Genetic Variability of Macedonian Tobacco Varieties Determined by Microsatellite Marker Analysis
Abstract
:1. Introduction
2. Results and Discussion
| Marker | No. of alleles | No. of rare alleles | Size range (bp) | Highest frequency allele | PIC Value | |
|---|---|---|---|---|---|---|
| Size (bp) | Frequency (%) | |||||
| PT 1193 | 1 | 0 | 228 | 228 | 100 | 0.00 |
| PT 20021 | 2 | 0 | 342–345 | 342 | 90 | 0.18 |
| PT 20165 | 2 | 1 | 202–205 | 202 | 95 | 0.10 |
| PT 20172 | 6 | 2 | 196–244 | 196 | 45 | 0.71 |
| PT 20176 | 2 | 0 | 255–261 | 255, 261 | 50 | 0.50 |
| PT 20242 | 5 | 1 | 195–216 | 210, 213 | 30 | 0.76 |
| PT 20388 | 3 | 0 | 182–191 | 191 | 80 | 0.34 |
| PT 20445 | 3 | 0 | 196–271 | 202, 271 | 40 | 0.64 |
| PT 30005 | 4 | 2 | 227–254 | 251 | 75 | 0.41 |
| PT 30021 | 6 | 2 | 222–251 | 241 | 40 | 0.73 |
| PT 30077 | 3 | 1 | 205–213 | 205 | 65 | 0.49 |
| PT 30087 | 2 | 0 | 177–180 | 180 | 90 | 0.18 |
| PT 30096 | 4 | 0 | 229–241 | 238 | 40 | 0.70 |
| PT 30138 | 3 | 1 | 216–222 | 216 | 85 | 0.27 |
| PT 30144 | 4 | 0 | 262–286 | 262 | 70 | 0.48 |
| PT 30150 | 2 | 0 | 186–189 | 189 | 65 | 0.46 |
| PT 30160 | 1 | 0 | 181 | 181 | 100 | 0.00 |
| PT 30164 | 2 | 0 | 152–155 | 152 | 80 | 0.32 |
| PT 30188 | 1 | 0 | 155 | 155 | 100 | 0.00 |
| PT 30231 | 1 | 0 | 180 | 180 | 100 | 0.00 |
| PT 30255 | 1 | 0 | 225 | 225 | 100 | 0.00 |
| PT 30274 | 5 | 2 | 193–208 | 202 | 45 | 0.67 |
| PT 30378 | 3 | 0 | 219–228 | 219 | 60 | 0.56 |
| PT 30392 | 5 | 1 | 253–271 | 253 | 35 | 0.70 |
| PT 30417 | 2 | 0 | 182–191 | 182 | 90 | 0.18 |
| PT 30463 | 1 | 0 | 149 | 149 | 100 | 0.00 |
| PT 30480 | 4 | 0 | 153–171 | 159 | 60 | 0.58 |
| PT 40015 | 6 | 0 | 156–172 | 168 | 40 | 0.76 |
| PT 40021 | 4 | 0 | 151–182 | 154 | 50 | 0.64 |
| PT 40035 | 2 | 0 | 196–199 | 199 | 80 | 0.32 |
| Total | 90 | 13 | ||||
| Mean | 3 | 0.43 | 70.00 | 0.39 | ||
| No. | Marker | Djebel | P 12-2/1 | P 23 | Jb-125/3 | Jk-48 | P-80pt | P-66-9/7 | NS-72 | B-2/93 | V MV1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PT1193 | 228 | 228 | 228 | 228 | 228 | 228 | 228 | 228 | 228 | 228 |
| 2 | PT20021 | 345 | 342 | 342 | 342 | 342 | 342 | 342 | 342 | 342 | 342 |
| 3 | PT20165 | 202 | 202 | 202 | 202 | 202 | 202 | 202 | 202 | 202/205 | 202 |
| 4 | PT20172 | 220 | 196 | 196 | 226 | 220 | 196 | 196 | 196/220 | 190/223 | 244 |
| 5 | PT20176 | 255 | 261 | 261 | 261 | 261 | 255 | 255 | 261 | 255 | 255 |
| 6 | PT20242 | 210 | 213 | 213 | 216 | 216 | 213 | 210 | 210 | 195/204 | 195 |
| 7 | PT20388 | 191 | 191 | 191 | 191 | 191 | 191 | 191 | 191 | 185 | 182 |
| 8 | PT20445 | 271 | 202 | 202 | 271 | 271 | 202 | 202 | 271 | 196 | 196 |
| 9 | PT30005 | 254 | 251 | 251 | 251 | 251 | 251 | 251 | 251 | 227/254 | 230/251 |
| 10 | PT30021 | 231 | 241 | 241 | 231 | 231 | 241 | 243 | 241 | 225/251 | 222 |
| 11 | PT30077 | 205 | 205 | 205 | 205/207 | 205/207 | 205 | 205 | 207 | 205/213 | 207 |
| 12 | PT30087 | 180 | 180 | 180 | 180 | 180 | 180 | 180 | 180 | 180 | 177 |
| 13 | PT30096 | 232 | 238 | 238 | 232 | 232 | 238 | 241 | 238 | 229 | 229 |
| 14 | PT30138 | 216 | 216 | 216 | 216 | 216 | 216 | 216 | 216 | 216/222 | 219/222 |
| 15 | PT30144 | 262 | 262 | 262 | 262 | 262 | 266 | 262 | 262 | 286 | 270 |
| 16 | PT30150 | 186 | 189 | 189 | 186 | 186 | 189 | 189 | 189 | 186/189 | 189 |
| 17 | PT30160 | 181 | 181 | 181 | 181 | 181 | 181 | 181 | 181 | 181 | 181 |
| 18 | PT30164 | 152 | 152 | 152 | 152 | 152 | 155 | 155 | 152 | 152 | 152 |
| 19 | PT30188 | 155 | 155 | 155 | 155 | 155 | 155 | 155 | 155 | 155 | 155 |
| 20 | PT30231 | 180 | 180 | 180 | 180 | 180 | 180 | 180 | 180 | 180 | 180 |
| 21 | PT30255 | 225 | 225 | 225 | 225 | 225 | 225 | 225 | 225 | 225 | 225 |
| 22 | PT30274 | 208 | 202 | 202 | 208 | 208 | 202 | 202 | 202/208 | 196/199 | 193 |
| 23 | PT30378 | 219 | 219 | 219 | 228 | 228 | 219 | 222 | 222 | 219 | 219 |
| 24 | PT30392 | 271 | 256 | 256 | 253 | 253 | 256 | 256 | 253 | 253/259 | 265 |
| 25 | PT30417 | 182 | 182 | 182 | 182 | 182 | 182 | 182 | 182 | 182 | 191 |
| 26 | PT30463 | 149 | 149 | 149 | 149 | 149 | 149 | 149 | 149 | 149 | 149 |
| 27 | PT30480 | 171 | 153 | 159 | 159 | 159 | 153 | 162 | 159 | 159 | 159 |
| 28 | PT40015 | 158 | 168 | 168 | 156 | 156 | 168 | 168 | 170 | 172 | 160 |
| 29 | PT40021 | 154 | 151 | 151 | 154 | 154 | 151 | 154 | 154 | 182 | 179 |
| 30 | PT40035 | 199 | 199 | 199 | 199 | 199 | 199 | 199 | 199 | 196 | 196 |
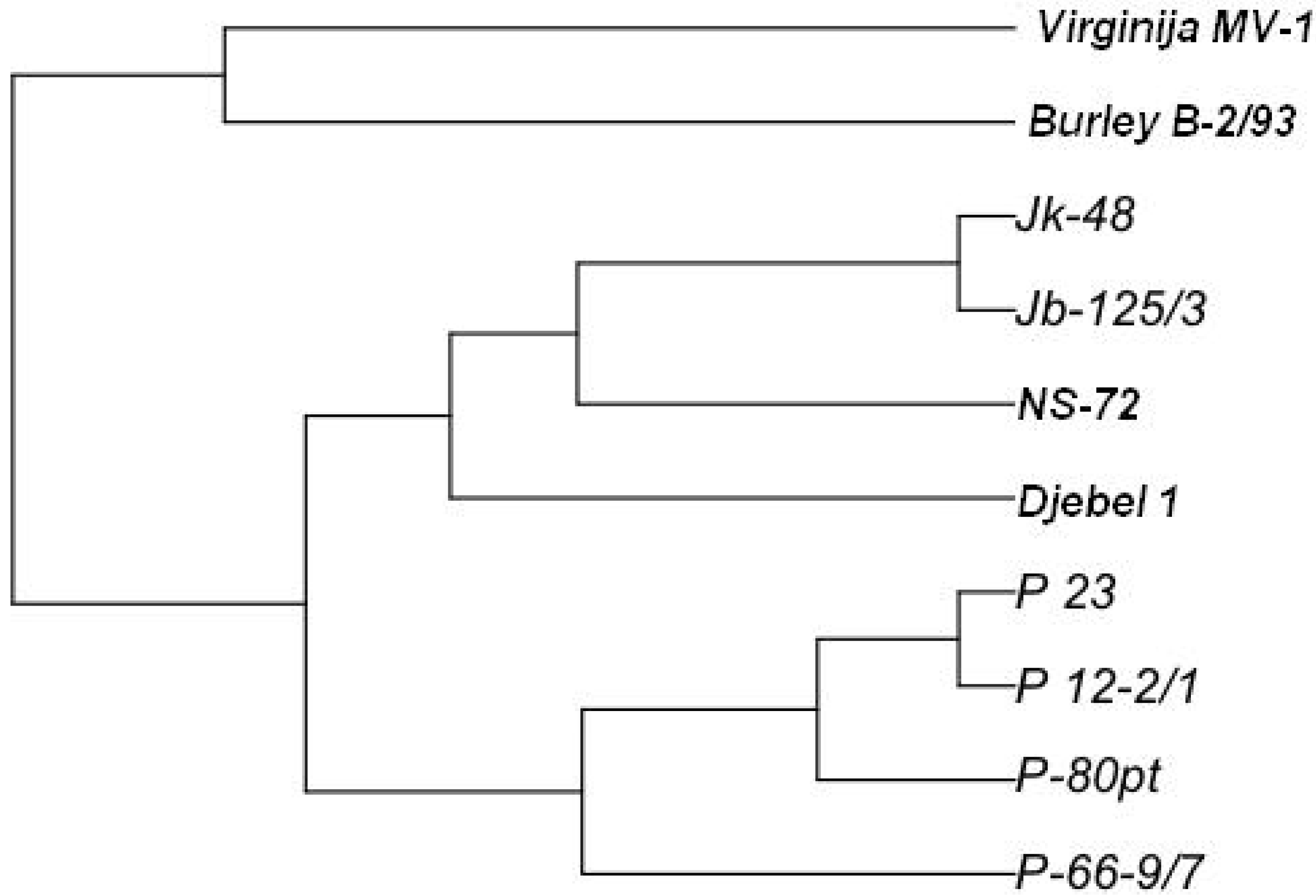
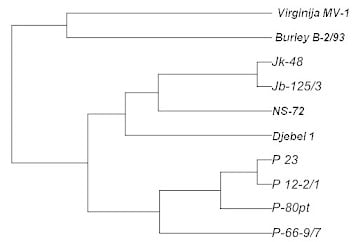
| B-2/93 | Djebel | NS- 72 | Jb 125/3 | Jk 48 | P 12-2/1 | P 23 | P 66-9/7 | P 80pt | V MB1 | |
|---|---|---|---|---|---|---|---|---|---|---|
| B-2/93 | 0.00 | |||||||||
| Djebel | 0.71 | 0.00 | ||||||||
| HC 72 | 0.73 | 0.49 | 0.00 | |||||||
| Jb 125/3 | 0.72 | 0.37 | 0.28 | 0.00 | ||||||
| Jk 48 | 0.72 | 0.32 | 0.26 | 0.03 | 0.00 | |||||
| P 12-2/1 | 0.75 | 0.63 | 0.34 | 0.53 | 0.53 | 0.00 | ||||
| P 23 | 0.67 | 0.63 | 0.29 | 0.48 | 0.48 | 0.03 | 0.00 | |||
| P 66-9/7 | 0.83 | 0.57 | 0.39 | 0.59 | 0.59 | 0.31 | 0.31 | 0.00 | ||
| P 80pt | 0.75 | 0.69 | 0.49 | 0.72 | 0.72 | 0.11 | 0.14 | 0.27 | 0.00 | |
| V MB1 | 0.54 | 1.08 | 0.84 | 0.98 | 0.98 | 0.94 | 0.86 | 1.03 | 0.94 | 0.00 |
3. Experimental Section
3.1. Plant Material and DNA Isolation
3.2. Molecular Markers
| Name | Repeat | Expected size (bp) |
|---|---|---|
| PT 1193 | AC/AT | 328 |
| PT 20021 | AAC | 345 |
| PT 20165 | CT/CTT | 208 |
| PT 20172 | CTT | 203 |
| PT 20176 | CTT | 258 |
| PT 20242 | AGG | 200 |
| PT 20388 | AAG | 185 |
| PT 20445 | AAG | 193 |
| PT 30005 | TAA | 230 |
| PT 30021 | TA | 224 |
| PT 30077 | CA | 210 |
| PT 30087 | CAA | 177 |
| PT 30096 | GAA | 234 |
| PT 30138 | GAA | 222 |
| PT 30144 | TA | 266 |
| PT 30150 | TA | 229 |
| PT 30160 | TAAAAA | 186 |
| PT 30164 | CAG | 153 |
| PT 30188 | GA | 159 |
| PT 30231 | CAT | 184 |
| PT 30255 | GAAA | 228 |
| PT 30274 | GGA | 213 |
| PT 30378 | CGA | 222 |
| PT 30392 | TAA | 270 |
| PT 30417 | CAA | 193 |
| PT 30463 | TAAA | 169 |
| PT 30480 | CAG | 163 |
| PT 40015 | GA | 170 |
| PT 40021 | CAG | 153 |
| PT 40035 | CAT | 199 |
3.3. PCR Conditions and Allele Detection
3.4. Diversity Analysis
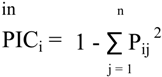
4. Conclusions
Acknowledgements
References and Notes
- Kenton, A.; Parokonny, A.S.; Gleba, Y.Y.; Bennett, M.D. Characterization of the Nicotiana tabacum L. genome by molecular cytogenetics. Mol. Gen. Genet. 1993, 240, 159–169. [Google Scholar] [CrossRef]
- Lim, K.Y.; Matyasek, R.; Kovarik, A.; Leitch, A.R. Genome evolution in allotetraploid Nicotiana. Biol. J. Linn. Soc. 2004, 82, 599–606. [Google Scholar] [CrossRef]
- Gadani, F.; Ayers, D.; Hempfling, W. Tobacco: a tool for plant genetic engineering research and molecular farming. Part I. Agro. Food Ind. Hi. Tech. 1995, 6, 19–24. [Google Scholar]
- Brandle, J.; Bai, D. Biotechnology: uses and applications in tobacco improvement. In Tobacco: Production, Chemistry and Technology; Davis, N., Ed.; Wiley-Blackwell: Oxford, UK, 1999; pp. 49–65. [Google Scholar]
- Korubin-Aleksoska, A. Tobacco Varieties from Tobacco Institute-Prilep; Trajkovski, F., Ed.; Alfa print: Skopje, Macedonia, 2004. [Google Scholar]
- Martz, F.; Maury, S.; Pincon, G.; Legrand, M. cDNA cloning, substrate specificity and expression study of tobacco caffeoyl-Coa 3-O-methyltransferase, a lignin biosynthetic enzyme. Annu. Rev. Plant. Physiol. 1998, 36, 427–437. [Google Scholar]
- Bai, D.; Reeleder, R.; Brandle, J.E. Identification of two RAPD markers tightly linked with the Nicotiana debneyi gene for resistance to black root rot of tobacco. Theor. Appl. Genet. 1995, 91, 1184–1189. [Google Scholar]
- Bai, D. Three novel Nicotiana debneyi specific repetitive DNA elements derived from a RAPD marker. Genome 1999, 42, 104–109. [Google Scholar] [CrossRef]
- Del Piano, L.; Abet, M.; Sorrentino, C.; Acanfora, F.; Cozzolino, E.; Di Muro, A. Genetic variability in Nicotiana tabacum and Nicotiana species as revealed by RAPD markers: Development of the RAPD procedure. Contrib. Tob. Res. 2000, 19, 1–15. [Google Scholar]
- Rossi, L.; Bindler, G.; Pijnenburg, H.; Isaac, P.G.; Giraud-Henry, I.; Mahe, M.; Orvain, C.; Gadani, F. Potential of molecular marker analysis for variety identification in processed tobacco. Plant Var. Seeds 2001, 14, 89–101. [Google Scholar]
- Tezuka, T.; Onosato, K.; Hijishita, S.; Marubashi, W. Development of Q-chromosome-specific DNA markers in tobacco and their use for identification of a tobacco monosomic line. Plant Cell Physiol. 2004, 45, 1863–1869. [Google Scholar] [CrossRef]
- Sarla, K.; Rao, R.V.S. Genetic diversity in Indian FCV and burley tobacco cultivars. J. Genet. 2008, 87, 159–163. [Google Scholar] [CrossRef]
- Ren, N.; Timko, M.P. AFLP analysis of genetic polymorphism and evolutionary relationships among cultivated and wild Nicotiana species. Genome 2001, 44, 559–571. [Google Scholar] [CrossRef]
- Bindler, G.; van der Hoeven, R.; Gunduz, I.; Plieske, J.; Ganal, M.; Rossi, L.; Gadani, F.; Donini, P. A microsatellite marker based linkage map of tobacco. Theor. Appl. Genet. 2007, 114, 341–349. [Google Scholar]
- Zhang, H.Y.; Liu, X.Z.; He, C.S.; Yang, Y.M. Genetic Diversity among flue-cured tobacco cultivars on the basis of RAPD and AFLP markers. Braz. Arch. Biol. Technol. 2008, 51, 1097–1101. [Google Scholar] [CrossRef]
- Siva-Raju, K.; Madhav, M.S.; Sharma, R.K.; Murthy, T.G.K.; Mohapatra, T. Genetic polymorphism of Indian tobacco types as revealed by amplified fragment length polymorphism. Curr. Sci. 2008, 94, 633–639. [Google Scholar]
- Moon, H.S.; Nicholson, J.S.; Heineman, A.; Lion, K.; van der Hoeven, R.; Hayes, A.J; Lewis, R.S. Changes in genetic diversity of U.S. flue-cured tobacco germplasm over seven decades of cultivar development. Crop Sci. 2009, 49, 498–508. [Google Scholar] [CrossRef]
- Moon, H.S.; Nifong, J.M.; Nicholson, J.S.; Heineman, A.; Lion, K.; van der Hoeven, R.; Hayes, A.J.; Lewis, R.S. Microsatellite-based analysis of tobacco (Nicotiana tabacum L.) genetic resources. Crop Sci. 2009, 49, 2149–2159. [Google Scholar] [CrossRef]
- Edwards, A.W. Distances between populations on the basis of gene frequencies. Biometrics 1971, 27, 873–81. [Google Scholar] [CrossRef]
- Nei, M. Genetic distances between populations. Amer. Naturalist 1972, 106, 283–292. [Google Scholar]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 23, 341–369. [Google Scholar]
- Rogers, J.S. Measures of genetic similarity and genetic distances. Stud. Gen. 1972, 7213, 145–153. [Google Scholar]
- Anderson, J.A.; Churchill, G.A.; Autrique, J.E.; Tanksley, S.D.; Sorrellis, M.E. Optimizing parental selection for genetic linkage maps. Genome 1993, 36, 181–186. [Google Scholar] [CrossRef]
- The R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2009. [Google Scholar]
- Chessel, D.; Dufour, A.B.; Thioulouse, J. The ade4 package-I- One-table methods. R. News 2004, 4, 5–10. [Google Scholar]
- Dray, S.; Dufour, A.B.; Chessel, D. The ade4 package-II: Two-table and K-table methods. R. News 2007, 7, 47–52. [Google Scholar]
- Paradis, E.; Claude, J.; Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef]
- Meirmans, P.G.; Van Tienderen, P.H. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 2004, 4, 792–794. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Davalieva, K.; Maleva, I.; Filiposki, K.; Spiroski, O.; Efremov, G.D. Genetic Variability of Macedonian Tobacco Varieties Determined by Microsatellite Marker Analysis. Diversity 2010, 2, 439-449. https://doi.org/10.3390/d2040439
Davalieva K, Maleva I, Filiposki K, Spiroski O, Efremov GD. Genetic Variability of Macedonian Tobacco Varieties Determined by Microsatellite Marker Analysis. Diversity. 2010; 2(4):439-449. https://doi.org/10.3390/d2040439
Chicago/Turabian StyleDavalieva, Katarina, Ivana Maleva, Kiril Filiposki, Ognen Spiroski, and Georgi D. Efremov. 2010. "Genetic Variability of Macedonian Tobacco Varieties Determined by Microsatellite Marker Analysis" Diversity 2, no. 4: 439-449. https://doi.org/10.3390/d2040439
APA StyleDavalieva, K., Maleva, I., Filiposki, K., Spiroski, O., & Efremov, G. D. (2010). Genetic Variability of Macedonian Tobacco Varieties Determined by Microsatellite Marker Analysis. Diversity, 2(4), 439-449. https://doi.org/10.3390/d2040439