Genetic Diversity of the Pm3 Powdery Mildew Resistance Alleles in Wheat Gene Bank Accessions as Assessed by Molecular Markers
Abstract
:1. Introduction
2. Experimental Section
2.1. Plant Material
2.2. Phenotypic Screening of the Wheat Accessions for Powdery Mildew Resistance
2.3. Pm3 haplotype specific STS marker
2.4. Pm3 Allele Specific PCRs
2.5. Isolation of the Full-Length Coding Sequence of Pm3 and Sequencing
3. Results and Discussion
3.1. Detection of Pm3 Alleles in Gene Bank Accessions Using Allele-Specific Markers
| Pm3 allele | Number of accessions carrying the tested Pm3 allele | Country of origin | Accession (s) in which the particular Pm3 allele was detected | Source of the accessions (gene bank) | Reference |
|---|---|---|---|---|---|
| Pm3b | 1 | France | TRI980 | IPK1 | This work |
| 1 | Kazakhstan | TRI7321 | IPK | This work | |
| 1 | Uzbekistan | TRI17549 | IPK | This work | |
| 1 | Tajikistan | TRI17561 | IPK | This work | |
| 2 | Russia | TRI18263; TRI18742 | IPK | This work | |
| 6 | Russia | VIR23918, VIR23922, VIR34986, VIR35021, VIR35030, VIR34984 | VIR2 | [27] | |
| 15 | Afghanistan | AUS9943, AUS9948, AUS10003, AUS10033, AUS13239, AUS13297, AUS13306, AUS13307, AUS13311, AUS14504, AUS14532, AUS14840, VIR45538, VIR49005, VIR49006 | AWCC3, VIR | [27] | |
| 6 | Iran | IG122348, IG122354, IG122361, IG122373, IG122502, VIR38613 | ICARDA4 | [27] | |
| 2 | Azerbaijan | VIR16766, VIR31595 | VIR | [27] | |
| 1 | Turkey | VIR35203 | VIR | [27] | |
| Pm3c | 8 | Nepal | TRI2437; TRI2439; TRI2448; TRI2748; TRI2765; TRI3255; TRI4029; TRI4091 | IPK | This work |
| 7 | India | TRI2799; TRI2804; TRI3375; TRI3542; TRI3552; TRI3986; TRI9986 | IPK | This work | |
| 1 | China | TRI4088 | IPK | This work | |
| 1 | Australia | TRI8320 | IPK | This work | |
| 3 | Iran | IG122491, IG122372, IG122346 | ICARDA | [27] | |
| 1 | Azerbaijan | VIR46301 | VIR | [27] | |
| Pm3d | 1 | Argentina | TRI11472 | IPK | This work |
| 1 | France | Oid HD4-266 | INRA5 | [19] | |
| Pm3e | 2 | India | TRI2554; TRI2782 | IPK | This work |
| 1 | Tajikistan | TA10381 | KSU6 | [19] | |
| 1 | France | Oid 91-35 | INRA | [19] | |
| Pm3f | 1 | Argentina | TRI7521 | IPK | This work |
| 1 | China | TRI16947 | IPK | This work | |
| Pm3g | 1 | France | Oid HD4-219 | INRA | [19] |
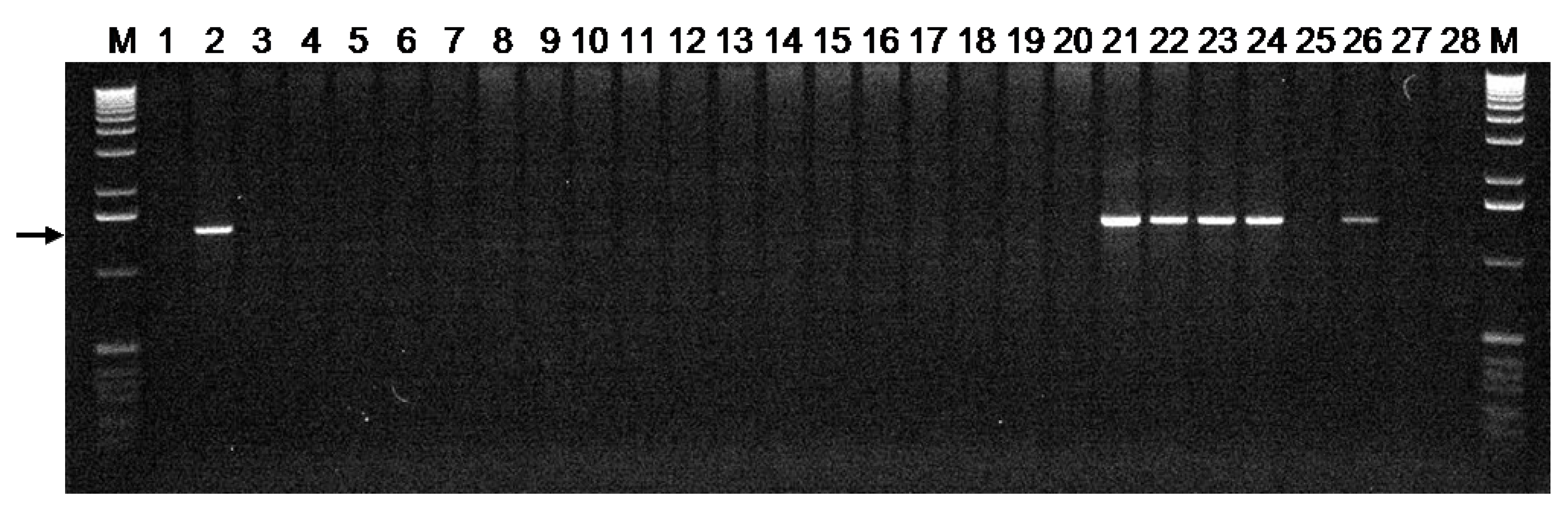
3.2. The Susceptible Pm3CS Allele is Present in Accessions From Diverse Geographical Origins
3.3. Abundant Presence of the Transitional, Susceptible Pm3Go/Jho Allele in Accessions from Nepal, India and Bhutan
| Pm3 allele | Origin | Number of accessions | Accession (s) | Type | Source of accessions | Reference |
|---|---|---|---|---|---|---|
| Pm3CS | Hexaploid wheat | |||||
| India | 2 | TRI2480, TRI2739 | unknown | IPK1 | This work | |
| Australia | 1 | TRI7243 | unknown | IPK | This work | |
| France | 1 | TRI7345 | unknown | IPK | This work | |
| Canada | 2 | TRI7736, TRI7741 | unknown | IPK | This work | |
| Ethiopia | 1 | TRI15026 | unknown | IPK | This work | |
| Tajikistan | 1 | TRI17510 | unknown | IPK | This work | |
| Pakistan | 2 | AUS 4856, IG41554 | Landrace | AWCC2 | [9] | |
| Afghanistan | 7 | AWCC9947, AWCC14695, AWCC14849, AUS13655, AUS13656, AUS13704, AUS14526 | Landrace | AWCC | [9] | |
| Turkey | 2 | IG42398, IG42869 | Landrace | ICARDA3 | [9] | |
| Iran | 1 | IG122584 | Landrace | ICARDA | [9] | |
| China | 1 | Chinese Spring | Landrace | ART4 | [19] | |
| Europe | 5 | Caribo, Greif, Obelisk, Kormoran, Monopol, | Cultivar | ART | [19] | |
| Belgium | 1 | Rouquin | Cultivar | ART | [19] | |
| Switzerland | 1 | Boval | Cultivar | ART | [19] | |
| Germany | 1 | Kanzler | Cultivar | ART | [19] | |
| France | 1 | Oid HD4-234 | Breeding line | INRA5 | [19] | |
| UK | 1 | Maris Huntsman | Cultivar | ART | [19] | |
| USA | 1 | Thatcher | Cultivar | ART | [19] | |
| Tajikistan | 1 | TA 10384 | Landrace | KSU6 | [19] | |
| Pm3CS | Tetraploid wheat | |||||
| Turkey | 5 | PI560872, PI560874, PI428145,PI428053, IG116184 | T. dicoccoides | USDA/ARS7/ ICARDA | [20] | |
| Ethiopia | 1 | PI58789 | T. dicoccum | USDA/ARS | [20] | |
| Ethiopia | 1 | CItr14846 | T. durum | USDA/ARS | [20] | |
| Pm3 Go/Jho | Hexaploid wheat | |||||
| India | 4 | TRI2596, TRI3197, TRI3535, TRI3992 | unknown | IPK | This work | |
| Nepal | 13 | TRI2611, TRI2889, TRI3232, TRI3628, TRI4359, TRI11131, TRI11132, TRI11133, TRI11135, TRI11136, TRI11137, TRI11139, TRI11151 | unknown | IPK | This work | |
| China | 1 | TRI14752 | unknown | IPK | This work | |
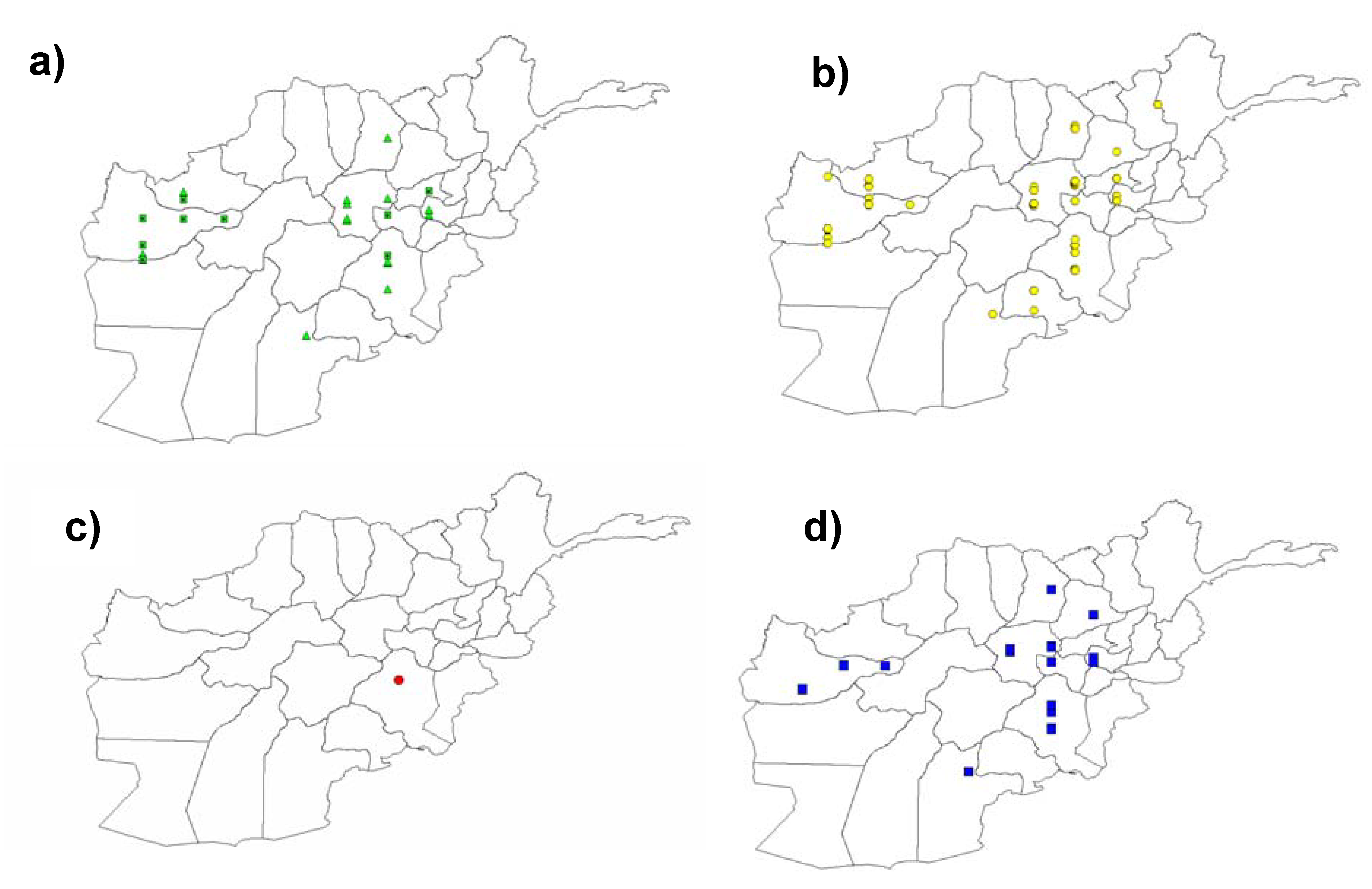
3.4. Widespread Existence of the Pm3b Resistance Allele in Landraces from Afghanistan
4. Conclusions
Acknowledgements
References
- Tanksley, S.D.; McCouch, S.R. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef]
- Hoisington, D.; Khairallah, M.; Reeves, T.; Ribaut, J.; Skovmand, B.; Taba, S.; Warburton, M. Plant genetic resources: What can they contribute toward increased crop productivity? Proc. Natl. Acad. Sci. USA 1999, 96, 5937–5943. [Google Scholar] [CrossRef]
- Huang, X.Q.; Börner, A.; Börner, M.S.; Ganal, M.W. Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor. Appl. Genet. 2002, 105, 699–707. [Google Scholar] [CrossRef]
- Mondini, L.; Noorani, A.; Pagnotta, M.A. Assessing plant genetic diversity by molecular tools. Diversity 2009, 1, 19–35. [Google Scholar] [CrossRef]
- Warburton, M.L.; Crossa, J.; Franco, J.; Kazi, M.; Trethowan, R.; Rajaram, S.; Pfeiffer, W.; Zhang, P.; Dreisigacker, S.; van Ginkel, M. Bringing wild relatives back into the family: Recovering genetic diversity of CIMMYT bread wheat germplasm. Euphytica. 2006, 149, 289–301. [Google Scholar] [CrossRef]
- Roussel, V.; Koenig, J.; Beckert, M.; Balfourier, F. Molecular diversity in french bread wheat accessions related to temporal trends and breeding programmes. Theor. Appl. Genet. 2004, 108, 920–930. [Google Scholar] [CrossRef]
- Hao, C.Y.; Zhang, X.Y.; Wang, L.F.; Dong, Y.S.; Shang, X.W.; Jia, J.Z. Genetic diversity and core collection evaluations in common wheat germplasm from the Northwestern Spring Wheat Region in China. Mol. Breed. 2006, 17, 69–77. [Google Scholar] [CrossRef]
- Landjeva, S.; Korzun, V.; Börner, A. Molecular markers: actual and potential contributions to wheat genome characterization and breeding. Euphytica 2007, 156, 271–296. [Google Scholar] [CrossRef]
- Bhullar, N.K.; Street, K.; Mackay, M.; Yahiaoui, N.; Keller, B. Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc. Natl. Acad. Sci. USA 2009, 106, 9519–9524. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J.A. NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef]
- Yan, L.; Loukoianov, A.; Tranquilli, G.; Helguera, M.; Fahima, T.; Dubcovsky, J. Positional cloning of the wheat vernalization gene Vrn1. Proc. Natl. Acad. Sci. USA 2003, 100, 6263–6268. [Google Scholar] [CrossRef]
- Yan, L.; Loukoianov, A.; Blechl, A.; Tranquilli, G.; Ramakrishna, W.; SanMiguel, P.; Bennetzen, J.L.; Echenique, V.; Dubcovsky, J. The wheat Vrn2 gene is a flowering repressor down-regulated by vernalization. Science 2004, 303, 1640–1644. [Google Scholar] [CrossRef]
- Faris, J.D.; Fellers, J.P.; Brooks, S.A.; Gill, B.S. A bacterial artificial chromosome contig spanning the major domestication locus Q in wheat and identification of a candidate gene. Genetics 2003, 164, 311–321. [Google Scholar]
- Huang, L.; Brooks, S.A.; Li, W.; Fellers, J.P.; Trick, H.N.; Gill, B.S. Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 2003, 164, 655–664. [Google Scholar]
- Feuillet, C.; Travella, S.; Stein, N.; Albar, L.; Nublat, A.; Keller, B. Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. USA 2003, 100, 15253–15258. [Google Scholar] [CrossRef]
- Cloutier, S.; McCallum, B.D.; Loutre, C.; Banks, T.W.; Wicker, T.; Feuillet, C.; Keller, B.; Jordan, M.C. Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol. Biol. 2007, 65, 93–106. [Google Scholar] [CrossRef]
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W; Singh, R.P.; Huerta-Espino, J.; McFadden, H.; Bossolini, E.; Selter, L.L.; Keller, B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 2009, 323, 1360–1363. [Google Scholar] [CrossRef]
- Yahiaoui, N.; Srichumpa, P.; Dudler, R.; Keller, B. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 2004, 37, 528–538. [Google Scholar] [CrossRef]
- Yahiaoui, N.; Brunner, S.; Keller, B. Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J. 2006, 47, 85–98. [Google Scholar] [CrossRef]
- Yahiaoui, N.; Kaur, N.; Keller, B. Independent evolution of functional Pm3 resistance genes in wild tetraploid wheat and domesticated bread wheat. Plant J. 2009, 57, 846–856. [Google Scholar] [CrossRef]
- Srichumpa, P.; Brunner, S.; Keller, B.; Yahiaoui, N. Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol. 2005, 139, 885–895. [Google Scholar] [CrossRef]
- Rong, J.K.; Millet, E.; Manisterski, J.; Feldman, M. A new powdery mildew resistance gene: Introgression from wild emmer into common wheat and RFLP-based mapping. Euphytica 2000, 115, 121–126. [Google Scholar] [CrossRef]
- Blanco, A.; Gadaleta, A.; Cenci, A.; Carluccio, A.V.; Abdelbacki, A.M.; Simeone, R. Molecular mapping of the novel powdery mildew resistance gene Pm36 introgressed from Triticum turgidum var. dicoccoides in durum wheat. Theor. Appl. Genet. 2008, 117, 135–142. [Google Scholar] [CrossRef]
- Perugini, L.D.; Murphy, J.P.; Marshal, D.; Brown-Guedira, G. Pm37, a new broadly effective powdery mildew resistance gene from Triticum timopheevii. Theor. Appl. Genet. 2008, 116, 417–425. [Google Scholar] [CrossRef]
- Bougot, Y.; Lemoine, J.; Pavoine, M.T.; Barloy, D.; Doussinault, G. Identification of a microsatellite marker associated with Pm3 resistance alleles to powdery mildew in wheat. Plant Breeding 2002, 121, 325–329. [Google Scholar] [CrossRef]
- Tommasini, L.; Yahiaoui, N.; Srichumpa, P.; Keller, B. Development of functional markers specific for seven Pm3 resistance alleles and their validation in the bread wheat gene pool. Theor. Appl. Genet. 2006, 114, 165–175. [Google Scholar] [CrossRef]
- Kaur, N.; Street, K.; Mackay, M.; Yahiaoui, N.; Keller, B. Molecular approaches for characterization and use of natural disease resistance in wheat. Eur. J. Plant Pathol. 2008, 121, 387–397. [Google Scholar] [CrossRef]
- Schweizer, P.; Christoffel, A.; Dudler, R. Transient expression of members of the germin-like gene family in epidermal cells of wheat confers disease resistance. Plant J. 1999, 20, 541–552. [Google Scholar] [CrossRef]
- Briggle, L.W. Three Loci in wheat involving resistance to Erysiphe graminis f. sp. tritici. Crop Sci. 1966, 6, 461–465. [Google Scholar] [CrossRef]
- Bennett, F.G.A. Resistance to powdery mildew in wheat: a review of its use in agriculture and breeding programmes. Plant Pathol. 1984, 33, 279–300. [Google Scholar] [CrossRef]
- Hsam, S.L.K.; Zeller, F.J. Breeding for powdery mildew resistance in common wheat (Triticum aestivum L.). In The Powdery Mildews: A Comprehensive Treatise; Be´langer, R.R., Bushnell, W.R., Dik, A.J., Carver, T.L.W., Eds.; APS Press: St Paul, MN, USA, 2002; pp. 219–238. [Google Scholar]
- Zeller, F.J.; Lutz, J.; Stephan, U. Chromosome location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L.) 1. Mlk and other alleles at the Pm3 locus. Euphytica 1993, 68, 223–229. [Google Scholar] [CrossRef]
- Santra, D.K.; Santra, M.; Allan, R.E.; Campbell, K.G.; Kidwell, K.K. Genetic and molecular characterization of vernalization genes Vrn-A1, Vrn-B1, and Vrn-D1 in spring wheat germplasm from the pacific northwest region of the USA. Plant breeding 2009, 128, 576–584. [Google Scholar] [CrossRef]
- Zhang, X.K.; Xiao, Y.G.; Zhang, Y.; Xia, X.C.; Dubcovsky, J.; He, Z.H. Allelic variation at the vernalization genes Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 in chinese wheat cultivars and their association with growth habit. Crop Sci. 2008, 48, 458–470. [Google Scholar] [CrossRef]
- Bhullar, N.K.; Zhang, Z.; Wicker, T.; Keller, B. Wheat gene bank accessions as a source of new alleles of the powdery mildew resistance gene Pm3: a large scale allele mining project. BMC Plant Biol. 2010, 10, 88. [Google Scholar] [CrossRef]
Appendix
| CODE | Accession Number | Origin | Pm3 haplotype | Pm3 allele detected when tested with allele-specific primers for Pm3a to Pm3g | |
| 1 | AUS | 9939 | Afghanistan | P | - |
| 2 | AUS | 9940 | Afghanistan | P | - |
| 3 | AUS | 9941 | Afghanistan | P | - |
| 4 | AUS | 9943 | Afghanistan | P | Pm3b |
| 5 | AUS | 9944 | Afghanistan | P | - |
| 6 | AUS | 9945 | Afghanistan | A | - |
| 7 | AUS | 9947 | Afghanistan | P | - |
| 8 | AUS | 9948 | Afghanistan | P | Pm3b |
| 9 | AUS | 9949 | Afghanistan | P | - |
| 10 | AUS | 9950 | Afghanistan | P | - |
| 11 | AUS | 9951 | Afghanistan | A | - |
| 12 | AUS | 9952 | Afghanistan | A | - |
| 13 | AUS | 9963 | Afghanistan | P | - |
| 14 | AUS | 9964 | Afghanistan | P | - |
| 15 | AUS | 9965 | Afghanistan | P | - |
| 16 | AUS | 9966 | Afghanistan | P | - |
| 17 | AUS | 9997 | Afghanistan | P | - |
| 18 | AUS | 9998 | Afghanistan | P | - |
| 19 | AUS | 9999 | Afghanistan | P | - |
| 20 | AUS | 10000 | Afghanistan | P | - |
| 21 | AUS | 10001 | Afghanistan | P | - |
| 22 | AUS | 10002 | Afghanistan | P | - |
| 23 | AUS | 10003 | Afghanistan | P | Pm3b |
| 24 | AUS | 10027 | Afghanistan | P | - |
| 25 | AUS | 10028 | Afghanistan | A | - |
| 26 | AUS | 10029 | Afghanistan | P | - |
| 27 | AUS | 10030 | Afghanistan | P | - |
| 28 | AUS | 10031 | Afghanistan | P | - |
| 29 | AUS | 10032 | Afghanistan | P | - |
| 30 | AUS | 10033 | Afghanistan | P | Pm3b |
| 31 | AUS | 10034 | Afghanistan | A | - |
| 32 | AUS | 10035 | Afghanistan | P | - |
| 33 | AUS | 10036 | Afghanistan | P | - |
| 34 | AUS | 10963 | Afghanistan | P | - |
| 35 | AUS | 13239 | Afghanistan | P | Pm3b |
| 36 | AUS | 13240 | Afghanistan | P | - |
| 37 | AUS | 13241 | Afghanistan | P | - |
| 38 | AUS | 13274 | Afghanistan | P | - |
| 39 | AUS | 13275 | Afghanistan | P | - |
| 40 | AUS | 13276 | Afghanistan | P | - |
| 41 | AUS | 13277 | Afghanistan | P | - |
| 42 | AUS | 13285 | Afghanistan | A | - |
| 43 | AUS | 13290 | Afghanistan | P | - |
| 44 | AUS | 13291 | Afghanistan | P | - |
| 45 | AUS | 13292 | Afghanistan | P | - |
| 46 | AUS | 13293 | Afghanistan | P | - |
| 47 | AUS | 13294 | Afghanistan | P | - |
| 48 | AUS | 13295 | Afghanistan | P | - |
| 49 | AUS | 13296 | Afghanistan | P | - |
| 50 | AUS | 13297 | Afghanistan | P | Pm3b |
| 51 | AUS | 13298 | Afghanistan | P | - |
| 52 | AUS | 13299 | Afghanistan | P | - |
| 53 | AUS | 13300 | Afghanistan | P | - |
| 54 | AUS | 13301 | Afghanistan | A | - |
| 55 | AUS | 13302 | Afghanistan | P | - |
| 56 | AUS | 13303 | Afghanistan | P | - |
| 57 | AUS | 13304 | Afghanistan | P | - |
| 58 | AUS | 13305 | Afghanistan | P | - |
| 59 | AUS | 13306 | Afghanistan | P | Pm3b |
| 60 | AUS | 13307 | Afghanistan | P | Pm3b |
| 61 | AUS | 13309 | Afghanistan | P | - |
| 62 | AUS | 13310 | Afghanistan | A | - |
| 63 | AUS | 13311 | Afghanistan | P | Pm3b |
| 64 | AUS | 13312 | Afghanistan | A | - |
| 65 | AUS | 13313 | Afghanistan | A | - |
| 66 | AUS | 13314 | Afghanistan | P | - |
| 67 | AUS | 13315 | Afghanistan | A | - |
| 68 | AUS | 13636 | Afghanistan | P | - |
| 69 | AUS | 13637 | Afghanistan | P | - |
| 70 | AUS | 13638 | Afghanistan | P | - |
| 71 | AUS | 13639 | Afghanistan | P | - |
| 72 | AUS | 13640 | Afghanistan | P | - |
| 73 | AUS | 13641 | Afghanistan | P | - |
| 74 | AUS | 13642 | Afghanistan | P | - |
| 75 | AUS | 13643 | Afghanistan | P | - |
| 76 | AUS | 13644 | Afghanistan | P | - |
| 77 | AUS | 13645 | Afghanistan | P | - |
| 78 | AUS | 13646 | Afghanistan | P | - |
| 79 | AUS | 13647 | Afghanistan | P | - |
| 80 | AUS | 13654 | Afghanistan | P | - |
| 81 | AUS | 13655 | Afghanistan | P | - |
| 82 | AUS | 13656 | Afghanistan | P | - |
| 83 | AUS | 13657 | Afghanistan | P | - |
| 84 | AUS | 13658 | Afghanistan | P | - |
| 85 | AUS | 13659 | Afghanistan | P | - |
| 86 | AUS | 13660 | Afghanistan | P | - |
| 87 | AUS | 13661 | Afghanistan | P | - |
| 88 | AUS | 13662 | Afghanistan | P | - |
| 89 | AUS | 13663 | Afghanistan | P | - |
| 90 | AUS | 13664 | Afghanistan | P | - |
| 91 | AUS | 13665 | Afghanistan | P | - |
| 92 | AUS | 13666 | Afghanistan | P | - |
| 93 | AUS | 13703 | Afghanistan | P | - |
| 94 | AUS | 13704 | Afghanistan | P | - |
| 95 | AUS | 13705 | Afghanistan | P | - |
| 96 | AUS | 13706 | Afghanistan | P | - |
| 97 | AUS | 13707 | Afghanistan | P | - |
| 98 | AUS | 13708 | Afghanistan | A | - |
| 99 | AUS | 13723 | Afghanistan | A | - |
| 100 | AUS | 13724 | Afghanistan | P | - |
| 101 | AUS | 13725 | Afghanistan | P | - |
| 102 | AUS | 13726 | Afghanistan | P | - |
| 103 | AUS | 13727 | Afghanistan | P | - |
| 104 | AUS | 13728 | Afghanistan | P | - |
| 105 | AUS | 13729 | Afghanistan | P | - |
| 106 | AUS | 13730 | Afghanistan | P | - |
| 107 | AUS | 13731 | Afghanistan | P | - |
| 108 | AUS | 13732 | Afghanistan | P | - |
| 109 | AUS | 13733 | Afghanistan | P | - |
| 110 | AUS | 13734 | Afghanistan | A | - |
| 111 | AUS | 13735 | Afghanistan | P | - |
| 112 | AUS | 13736 | Afghanistan | A | - |
| 113 | AUS | 13737 | Afghanistan | P | - |
| 114 | AUS | 13738 | Afghanistan | P | - |
| 115 | AUS | 14442 | Afghanistan | P | - |
| 116 | AUS | 14443 | Afghanistan | P | - |
| 117 | AUS | 14444 | Afghanistan | P | - |
| 118 | AUS | 14446 | Afghanistan | P | - |
| 119 | AUS | 14447 | Afghanistan | P | - |
| 120 | AUS | 14448 | Afghanistan | P | - |
| 121 | AUS | 14449 | Afghanistan | P | - |
| 122 | AUS | 14450 | Afghanistan | A | - |
| 123 | AUS | 14451 | Afghanistan | P | - |
| 124 | AUS | 14452 | Afghanistan | P | - |
| 125 | AUS | 14454 | Afghanistan | P | - |
| 126 | AUS | 14455 | Afghanistan | P | - |
| 127 | AUS | 14456 | Afghanistan | P | - |
| 128 | AUS | 14457 | Afghanistan | P | - |
| 129 | AUS | 14458 | Afghanistan | P | - |
| 130 | AUS | 14459 | Afghanistan | P | - |
| 131 | AUS | 14461 | Afghanistan | P | - |
| 132 | AUS | 14463 | Afghanistan | P | - |
| 133 | AUS | 14474 | Afghanistan | P | - |
| 134 | AUS | 14475 | Afghanistan | P | - |
| 135 | AUS | 14476 | Afghanistan | P | - |
| 136 | AUS | 14480 | Afghanistan | P | - |
| 137 | AUS | 14481 | Afghanistan | P | - |
| 138 | AUS | 14482 | Afghanistan | P | - |
| 139 | AUS | 14483 | Afghanistan | P | - |
| 140 | AUS | 14484 | Afghanistan | P | - |
| 141 | AUS | 14485 | Afghanistan | P | - |
| 142 | AUS | 14486 | Afghanistan | P | - |
| 143 | AUS | 14487 | Afghanistan | P | - |
| 144 | AUS | 14488 | Afghanistan | P | - |
| 145 | AUS | 14489 | Afghanistan | P | - |
| 146 | AUS | 14490 | Afghanistan | P | - |
| 147 | AUS | 14491 | Afghanistan | P | - |
| 148 | AUS | 14492 | Afghanistan | P | - |
| 149 | AUS | 14493 | Afghanistan | P | - |
| 150 | AUS | 14494 | Afghanistan | P | - |
| 151 | AUS | 14495 | Afghanistan | P | - |
| 152 | AUS | 14496 | Afghanistan | P | - |
| 153 | AUS | 14497 | Afghanistan | P | - |
| 154 | AUS | 14498 | Afghanistan | P | - |
| 155 | AUS | 14499 | Afghanistan | P | - |
| 156 | AUS | 14501 | Afghanistan | P | - |
| 157 | AUS | 14502 | Afghanistan | P | - |
| 158 | AUS | 14503 | Afghanistan | P | - |
| 159 | AUS | 14504 | Afghanistan | P | Pm3b |
| 160 | AUS | 14505 | Afghanistan | P | - |
| 161 | AUS | 14506 | Afghanistan | P | - |
| 162 | AUS | 14513 | Afghanistan | P | - |
| 163 | AUS | 14514 | Afghanistan | P | - |
| 164 | AUS | 14515 | Afghanistan | P | - |
| 165 | AUS | 14516 | Afghanistan | P | - |
| 166 | AUS | 14517 | Afghanistan | P | - |
| 167 | AUS | 14518 | Afghanistan | P | - |
| 168 | AUS | 14519 | Afghanistan | P | - |
| 169 | AUS | 14520 | Afghanistan | P | - |
| 170 | AUS | 14521 | Afghanistan | P | - |
| 171 | AUS | 14522 | Afghanistan | P | - |
| 172 | AUS | 14523 | Afghanistan | P | - |
| 173 | AUS | 14524 | Afghanistan | P | - |
| 174 | AUS | 14525 | Afghanistan | P | - |
| 175 | AUS | 14526 | Afghanistan | P | - |
| 176 | AUS | 14527 | Afghanistan | P | - |
| 177 | AUS | 14528 | Afghanistan | P | - |
| 178 | AUS | 14531 | Afghanistan | P | - |
| 179 | AUS | 14532 | Afghanistan | P | Pm3b |
| 180 | AUS | 14535 | Afghanistan | P | - |
| 181 | AUS | 14546 | Afghanistan | A | - |
| 182 | AUS | 14547 | Afghanistan | P | - |
| 183 | AUS | 14565 | Afghanistan | P | - |
| 184 | AUS | 14566 | Afghanistan | P | - |
| 185 | AUS | 14567 | Afghanistan | P | - |
| 186 | AUS | 14568 | Afghanistan | P | - |
| 187 | AUS | 14569 | Afghanistan | P | - |
| 188 | AUS | 14605 | Afghanistan | A | - |
| 189 | AUS | 14606 | Afghanistan | P | - |
| 190 | AUS | 14607 | Afghanistan | P | - |
| 191 | AUS | 14608 | Afghanistan | P | - |
| 192 | AUS | 14609 | Afghanistan | P | - |
| 193 | AUS | 14610 | Afghanistan | P | - |
| 194 | AUS | 14611 | Afghanistan | P | - |
| 195 | AUS | 14612 | Afghanistan | P | - |
| 196 | AUS | 14613 | Afghanistan | P | - |
| 197 | AUS | 14614 | Afghanistan | P | - |
| 198 | AUS | 14624 | Afghanistan | P | - |
| 199 | AUS | 14625 | Afghanistan | P | - |
| 200 | AUS | 14626 | Afghanistan | P | - |
| 201 | AUS | 14627 | Afghanistan | P | - |
| 202 | AUS | 14628 | Afghanistan | P | - |
| 203 | AUS | 14629 | Afghanistan | A | - |
| 204 | AUS | 14630 | Afghanistan | A | - |
| 205 | AUS | 14631 | Afghanistan | P | - |
| 206 | AUS | 14632 | Afghanistan | P | - |
| 207 | AUS | 14633 | Afghanistan | A | - |
| 208 | AUS | 14634 | Afghanistan | P | - |
| 209 | AUS | 14635 | Afghanistan | A | - |
| 210 | AUS | 14636 | Afghanistan | P | - |
| 211 | AUS | 14637 | Afghanistan | P | - |
| 212 | AUS | 14638 | Afghanistan | P | - |
| 213 | AUS | 14639 | Afghanistan | P | - |
| 214 | AUS | 14640 | Afghanistan | P | - |
| 215 | AUS | 14641 | Afghanistan | P | - |
| 216 | AUS | 14642 | Afghanistan | A | - |
| 217 | AUS | 14643 | Afghanistan | P | - |
| 218 | AUS | 14644 | Afghanistan | P | - |
| 219 | AUS | 14645 | Afghanistan | P | - |
| 220 | AUS | 14646 | Afghanistan | P | - |
| 221 | AUS | 14647 | Afghanistan | P | - |
| 222 | AUS | 14648 | Afghanistan | P | - |
| 223 | AUS | 14649 | Afghanistan | P | - |
| 224 | AUS | 14650 | Afghanistan | P | - |
| 225 | AUS | 14689 | Afghanistan | A | - |
| 226 | AUS | 14690 | Afghanistan | P | - |
| 227 | AUS | 14691 | Afghanistan | P | - |
| 228 | AUS | 14692 | Afghanistan | P | - |
| 229 | AUS | 14693 | Afghanistan | P | - |
| 230 | AUS | 14694 | Afghanistan | P | - |
| 231 | AUS | 14695 | Afghanistan | P | - |
| 232 | AUS | 14696 | Afghanistan | P | - |
| 233 | AUS | 14697 | Afghanistan | P | - |
| 234 | AUS | 14698 | Afghanistan | P | - |
| 235 | AUS | 14699 | Afghanistan | A | - |
| 236 | AUS | 14700 | Afghanistan | P | - |
| 237 | AUS | 14701 | Afghanistan | P | - |
| 238 | AUS | 14702 | Afghanistan | P | - |
| 239 | AUS | 14703 | Afghanistan | P | - |
| 240 | AUS | 14704 | Afghanistan | A | - |
| 241 | AUS | 14705 | Afghanistan | P | - |
| 242 | AUS | 14706 | Afghanistan | A | - |
| 243 | AUS | 14707 | Afghanistan | P | - |
| 244 | AUS | 14708 | Afghanistan | P | - |
| 245 | AUS | 14709 | Afghanistan | P | - |
| 246 | AUS | 14710 | Afghanistan | P | - |
| 247 | AUS | 14711 | Afghanistan | A | - |
| 248 | AUS | 14713 | Afghanistan | P | - |
| 249 | AUS | 14714 | Afghanistan | P | - |
| 250 | AUS | 14715 | Afghanistan | P | - |
| 251 | AUS | 14840 | Afghanistan | P | Pm3b |
| 252 | AUS | 14841 | Afghanistan | P | - |
| 253 | AUS | 14842 | Afghanistan | P | - |
| 254 | AUS | 14843 | Afghanistan | A | - |
| 255 | AUS | 14844 | Afghanistan | A | - |
| 256 | AUS | 14845 | Afghanistan | P | - |
| 257 | AUS | 14846 | Afghanistan | P | - |
| 258 | AUS | 14847 | Afghanistan | P | - |
| 259 | AUS | 14848 | Afghanistan | P | - |
| 260 | AUS | 14849 | Afghanistan | P | - |
| 261 | AUS | 14850 | Afghanistan | A | - |
| 262 | AUS | 14851 | Afghanistan | A | - |
| 263 | AUS | 14852 | Afghanistan | P | - |
| 264 | AUS | 15209 | Afghanistan | A | - |
| 265 | AUS | 15210 | Afghanistan | A | - |
| 266 | AUS | 15212 | Afghanistan | P | - |
| 267 | AUS | 15218 | Afghanistan | P | - |
| 268 | AUS | 15320 | Afghanistan | A | - |
| 269 | AUS | 15321 | Afghanistan | P | - |
| 270 | AUS | 15624 | Afghanistan | P | - |
| 271 | AUS | 17502 | Afghanistan | P | - |
| 272 | AUS | 17503 | Afghanistan | A | - |
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an Open Access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Bhullar, N.K.; Mackay, M.; Keller, B. Genetic Diversity of the Pm3 Powdery Mildew Resistance Alleles in Wheat Gene Bank Accessions as Assessed by Molecular Markers. Diversity 2010, 2, 768-786. https://doi.org/10.3390/d2050768
Bhullar NK, Mackay M, Keller B. Genetic Diversity of the Pm3 Powdery Mildew Resistance Alleles in Wheat Gene Bank Accessions as Assessed by Molecular Markers. Diversity. 2010; 2(5):768-786. https://doi.org/10.3390/d2050768
Chicago/Turabian StyleBhullar, Navreet K., Michael Mackay, and Beat Keller. 2010. "Genetic Diversity of the Pm3 Powdery Mildew Resistance Alleles in Wheat Gene Bank Accessions as Assessed by Molecular Markers" Diversity 2, no. 5: 768-786. https://doi.org/10.3390/d2050768
APA StyleBhullar, N. K., Mackay, M., & Keller, B. (2010). Genetic Diversity of the Pm3 Powdery Mildew Resistance Alleles in Wheat Gene Bank Accessions as Assessed by Molecular Markers. Diversity, 2(5), 768-786. https://doi.org/10.3390/d2050768




