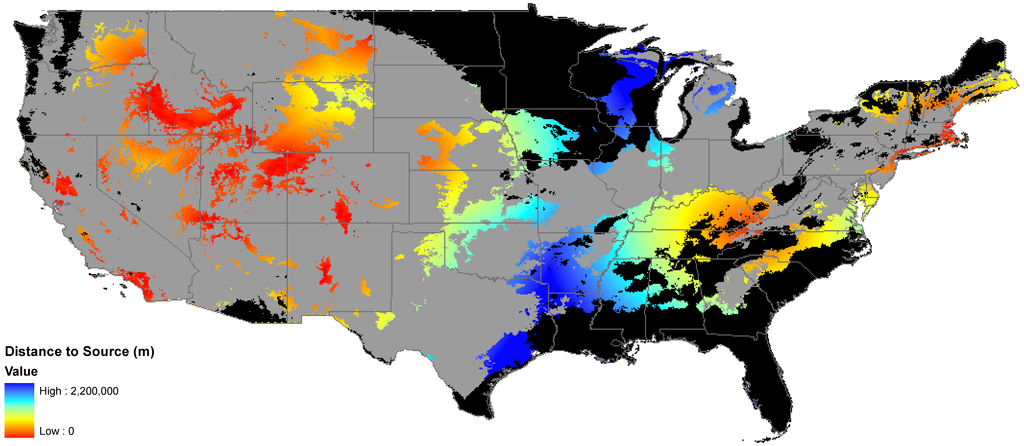
Figure 1.
Invasibility Index. Colors on the red end of the spectrum are closer to known seed sources, colors on the blue end of the spectrum are farther away. Grey represents unsuitable habitat, black represents areas of clamping, or where the model was extended beyond the environmental space it was trained on.
2.3. Utility of This Approach
This approach provides a methodology to conduct a triage of invasive species. It has been established that it is cost effective and efficient to control small invasions early in the process [8], and this method gives resource managers the ability to assess where an invasion may move in 10 to 25 years time to determine what may happen in the short term [31], instead of looking at the year 2100. These models help to identify the leading edge of the invasion, the areas of new potentially suitable habitat [47]. The leading edge is extremely important for watch lists, natural areas, ranches and farms.
This method can be useful for targeted surveys [22] and monitoring to better track actual spread of species. These models also identify areas where suitable habitat is receding, changing to less suitable habitat, although there is little evidence that, once established, plants ever leave a county-sized area [14]. Pearson and Dawson [19] discuss that bioclimatic envelope modeling has its limitations, but it works well as a first approximation, especially applicable for an early detection and rapid assessment program.
3. Experimental Section
Space prohibits us from discussing each of the 12 species modeled individually (but see Appendix B.), so we will focus on one example, Lepidium latifolium (perennial pepperweed, Figure 2). The main concentration of the 1015 data points that we were able to gather on L. latifolium were in the intermountain west of the United States, with some additional data points in California, and a smattering of points in the northeast. The current potentially suitable habitat was well distributed throughout the US with large areas of potential habitat in the west, throughout the plains states, and a large amount of potential habitat in the southeastern United States in an area that was sparse of data. The scenario model, showing potential habitat suitability change between current and 2035, suggests that there are modest areas of change throughout the United States with potential habitat in the west becoming more dispersed and increasing and the habitat in the southeast remaining stable for the most part.
In particular, the models show an overall increase in potential suitable habitat of about 95,000 km2 in the US between now and 2035 (Table 1). The areas in red on the scenario model (Figure 2C) have a high potential to be the leading edge of an invasion by this species, just as the areas in blue that go from being suitable to unsuitable have the potential to be the trailing edge of the invasion ( [12]; Table 1). This scenario map shows that areas of increased potential suitable habitat of the species are not confined to any particular part of the country.
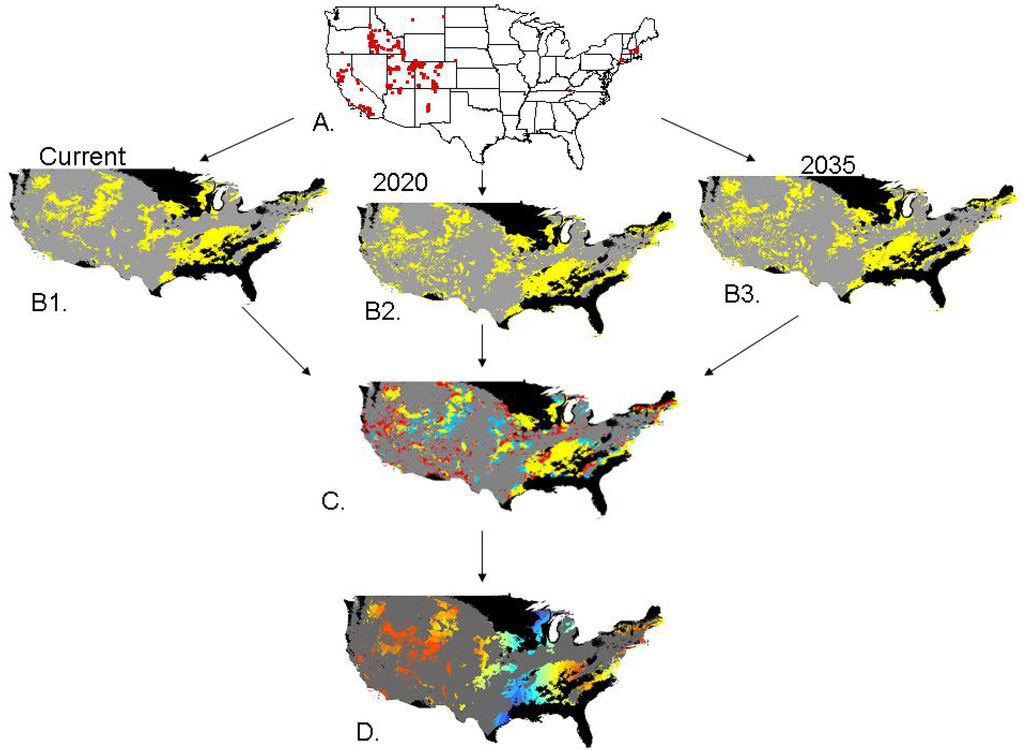
Figure 2.
Potential habitat suitability modeling process for Lepidium latifolium. (A) Distribution of data points. (B1) Current potential habitat suitability, (B2) Potential habitat suitability in 2020, (B3) Potential habitat suitability in 2035. (C) Scenario model showing stable potential suitable habitat in yellow, decreasing potential habitat suitability in blue, and increasing potential habitat suitability in red. (D) Invasion index with colors on the red end of the spectrum closer to potential seed source and colors on the blue end of the spectrum farther away. In the entire figure grey represents unsuitable habitat and black represents clamping, or areas the model was extended beyond the environmental space it was trained on.
For each variable included in the models, Maxent provides a response curve allowing for interpretation of environmental relationships to the distribution of the species’ suitable habitat (Figure 3).
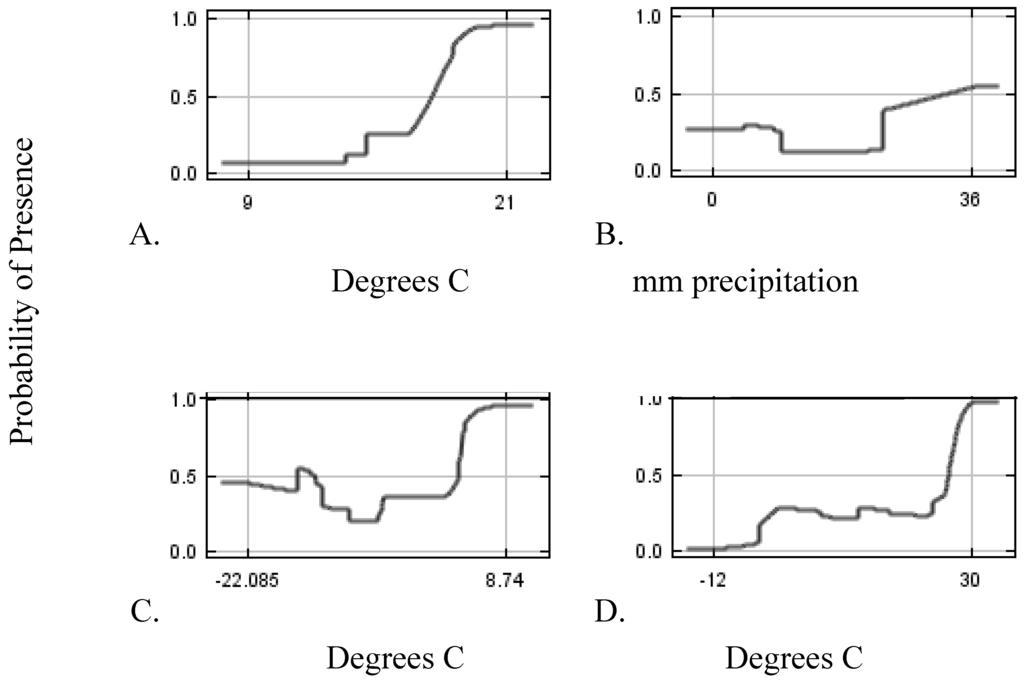
Figure 3.
Response curves of the most influential predictors. (A) Mean Diurnal Range, (B) Precipitation of warmest quarter, (C) Minimum temperature of coldest month, (D) Mean temperature of wettest quarter.
For L. latifolium, the major contributing factors associated with distribution include mean diurnal temperature range (24%), precipitation of the warmest quarter (16%), minimum temperature of the coldest month (12%), and mean temperature of the wettest quarter (10%; Table 2). The response curves associated with these factors show that there may be environmental thresholds for the ideal growth of L. latifolium (Figure 3). For example, for mean diurnal range, habitat suitability was low until the range begins to increase around 15 degrees C and steadily increases to around 21 degrees C showing L. latifolium to have a stronger relationship to a larger diurnal temperature range. L. latifolium also has higher habitat suitability with low or high precipitation in the warmest quarter, with high minimum temperature of the coldest month, and with higher mean temperature of the wettest quarter. Almost all factors in Table 2 are related to temperature. Using these relationships between variables, we can learn about the environmental drivers of the systems that we are modeling.

Table 2.
Top predictors by percent contributed to the model.
| Variable | Percent Contribution |
|---|---|
| Mean Diurnal Range | 24 |
| Precipitation of Warmest Quarter | 16 |
| Min Temperature of Coldest Month | 12 |
| Mean Temperature of Driest Quarter | 10 |
| Mean Temperature of Wettest Quarter | 10 |
| Mean Temperature of Warmest Quarter | 8 |
| Precipitation of Wettest Month | 7 |
| Precipitation Seasonality | 4 |
| Precipitation of Driest Month | 4 |
| Isothermality | 4 |
3.1. Invasibility Index Map
The invasibility map of L. latifolium shows this species to have less distance between seed sources in the west where most of the presence points were located (Figure 2D). The midwest contains potential suitable habitat but very few data points, so potentially minimal propagule pressure, and is therefore less invasible due to distance from nearest propagule pressure according to this model. There are a few data points at the Virginia-Kentucky border that are creating a hot spot. There are also potential hot spots in the northeast. From our data, it appears that the bulk of the propagule pressure is in the western United States. Across all 12 species, the invasibility index ranges from species with a concentrated distribution such as Pennisetum ciliare (Buffelgrass) to a wider distribution such as Microstegium vimineum (Japanese stiltgrass).
4. Conclusions
Our strategy was to assess plant invasion at a broad spatial scale. These same techniques are applicable to natural area, county, and state scales. We recognize that this is an iterative process of invasive species mapping and modeling. Models improve with more data, finer resolution prediction variables, and refined climate models. Maxent has proven to be an effective model for mapping species distributions despite small sample sizes [48], or scattered species distributions [49]. Different modeling techniques such as ensemble models [50,51] may also improve modeling efforts. We relied on Maxent, but other models may have done as good or better job of modeling these species [43]. We have provided a first approximation model of continental US potential habitat distribution maps for 12 species at a fine temporal scale. We hope it will be useful.
Acknowledgements
We would like to thank United States Geological Survey, Fort Collins Science Center and the Natural Resource Ecology Laboratory at Colorado State University for logistic support. Thank you to Sharon Gross at the USGS for providing funding for this work. A grant from USDA CSREES/NRI 2008-35615-04666 also contributed support. Two reviewers provided helpful comments and insightful suggestion. Sunil Kumar and Paul Evangelista have been invaluable with their modeling help. To all we are grateful. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
References
- Anderson, L.W.J. Control of invasive seaweeds. Bot. Mar. 2007, 50, 418–437. [Google Scholar]
- Noonburg, E.G.; Byers, J.E. More harm than good: When invader vulnerability to predators enhances impact on native species. Ecology 2005, 86, 2555–2560. [Google Scholar] [CrossRef]
- Snyder, W.E.; Evans, E.W. Ecological effects of invasive arthropod generalist predators. Annu. Rev. Ecol. Evol. S. 2006, 37, 95–122. [Google Scholar] [CrossRef]
- Crowl, T.A.; Crist, T.O.; Parmenter, R.R.; Belovsky, G.; Lugo, A.E. The spread of invasive species and infectious disease as drivers of ecosystem change. Front. Ecol. Environ. 2008, 6, 238–246. [Google Scholar] [CrossRef]
- Wilcove, D.S.; Rothstein, D.; Dubow, J.; Phillips, A.; Losos, E. Quantifying threats to imperiled species in the United States. Bioscience 1998, 48, 607–615. [Google Scholar] [CrossRef]
- Pimentel, D.; Zuniga, R.; Morrison, D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol. Econ. 2005, 52, 273–288. [Google Scholar] [CrossRef]
- Leung, B.; Lodge, D.M.; Finnoff, D.; Shogren, J.F.; Lewis, M.A.; Lamberti, G. An ounce of prevention or a pound of cure: bioeconomic risk analysis of invasive species. Proc. R. Soc. B 2002, 269, 2407–2413. [Google Scholar] [CrossRef]
- Moody, M.E.; Mack, R.N. Controlling the spread of plant invasions: The importance of nascent foci. J. App. Ecol. 1988, 25, 1009–1021. [Google Scholar] [CrossRef]
- Rejmanek, M.; Pitcairn, M.J. When is eradication of exotic pest plants a realistic goal? In Turning the Tide: The Eradication of Invasive Species; Veitch, C.R., Clout, M.N., Eds.; IUCN SSC Invasive Species Specialist Group: IUCN, Gland, Switzerland and Cambridge, UK, 2002; pp. 249–253. [Google Scholar]
- Thuiller, W.; Albert, C.; Araujo, M.B.; Berry, P.M.; Cabeza, M.; Guisan, A.; Hickler, T.; Midgely, G.F.; Paterson, J.; Schurr, F.M.; Sykes, M.T.; Zimmermann, N.E. Predicting global change impacts on plant species’ distributions: Future challenges. Perspect. Plant Ecol. 2008, 9, 137–152. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Dantonio, C.M.; Loope, L.L.; Westbrooks, R. Biological invasions as global environmental change. Am. Sci. 1996, 84, 468–478. [Google Scholar]
- Bradley, B.A.; Oppenheimer, M.; Wilcove, D.S. Climate Change and plant invasions: restoration opportunities ahead? Global Change Biol. 2009. [Google Scholar] [CrossRef]
- Hampe, A.; Petit, R.J. Conserving biodiversity under climate change: the rear edge matters. Ecol. Lett. 2005, 8, 461–467. [Google Scholar] [CrossRef]
- Stohlgren, T.J.; Barnett, D.T.; Jarnevich, C.S.; Flather, C.; Kartesz, J. The myth of plant species saturation. Ecol. Lett. 2008, 11, 313–322. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Graham, C.H. The ability of climate envelope models to predict the effect of climate change on species distributions. Global Change Biol. 2006, 12, 2272–2281. [Google Scholar] [CrossRef]
- Guisan, A.; Zimmermann, N.E. Predictive habitat distribution models in ecology. Ecol. Model. 2000, 135, 147–186. [Google Scholar] [CrossRef]
- Stohlgren, T.J. Measuring Plant Diversity: Lessons from the Field; Oxford University Press: Oxford, UK; New York, NY, USA, 2007. [Google Scholar]
- Stohlgren, T.J.; Schnase, J.L. Risk analysis for biological hazards: What we need to know about invasive species. Risk Anal. 2006, 26, 163–173. [Google Scholar] [CrossRef]
- Pearson, R.G.; Dawson, T.P. Predicting the impacts of climate change on the distribution of species: are bioclimate envelope models useful? Global Ecol. Biogeogr. 2003, 12, 361–371. [Google Scholar] [CrossRef]
- Guisan, A.; Thuiller, W. Predicting species distribution: offering more than simple habitat models. Ecol. Lett. 2005, 8, 993–1009. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Morisette, J.T.; Jarnevich, C.S.; Ullah, A.; Cai, W.J.; Pedelty, J.A.; Gentle, J.E.; Stohlgren, T.J.; Schnase, J L. A tamarisk habitat suitability map for the continental United States. Front. Ecol. Environ. 2006, 4, 11–17. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Thuiller, W.; Miaud, C. Prediction and validation of the potential global distribution of a problematic alien invasive species—the American bullfrog. Divers. Distrib. 2007, 13, 476–485. [Google Scholar] [CrossRef]
- Galatowitsch, S.M.; Anderson, N.O.; Ascher, P.D. Invasiveness in wetland plants in temperate North America. Wetlands 1999, 19, 733–755. [Google Scholar] [CrossRef]
- Ibarraf, F.A.; Cox, J.R.; Martinr, M.H.; Crowl, T.A.; Call, C.A. Predicting buffelgrass survival across a geographical and environmental gradient. J. Range Manag. 1995, 48, 53–59. [Google Scholar] [CrossRef]
- NIISS National Institute of Invasive Species Science. Available online: http://www.niiss.org/ (accessed February 29, 2010).
- Graham, J.; Newman, G.; Jarnevich, C.; Shory, R.; Stohlgren, T.J. A global organism detection and monitoring system for non-native species. Ecol. Inform. 2007, 2, 177–183. [Google Scholar]
- Nix, H.A. A biogeographic analysis of Australian elapid snakes. In Atlas of Elapid Snakes of Australia; Longmore, R., Ed.; Australian Government Publishing Service: Canberra, Australia, 1986. [Google Scholar]
- DAYMET. Climatological summaries for the conterminous United States, 1980–1997. Daily Surface Weather and Climatological Summaries. 2006. Available online: http://daymet.org (accessed March 31, 2009).
- Daly, C.; Gibson, W.P.; Taylor, G.H.; Johnson, G.L.; Pasteris, P. A knowledge-based approach to the statistical mapping of climate. Clim. Res. 2002, 22, 99–113. [Google Scholar] [CrossRef]
- Jarnevich, C.S.; Stohlgren, T.J. Near term climate projections for invasive species distributions. Biol. Invasions 2009, 11, 1373–1379. [Google Scholar] [CrossRef]
- SSI. SYSTAT 12.0; Systat Software, Inc.: Chicago, IL, USA, 2007. [Google Scholar]
- Phillips, S.J.; Dudik, M.; Elith, J.; Graham, C.H.; Lehmann, A.; Leathwick, J.; Ferrier, S. Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data. Ecol. Appl. 2009, 19, 181–197. [Google Scholar] [CrossRef]
- ESRI. ArcGIS 9.3; Environmental Systems Research Institute, Inc.: Redlands, CA, USA, 2008. [Google Scholar]
- Swets, J.A. Measuring the accuracy of diagnostic systems. Science 1988, 240, 1285–1293. [Google Scholar]
- Scott, J.M.; Heglund, P.; Morrison, M.L.; Raven, P.H. Predicting Species Occurrences: Issues of Accuracy and Scale; Island Press: Washington, DC, USA, 2002. [Google Scholar]
- Lobo, J.M.; Jimenez-Valverde, A.; Real, R. AUC: a misleading measure of the performance of predictive distribution models. Global Ecol. Biogeogr. 2008, 17, 145–151. [Google Scholar] [CrossRef]
- Hodkinson, D.J.; Thompson, K. Plant dispersal: the role of man. J. App. Ecol. 1997, 34, 1484–1496. [Google Scholar] [CrossRef]
- Mack, R.N.; Lonsdale, W.M. Humans as global plant dispersers: Getting more than we bargained for. Bioscience 2001, 51, 95–102. [Google Scholar] [CrossRef]
- Reichard, S.H.; White, P. Horticulture as a pathway of invasive plant introductions in the United States. Bioscience 2001, 51, 103–113. [Google Scholar] [CrossRef]
- Pearson, R.G.; Raxworthy, C.J.; Nakamura, M.; Peterson, A.T. Predicting species distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. J. Biogeogr. 2007, 34, 102–117. [Google Scholar]
- Beaumont, L.J.; Pitman, A.J.; Poulsen, M.; Hughes, L. Where will species go? Incorporating new advances in climate modelling into projections of species distributions. Global Change Biol. 2007, 13, 1368–1385. [Google Scholar] [CrossRef]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dudik, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A.; Li, J.; Lohmann, L.G.; Loiselle, B.A.; Manion, G.; Moritz, C.; Nakamura, M.; Nakazawa, Y.; Overton, J.M.; Peterson, A.T.; Phillips, S.J.; Richardson, K.; Scachetti-Pereira, R.; Schapire, R.E.; Soberon, J.; Williams, S.; Wisz, M.S.; Zimmermann, N.E. Novel methods improve prediction of species' distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef]
- Evangelista, P.H.; Kumar, S.; Stohlgren, T.J.; Jarnevich, C.S.; Crall, A.W.; Norman, J.B.; Barnett, D.T. Modelling invasion for a habitat generalist and a specialist plant species. Divers. Distrib. 2008, 14, 808–817. [Google Scholar] [CrossRef]
- Leathwick, J.R.; Austin, M.P. Competitive interactions between tree species in New Zealand's old-growth indigenous forests. Ecology 2001, 82, 2560–2573. [Google Scholar] [CrossRef]
- Anderson, R.P.; Peterson, A.T.; Gomez-Laverde, M. Using niche-based GIS modeling to test geographic predictions of competitive exclusion and competitive release in South American pocket mice. Oikos 2002, 98, 3–16. [Google Scholar] [CrossRef]
- Anderson, B.J.; Akcakaya, H.R.; Araujo, M.B.; Fordham, D.A.; Martinez-Meyer, E.; Thuiller, W.; Brook, B.W. Dynamics of range margins for metapopulations under climate change. Proc. R. Soc. B 2009, 276, 1415–1420. [Google Scholar] [CrossRef]
- Kumar, S.; Stohlgren, T.J. Maxent modeling for predicting suitable habitat for threatened and endangered tree Canacomyrica monticola in New Caledonia. JENE. 2009, 1, 94–98. [Google Scholar]
- Kumar, S.; Spaulding, S.A.; Stohlgren, T.J.; Hermann, K.A.; Schmidt, T.S.; Bahls, L.L. Potential habitat distribution for the freshwater diatom Didymosphenia geminata in the continental US. Front. Ecol. Environ. 2009, 7, 415–420. [Google Scholar]
- Araujo, M.B.; New, M. Ensemble forecasting of species distributions. Trends Ecol.Evol. 2007, 22, 42–47. [Google Scholar] [CrossRef]
- Stohigren, T.J.; Ma, P.; Kumar, S.; Rocca, M.; Morisette, J.T.; Jarnevich, C.S.; Benson, N. Ensemble habitat mapping of invasive plant species. Risk Anal. 2009, 30, 224–235. [Google Scholar]
- Crosier, C.S. Synergistic Methods to Generate Predictive Models at Large Spatial Extents and Fine Resolution; Colorado State University: Fort Collins, CO, USA, 2004. [Google Scholar]
- Stohlgren, T.J.; Jarnevich, C.; Chong, G.W.; Evangelista, P.H. Scale and plant invasions: a theory of biotic acceptance. Preslia 2006, 78, 405–426. [Google Scholar]

Appendix A.
List of data sources. Data sources used to model the twelve invasive plant species.
| Data source | Citation |
|---|---|
| Crosier PhD - Department of Transportation | [52] |
| Crosier PhD - Larimer County | [52] |
| Crosier PhD - San Luis Valley | [52] |
| Crosier PhD - The Nature Conservancy | [52] |
| Crosier PhD - Jackson County | [52] |
| Crosier PhD - Larimer County | [52] |
| Crosier PhD - Otero | [52] |
| Crosier PhD - Royal Gorge | [52] |
| Crosier PhD - San Luis Valley | [52] |
| Crosier PhD - Colorado State Parks | [52] |
| Crosier PhD - CNHP | [52] |
| Florida Natural Areas Inventory | http://fnai.org/invasivespecies.cfm |
| Florida Natural Areas Inventory | http://fnai.org/invasivespecies.cfm |
| The Great Lakes Indian Fish & Wildlife Commission | http://www.glifwc.org/ |
| Idaho State Department of Agriculture | Invasive Species Coordinator |
| Invasive Plant Atlas of the MidSouth | http://www.gri.msstate.edu/ipams/ |
| Invasive Plant Atlas of New England | http://nbii-nin.ciesin.columbia.edu/ipane/ |
| Montana Fish, Wildlife, and Parks | http://fwp.mt.gov/insidefwp/gis/shapefiles/fasweeds.zip |
| Modified Whittaker Plot Information | [53] |
| National Institute of Invasive Species Science project - Air Force Academy Weed Mapping | www.NIISS.org |
| National Institute of Invasive Species Science project - Bohemian Foundation | www.NIISS.org |
| National Institute of Invasive Species Science project - Colorado | www.NIISS.org |
| National Institute of Invasive Species Science project - ELK | www.NIISS.org |
| National Institute of Invasive Species Science project - Grand Staircase Escalante National Monument | www.NIISS.org |
| National Institute of Invasive Species Science project - Grazing effects | www.NIISS.org |
| National Institute of Invasive Species Science project - GVM Weed Test | www.NIISS.org |
| National Institute of Invasive Species Science project - Hart Mountain National Antelope | www.NIISS.org |
| National Institute of Invasive Species Science project - Highway 24 Weed Mapping | www.NIISS.org |
| National Institute of Invasive Species Science project - Invasive Carduus Thistles | www.NIISS.org |
| National Institute of Invasive Species Science project - National Elk Refuge | www.NIISS.org |
| National Institute of Invasive Species Science project - National Wildlife Refuge - USGS | www.NIISS.org |
| National Institute of Invasive Species Science project - Nevada Cheatgrass | www.NIISS.org |
| National Institute of Invasive Species Science project - New Invaders Watch List | www.NIISS.org |
| National Institute of Invasive Species Science project - Peterson Air Force Base Weed Mapping | www.NIISS.org |
| National Institute of Invasive Species Science project - Plains Riparian study | www.NIISS.org |
| National Institute of Invasive Species Science project - Plants of Concern | www.NIISS.org |
| National Institute of Invasive Species Science project - Pondicherry National Wildlife | www.NIISS.org |
| National Institute of Invasive Species Science project - Rocky Mountain NP LANDGAP | www.NIISS.org |
| National Institute of Invasive Species Science project - SAIN Invasive Plants | www.NIISS.org |
| National Institute of Invasive Species Science project - SE-EPPC EDDMaps | www.NIISS.org |
| National Institute of Invasive Species Science project - September 2007 Training at the ELC | www.NIISS.org |
| National Institute of Invasive Species Science project - Wisconsin Invasive Plants of the Future | www.NIISS.org |
| National Institute of Invasive Species Science project - Colorado Department of Transportation | www.NIISS.org |
| National Institute of Invasive Species Science project - Hart Mountain National Antelope | www.NIISS.org |
| National Institute of Invasive Species Science project - National Bison Range | www.NIISS.org |
| Personal Collection of Robert K. Peet | The University of North Carolina at Chapel Hill |
| Personal Collection of James F. Quinn | University of California, Davis |
| Southwest Exotic Mapping Program | http://sbsc.wr.usgs.gov/research/projects/swepic/swemp/swempA.asp |
| Bureau of Land Management, Utah State Office | Salt Lake City, UT |
| TexasInvasives.org | http://www.texasinvasives.org/ |
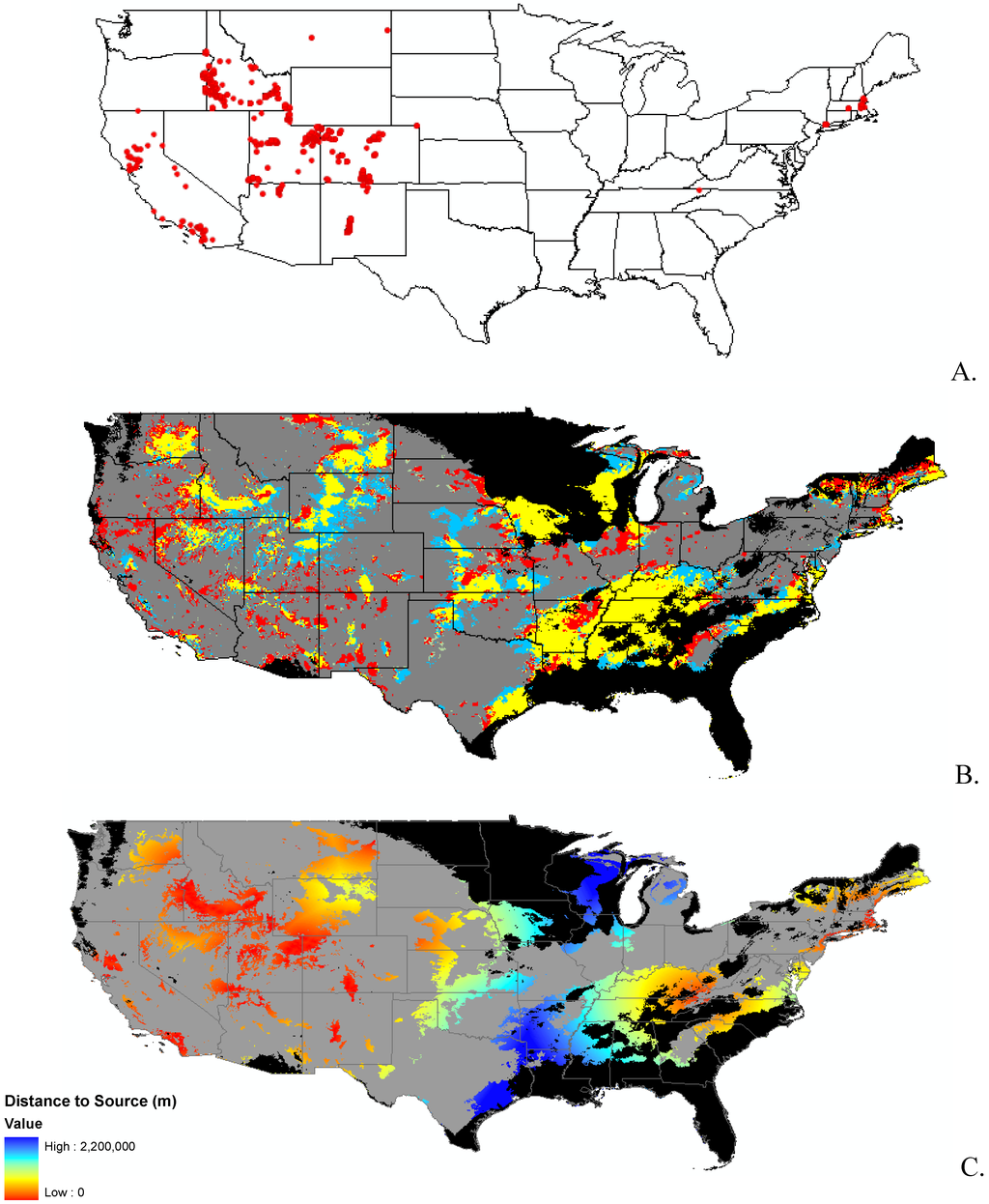
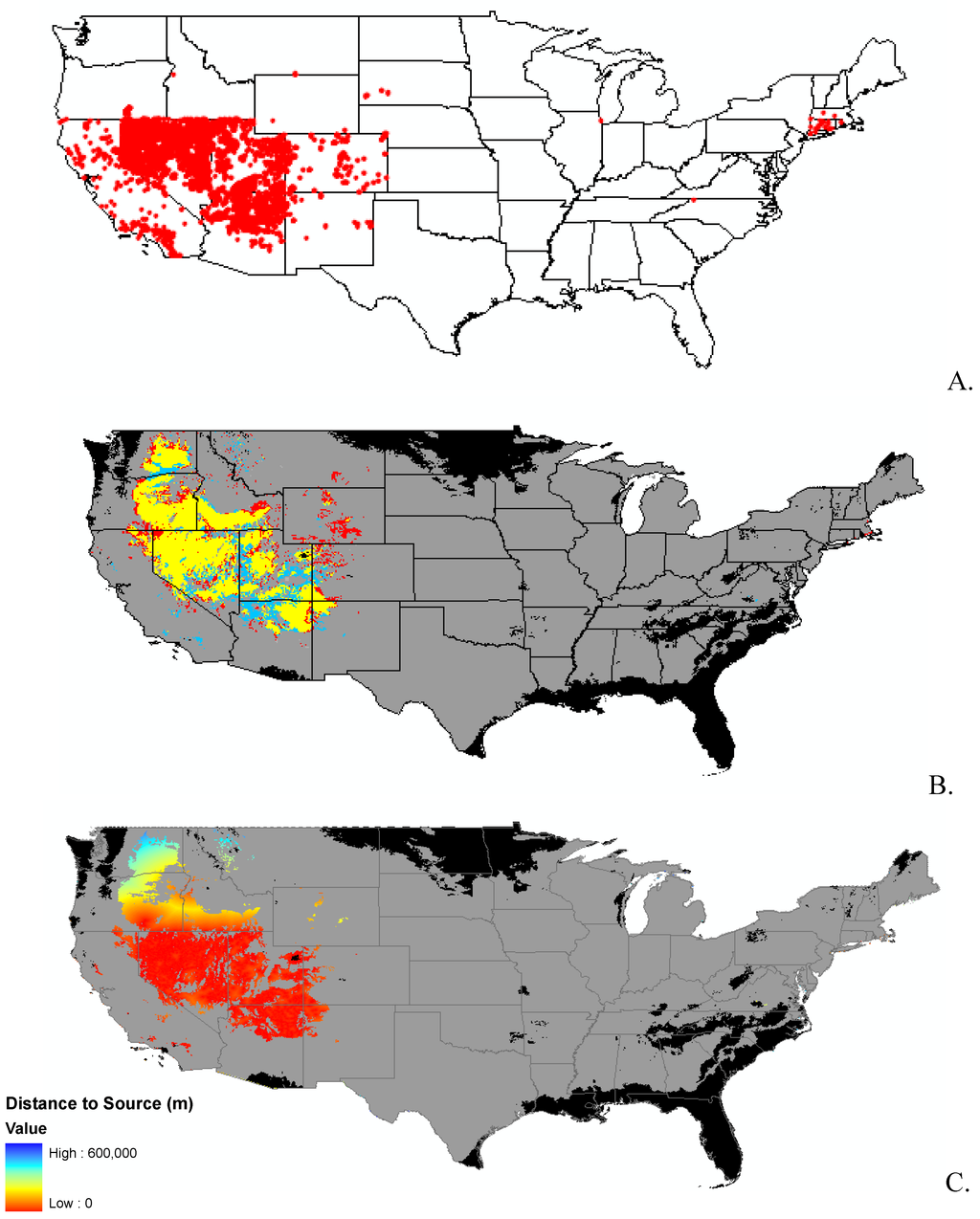
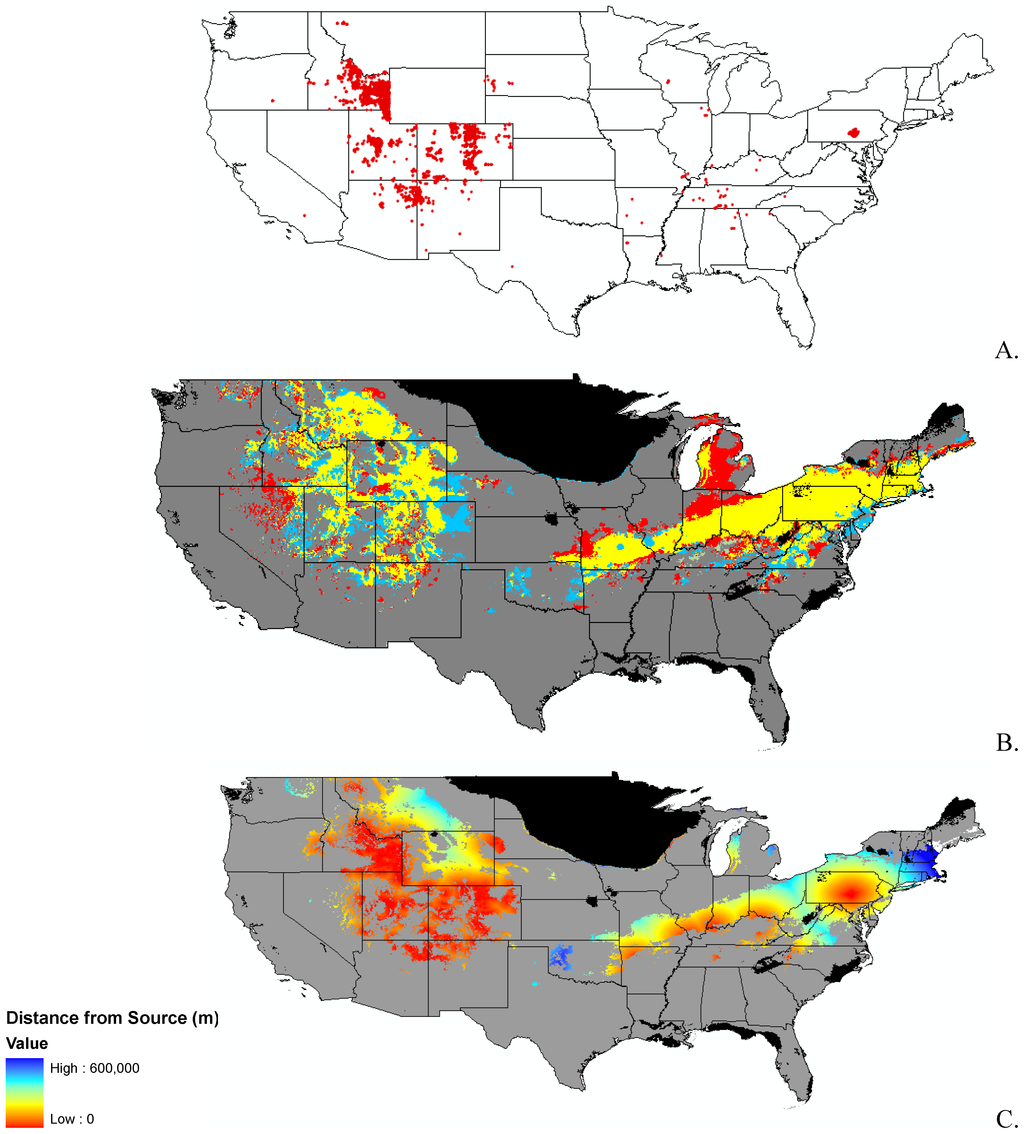
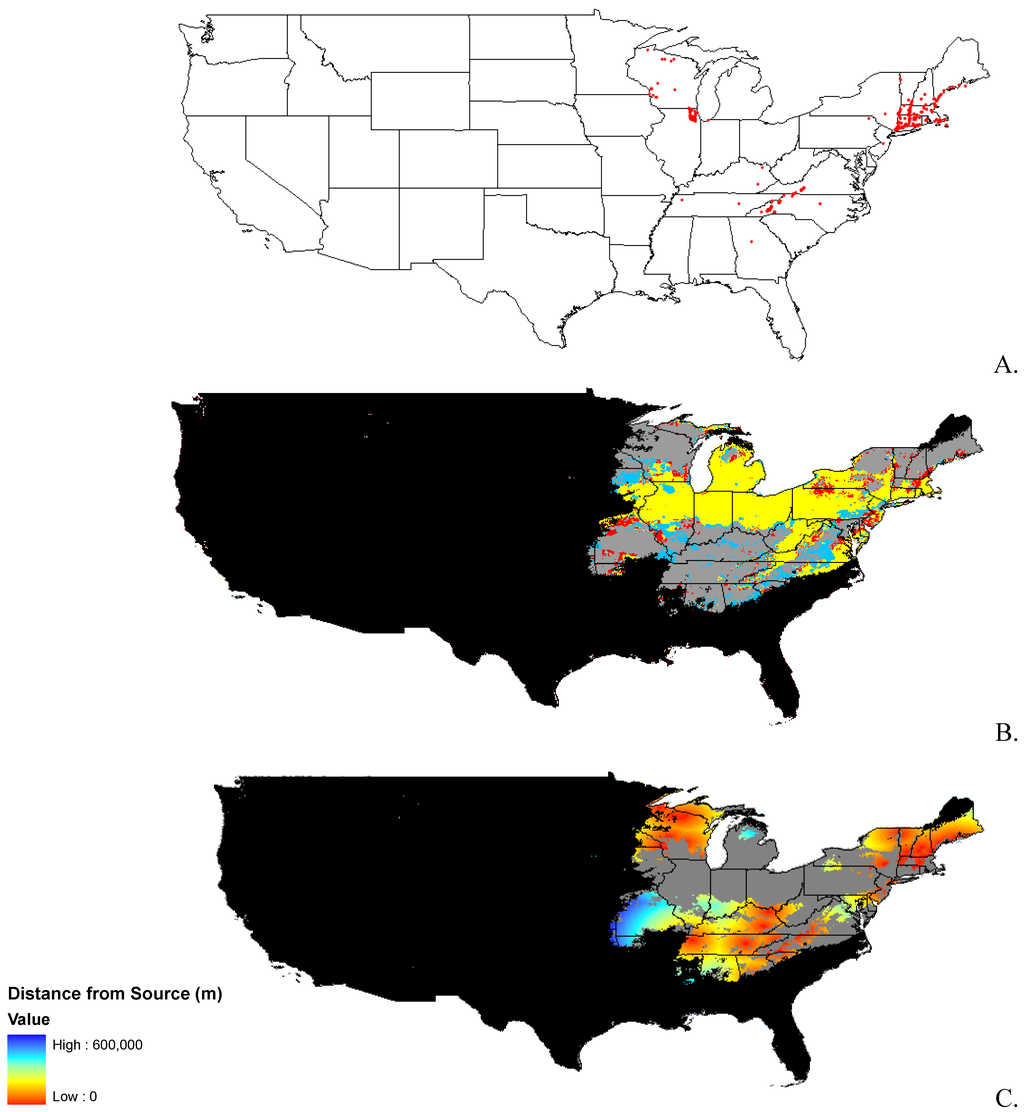
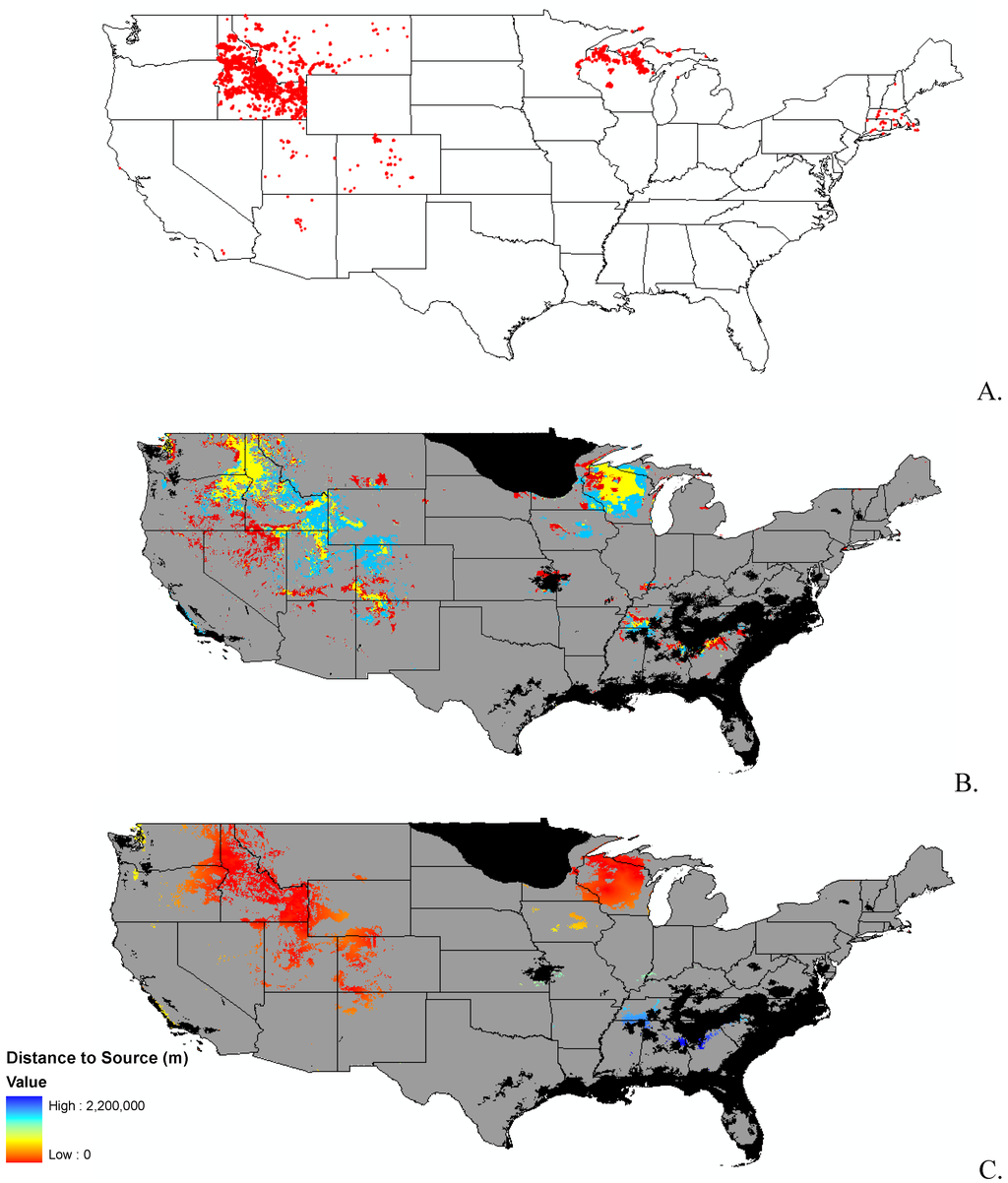
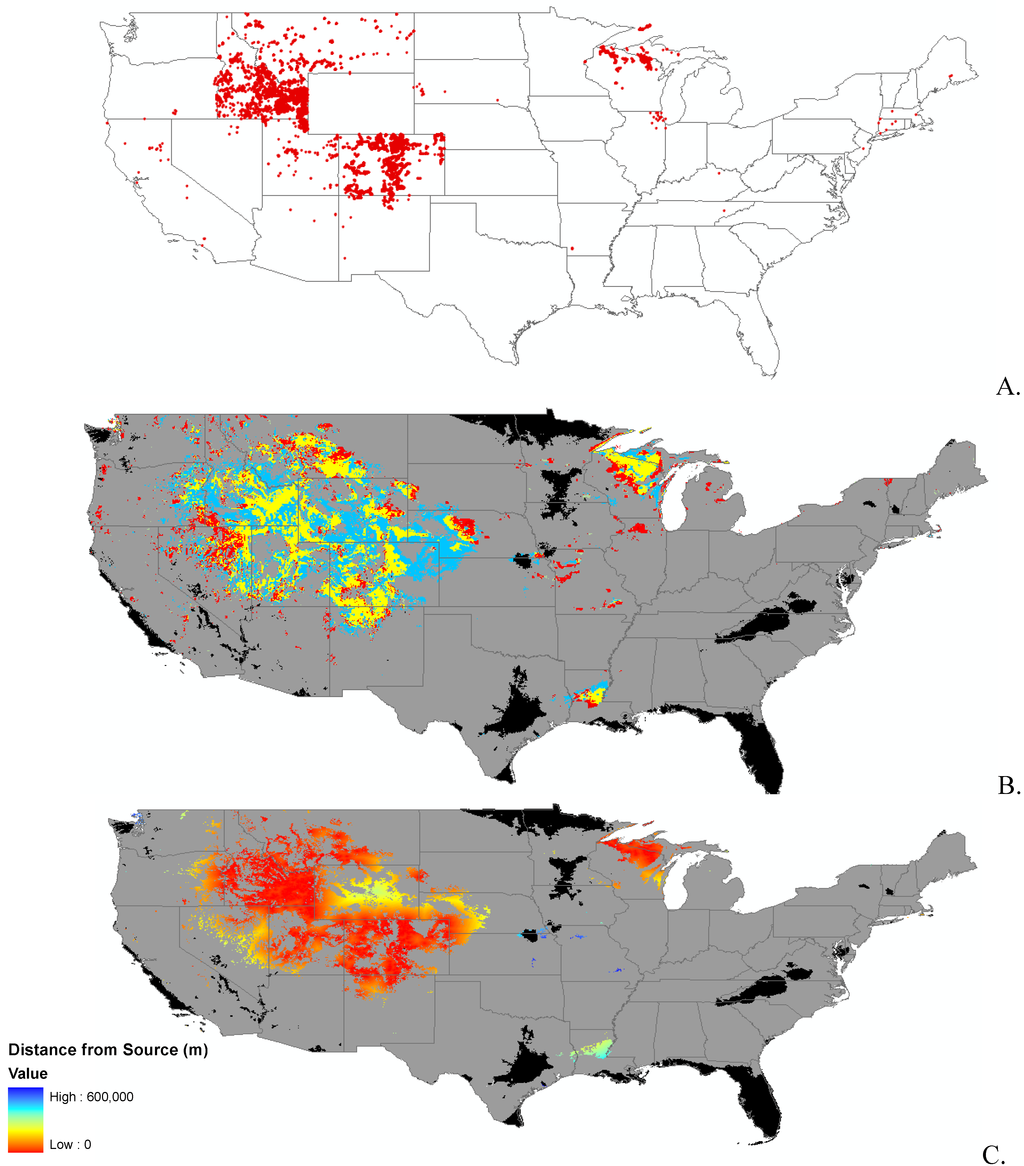
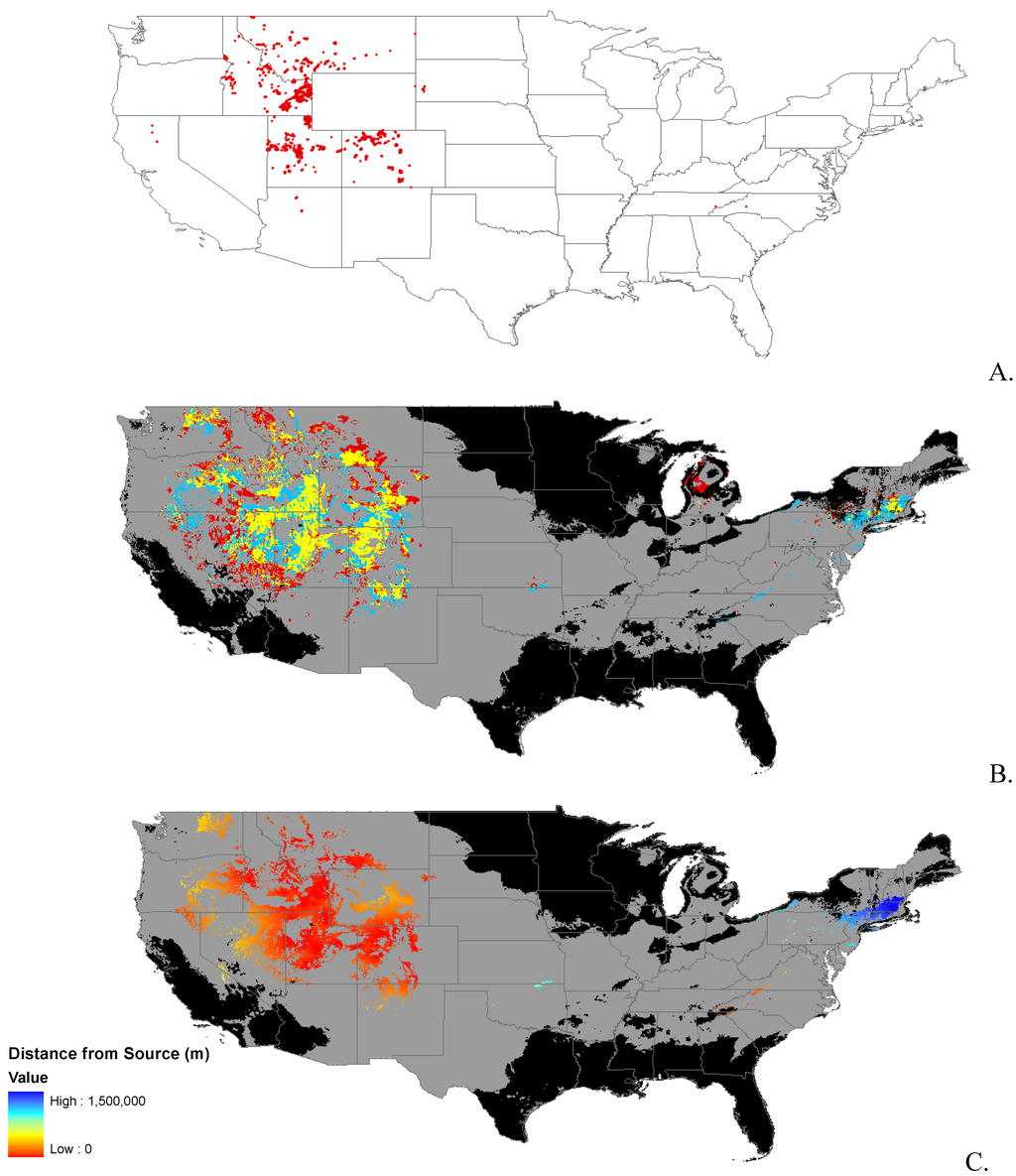
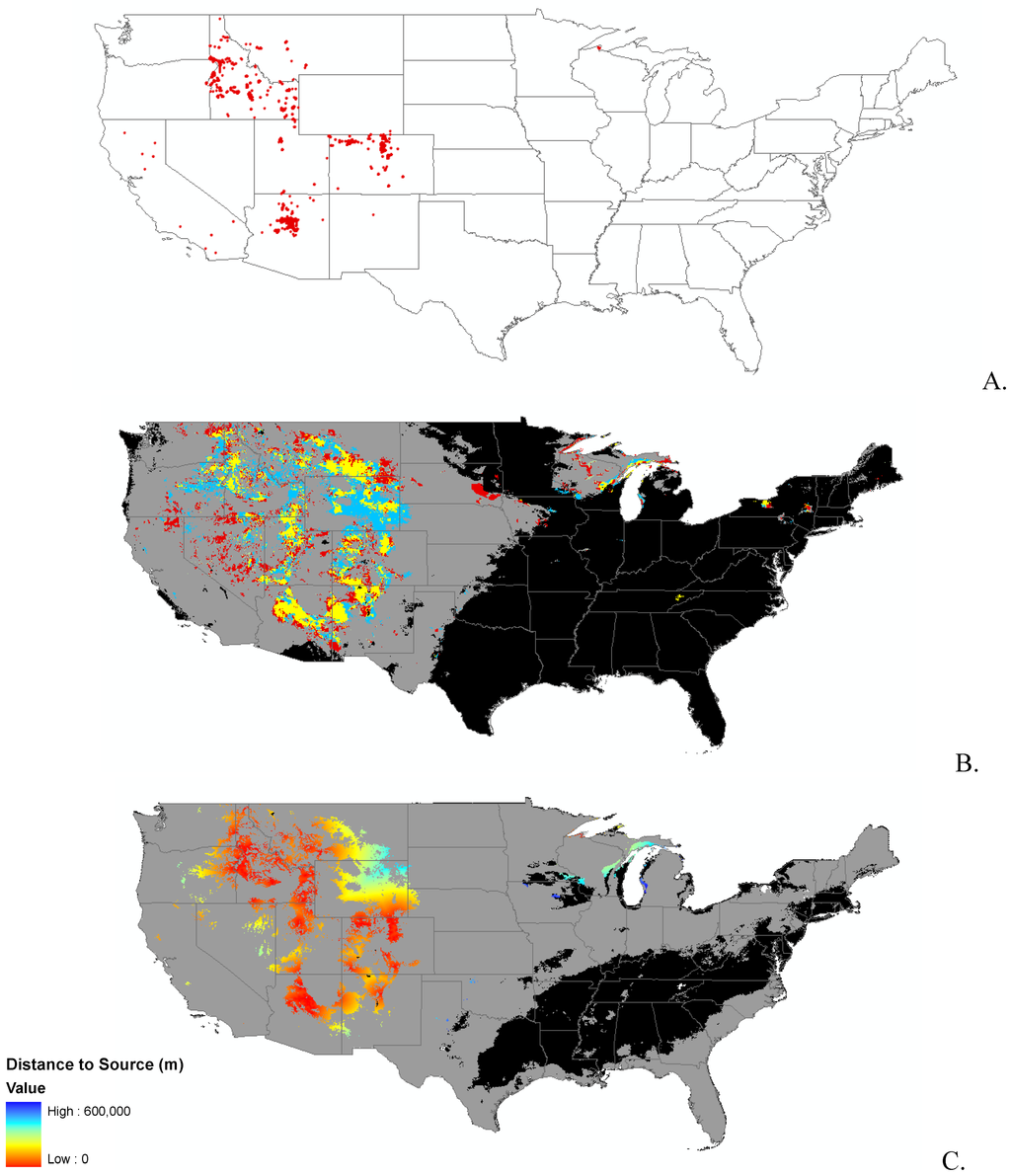
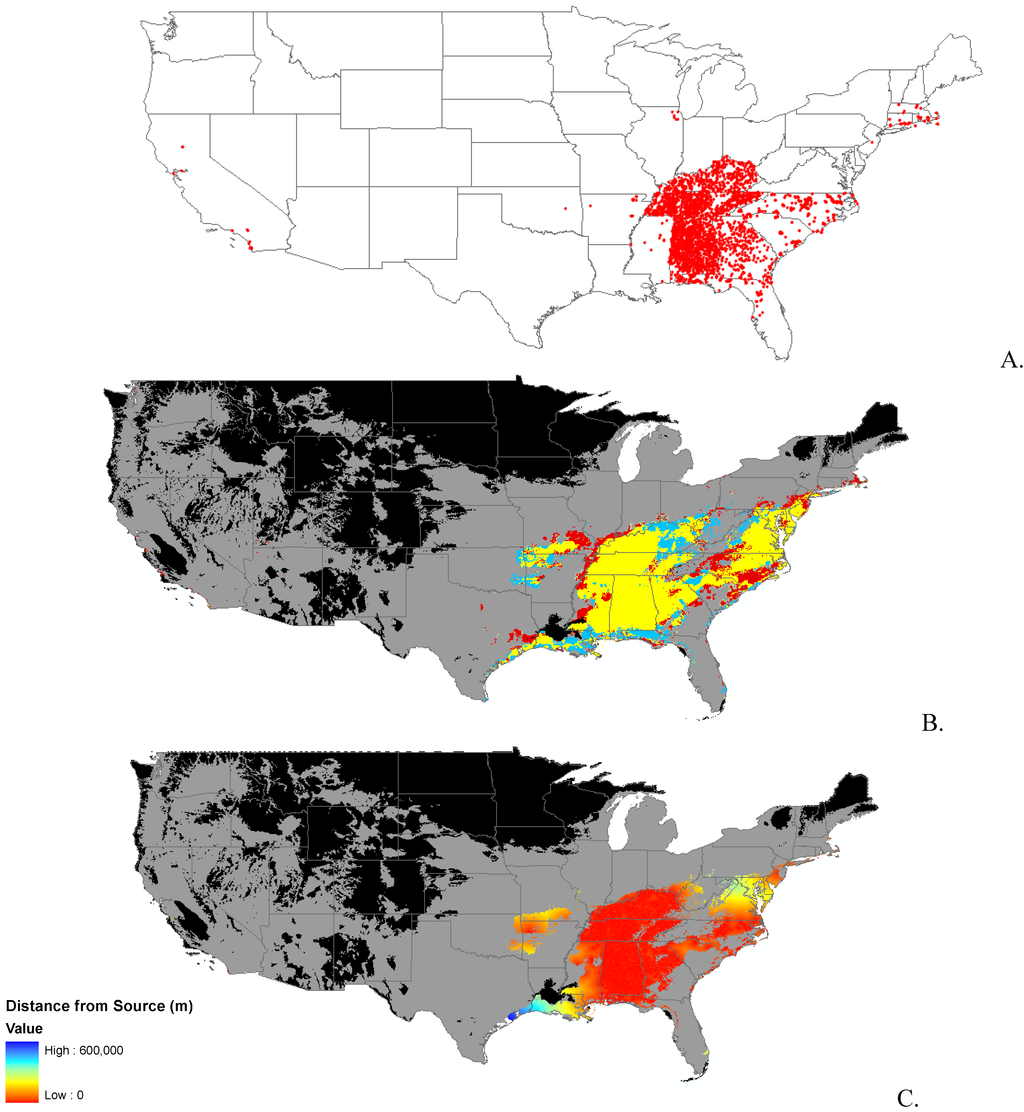
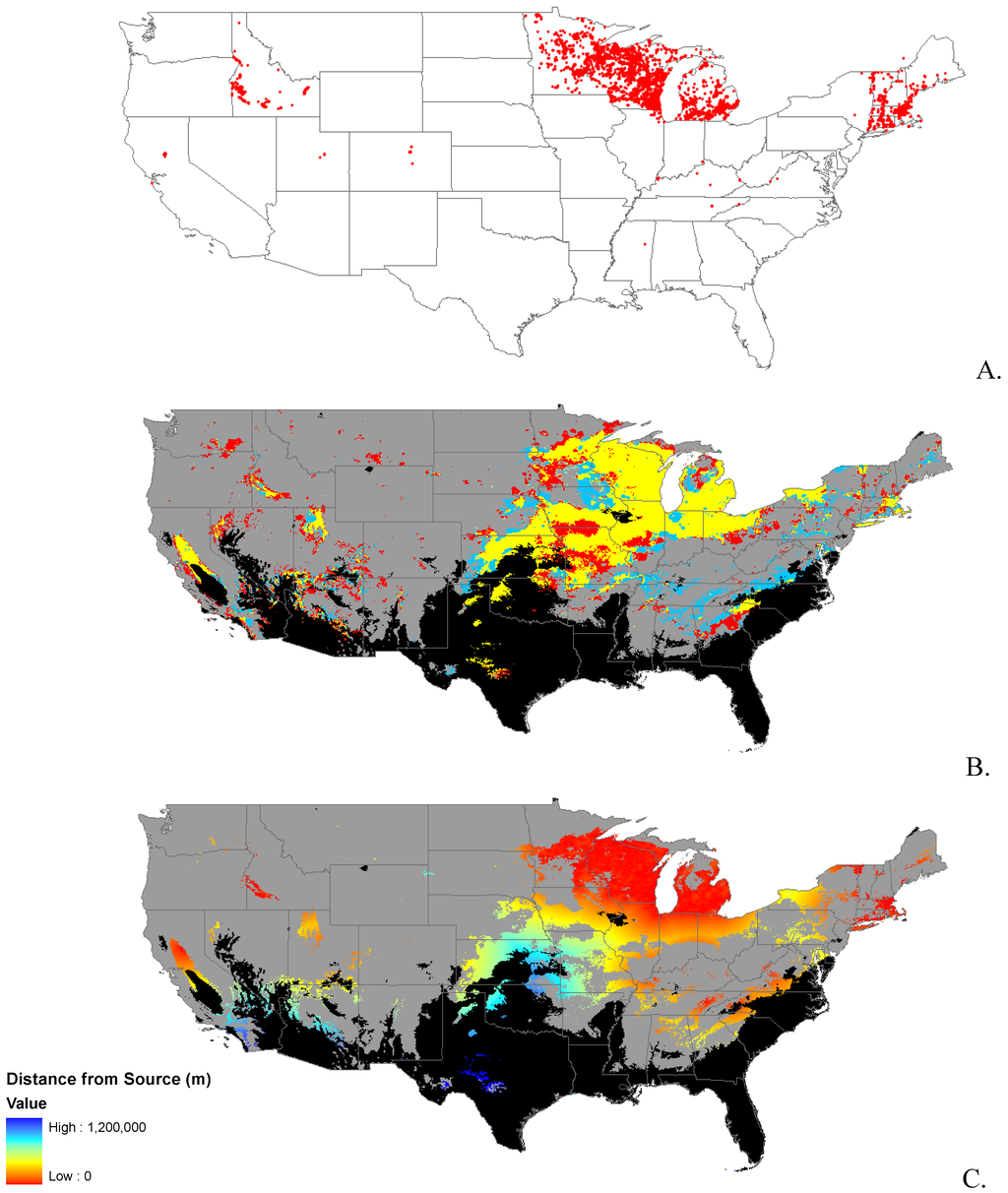
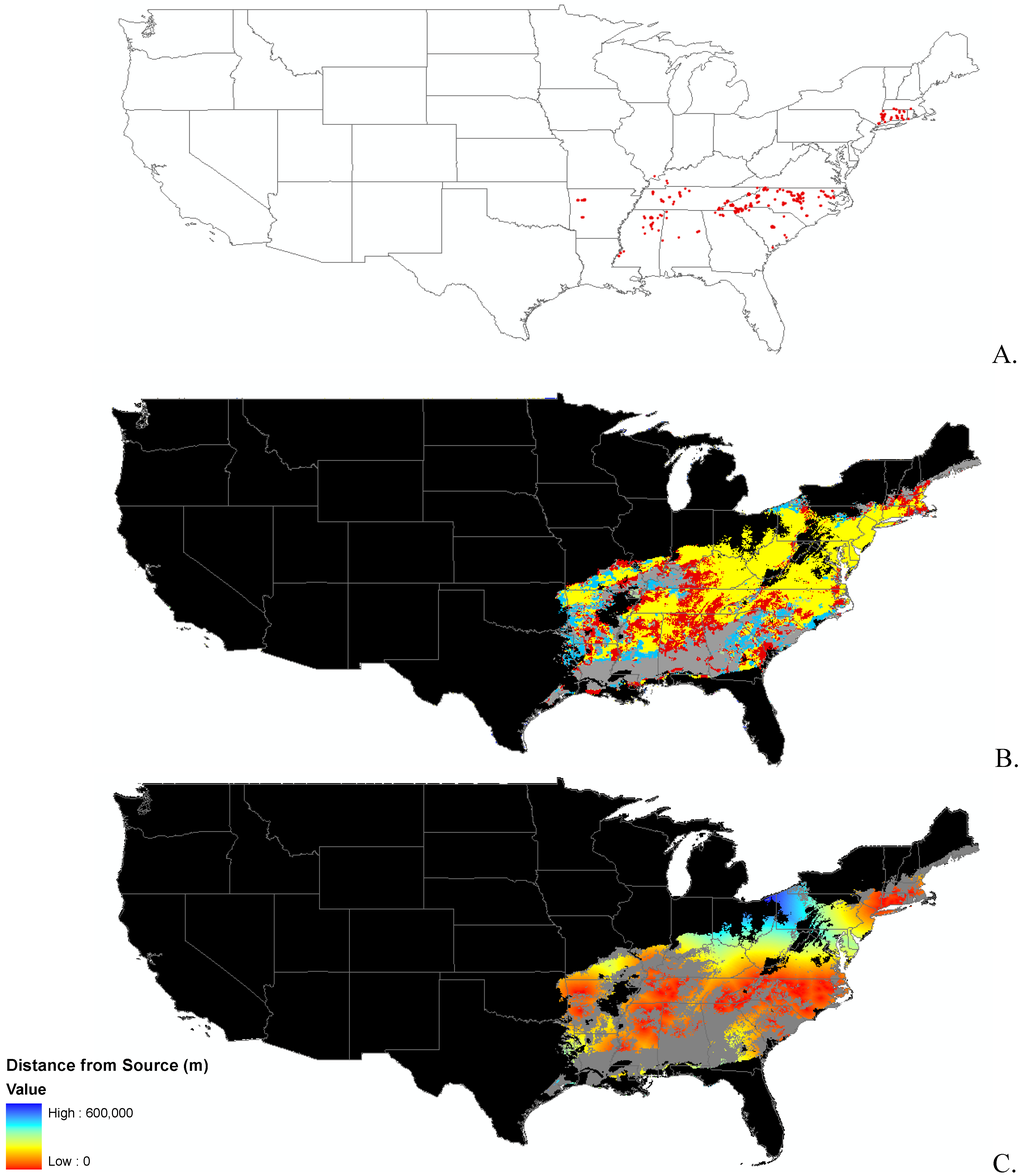
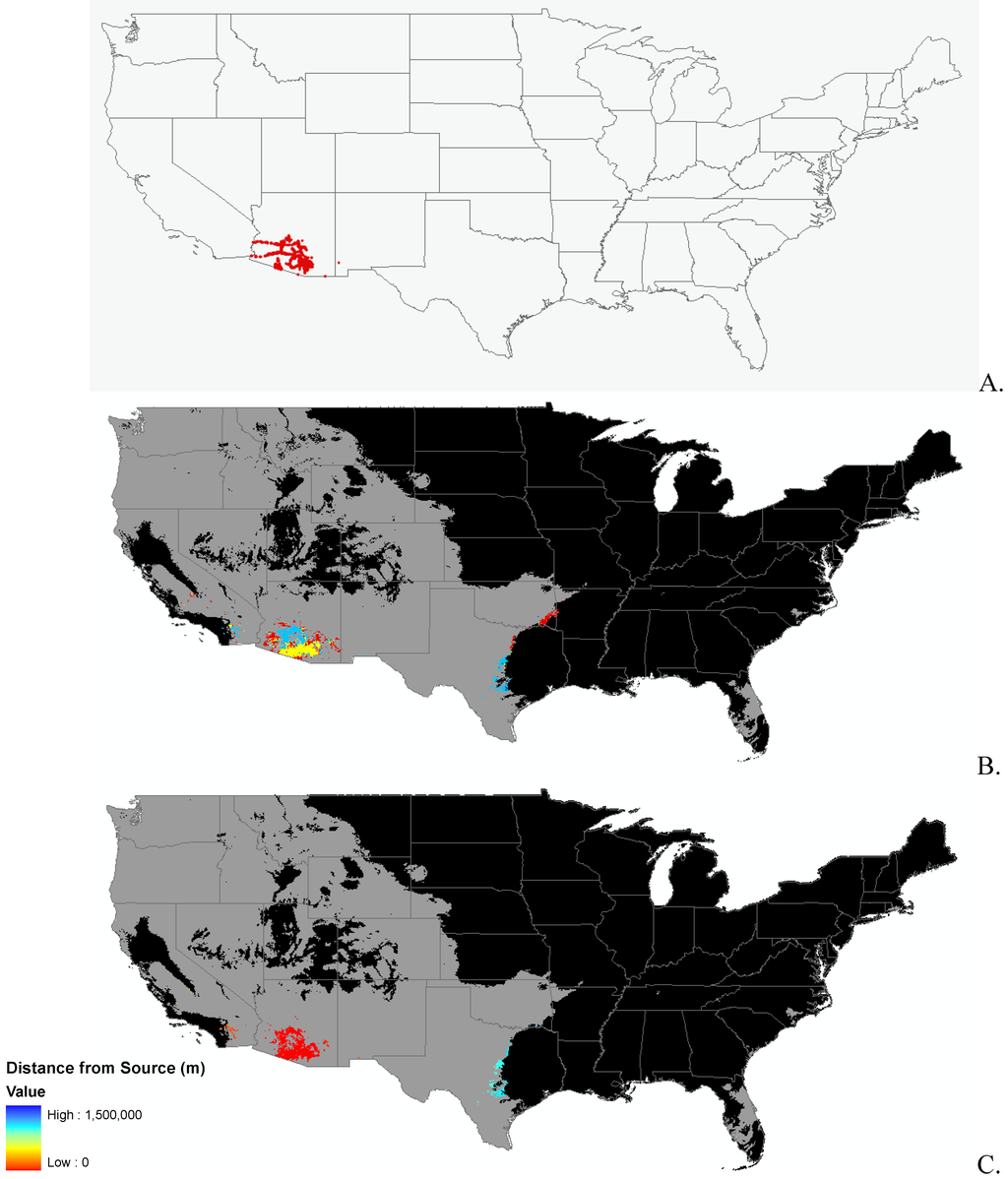
Appendix B.
Data distribution, spread model, and invasibility for 12 invasive plant species.
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an Open Access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).