Abstract
Neospora caninum is a parasite regarded a major cause of foetal loss in cattle. A key requirement to an understanding of the epidemiology and pathogenicity of N. caninum is knowledge of the biological characteristics of the species and the genetic diversity within it. Due to the broad intermediate host range of the species, worldwide geographical distribution and its capacity for sexual reproduction, significant biological and genetic differences might be expected to exist. N. caninum has now been isolated from a variety of different host species including dogs and cattle. Although isolates of this parasite show only minor differences in ultrastructure, considerable differences have been reported in pathogenicity using mainly mouse models. At the DNA level, marked levels of polymorphism between isolates were detected in mini- and microsatellites found in the genome of N. caninum. Knowledge of what drives the biological differences that have been observed between the various isolates at the molecular level is crucial in aiding our understanding of the epidemiology of this parasite and, in turn, the development of efficacious strategies, such as live vaccines, for controlling its impact. The purpose of this review is to document and discuss for the first time, the nature of the diversity found within the species Neospora caninum.
1. Introduction
Neospora caninum is a cyst-forming apicomplexan parasite [1], which infects many different hosts, cell types and tissues. It causes neosporosis, namely stillbirth and abortion in cattle and neonatal neuromuscular disease in dogs, and has been found in several other animal species [1,2,3]. Neospora caninum was originally described as a parasite of the domestic dog [1] and was initially recognised in Norway as a cause of hind limb Toxoplasma-like illness in dogs [4,5], resulting in paralysis in pups and early death. Neosporosis was first reported as a cause of abortion in dairy cows in 1989 in the United States [6] and infection was reported retrospectively from stored tissues of dogs and cattle dating back to 1957, 1958 [1,7] and 1974 [8], respectively. In addition, a new species of Neospora, named Neospora hughesi, was subsequently described in the brain and spinal cord of an adult horse in California [9]. Neospora caninum has a substantial economic impact in the dairy industry through losses of production estimated to reach millions of dollars [10,11,12].
Neospora caninum has been isolated from a variety of different host species such as the dog, cattle, sheep and water buffalo (Table 1). The first canine isolation (NC1) was by Dubey et al. [13] and the first bovine isolations (BPA1 and BPA2) were by Conrad et al. [14]; both from the United States. It is now clear that the phenotypic and genotypic characteristics of many of these isolates are not strictly conserved within the species, leading to significant levels of variation detectable amongst populations of parasites. A key requirement to the understanding of the epidemiology and pathogenicity of N. caninum is knowledge of the biological and genetic diversity found within the species. Due to the broad intermediate host range of the species, worldwide geographical distribution and its capacity for sexual reproduction, significant biological and genetic differences might be expected to exist [15]. The purpose of this review is to document for the first time, the nature of the diversity found within the species Neospora caninum.

Table 1.
Summary of Neospora caninum isolates from different host species.
| Isolates | Source | Country | Reference |
|---|---|---|---|
| NC1 | Brain of congenitally infected dog | United States | [13] |
| NC-2 | Muscle biopsy of naturally infected dog | United States | [16] |
| NC-3 | Brain and spinal cord of dog | United States | [17] |
| NC-4 & NC-5 | Brain of a congenitally dog | United States | [18] |
| NC-6, 7 & 8 | Brain and striated muscle of puppy | United States | [19] |
| NC-9 | Brain of naturally infected puppy | United States | [20] |
| CN1 | Brain and spinal cord of a congenitally infected dog | United States | [9] |
| NC-Liverpool | Cerebrum of congenitally infected puppy | United Kingdom | [21,22] |
| NC-Bahia | Brain of naturally infected adult dog | Brazil | [23] |
| NC-6 Argentina | Oocysts from naturally infected dog | Argentina | [24] |
| WA-K9 | Skin lesions of naturally infected dog | Australia | [25] |
| CZ-4 * | Oocysts from naturally infected dog | Czech Republic | [26] |
| NC-GER1 | Brain and spinal cord of congenitally infected puppy | Germany | [27] |
| Hammondia heydorni-Berlin-1996 | Oocysts from naturally infected dog | Germany | [28] |
| NC-GER2, 3, 4, 5 and NC-GER-6 | Oocystes from naturally infected dog | Germany | [29] |
| NC-GER7, 8 and NC-GER-9 | Oocysts from naturally infected dog | Germany | [30] |
| NC-P1 | Oocysts from naturally infected dog | Portugal | [31] |
| BPA1 & BPA2 | Brain of aborted bovine foetuses | United States | [14] |
| BPA3 & BPA4 | Brain of congenitally infected calf | United States | [32] |
| BPA6 | Brain and/or spinal cord of an aborted bovine foetus | United States | [33] |
| NC-Beef | Naturally infected calf | United States | [34] |
| NC-Illinois | Brain of naturally infected calf | United States | [35] |
| VMDL1 | Brain of aborted beef calf | United States | [36] |
| NC-LivB1 | Brain of stillborn calf | United Kingdom | [37,38] |
| NC-LivB2 | Brain of aborted bovine foetus | United Kingdom | [39] |
| NC-Porto1 | Brain of aborted bovine foetus | Portugal | [40] |
| NC-Sp1 | Brain of aborted bovine foetus | Spain | [41] |
| Five isolates | Brain of four aborted bovine foetuses and one stillborn calf | Spain | [42] |
| NC-Spain 1H | Brain of naturally infected and healthy calf | Spain | [43] |
| NC-Spain 2H, 3H, 4H and 5H, NC-Spain6, 7, 8, 9, 10 | Brain of naturally infected and healthy calf | Spain | [44] |
| JAP1 | Brain and spinal cord of congenitally infected calf | Japan | [45,46] |
| BT-2 & JAP-2 JAP-5 JAP-4 | Brain of congenitally infected calf Brain and spinal cord of congenitally infected calf Brain of stillborn calf | Japan | [47] |
| BT-3 | Brain of an infected adult cow | Japan | [48] |
| NC-Sheep+ | Brain of naturally infected pregnant sheep | Japan | [49] |
| NC-SweB1 | Brain of stillborn calf | Sweden | [50] |
| NC-VP1 | Brain of a congenitally infected calf | Italy | [51] |
| NCPG1 | Brain and placenta cotyledonary villi of clinically naturally infected calf | Italy | [52] |
| NC-MalB1 | Brain of congenitally infected calf | Malaysia | [53] |
| KBA-1 | Brain of a congenitally infected calf | South Korea | [54] |
| KBA-2 | Brain of aborted bovine foetus | ||
| NC-Kr2 | Brain of naturally infected cow | South Korea | Jeong et al. unpublished |
| BNC-PRI | Brain of congenitally blind calf | Brazil | [55] |
| BCN-PR3 | Brain of aborted bovine foetus | Brazil | [56] |
| No name | Brain of naturally infected sheep | Brazil | [57] |
| Nc-Goiás 1 | Brain of clinically healthy calf | Brazil | [58] |
| NC-Nowra | Brain and spinal cord of congenitally infected calf | Australia | [59] |
| NcNZ1 NcNZ2 NcNZ3 | Brain of naturally infected cow Brain of infected 2-days old calf Brain of stillborn calf | New Zealand | [60] |
| NcIs491 NcIs580 | Brain of aborted bovine foetus | Israel | [61] |
| NC-PolB1 | Brain of naturally infected calf | Poland | [62,63] |
| NcBrBuf-1, 2, 3, 4, 5 | Brain of naturally infected water buffalo | Brazil | [64] |
| Nc-Iran1 | Brain of aborted bovine foetus | Iran | Salehi et al. unpublished |
| NC-deer1 * | Brain of naturally infected white-tailed deer ( Odocoileus virginianus ) | United States | [65] |
| NC-WTDVA-1 NC-WTDVA-2* and 3* | Brain of naturally infected white-tailed deer ( Odocoileus virginianus) | United States | [66] |
* Neospora caninum was not established into a cell culture.+ Our designate.
2. Comparison of Biological Characteristics between Neospora caninum Isolates
2.1. Life Cycle of N caninum
Neospora caninum is an obligate intracellular parasite. The life cycle of this parasite (see Figure 1) involves two hosts: an intermediate and a definitive host [3]. Canids, such as the dog, coyote and dingo, are known to be definitive hosts [34,67,68,69]; and they shed unsporulated oocysts in their faeces. These sporulate within a couple of days to become infective to an intermediate host (such as cow, sheep, goat or deer) when they consume food or water contaminated with the oocysts. Sporozoites excyst from the oocysts in the gut, invade and begin to develop inside host cells where they multiply by binary fission (endodyogeny) to form more rod- or banana shaped cells called tachyzoites (“tachy” means fast—because they replicate fast). These undergo a number of replications and can invade and infect other tissues in the body, such as neural cells, macrophages, fibroblasts, vascular endothelial cells, muscles cells, liver cells and renal tubular epithelial cells. Eventually, under the influence of a strong, γ interferon dominated, cell mediated immune response, the tachyzoites transform into bradyzoites (“brady” meaning slow), which also divide by endodyogeny. Bradyzoites develop into large, intracellular cyst-like structures with a solid cyst wall surrounding them. The cyst itself is called a tissue cyst, usually up to 100 µm in size, and is typically found in neural or skeletal muscle cells. The tissue cyst can persist within an infected host for several month or years without causing significant clinical manifestations [70,71].
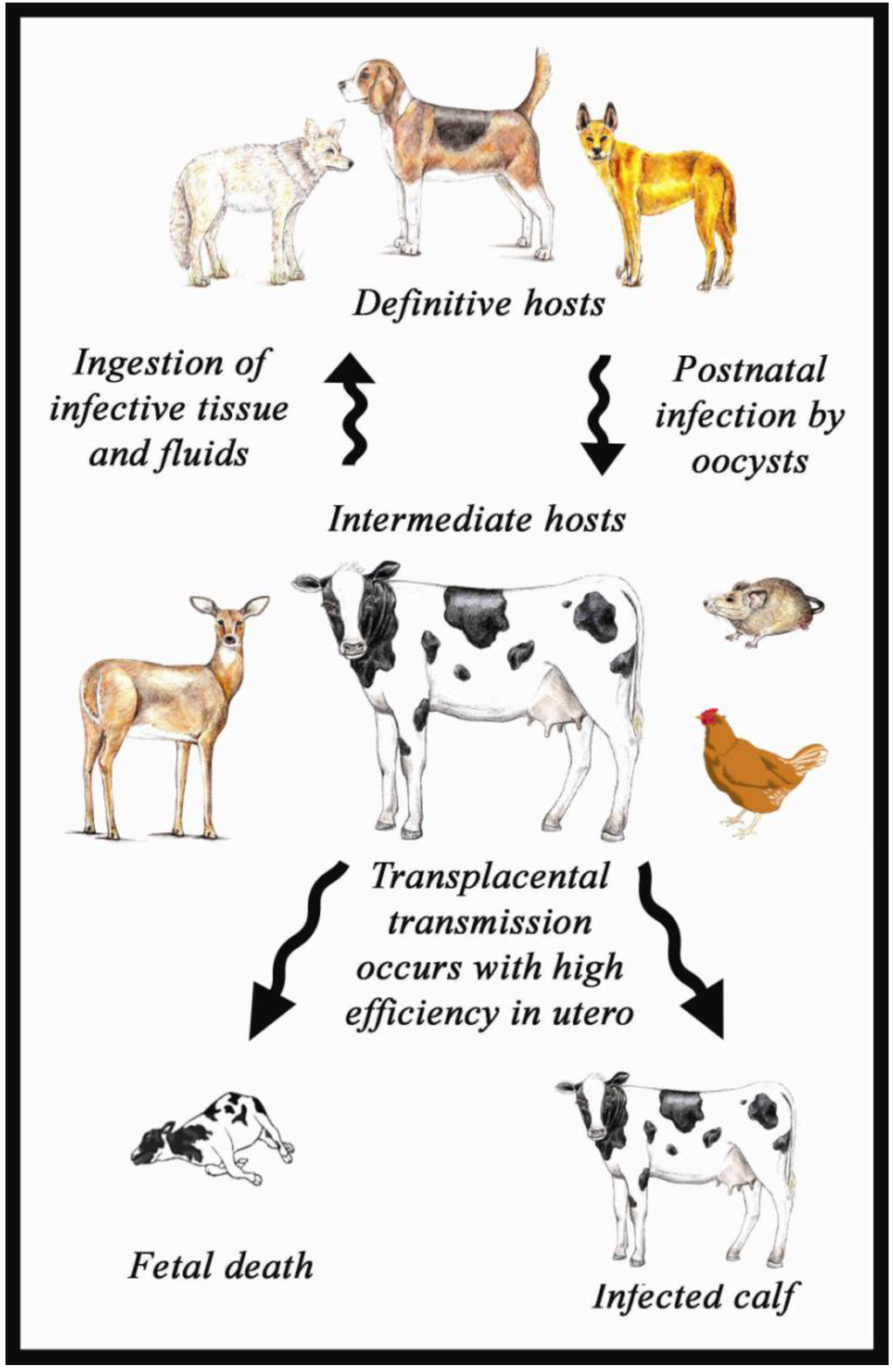
Figure 1.
Life cycle of N caninum.
Tachyzoites can also cross the placenta to cause foetal infection [13,70]. Congenitally infected foetuses may die in utero, be resorbed, mummified, autolysed or a calf may be stillborn. In cattle, however, calves infected later in pregnancy, after the onset of development of the foetal immune system, tend to survive. They may be born clinically normal, but chronically infected [72,73,74]. Dams infected in this way can also pass on this infection during a subsequent pregnancy, and so infections are commonly found in family lines.
A definitive host such as the dog becomes infected when it eats tissue containing bradyzoites or, rarely, tachyzoites [10,75,76] but whether definitive hosts can be infected by swallowing a sporulated oocyst is still unknown [10]. When tissue cysts are ingested by a definitive host, bradyzoites are released and subsequently they invade neural and skeletal muscle fiber cells and undergo schizogony (asexual reproduction). The schizont contains a highly vesicular nucleus plus a deeply basophilic cytoplasm and during schizogony the nucleus undergoes several rounds of division without any corresponding division of cytoplasm. Subsequently the schizonts become mature and result in the release of numerous zoites into the body [19].
The life cycle stages of N. caninum occurring during gametogony are unknown; no gamonts (macrogametocytes and microgametocytes) or zygotes have yet been observed within the canine alimentary canal, presumably because they are few in number.
2.2. Life Cycle Stages and Ultrastructure
Using the light microscope, little variation amongst isolates can be observed in the structure of N. caninum. Ultrastructurally, no major variation in tachyzoites and tissue cysts was detected between bovine and canine isolates of this parasite, although minor differences have been noted in several studies.
2.2.1. Tachyzoite size
Some minor differences are reported in tachyzoite size. Tachyzoites are usually within the (4.3–8.4 × 1.3–2.5 µm) range (see Table 2). The variation in tachyzoite size might reflect the stage of growth and division [70].

Table 2.
Size of tachyzoites from different Neospora caninum isolates.
| Isolate name | Tachyzoite size (µm) | References |
|---|---|---|
| NC1 | 4.3–4.6 × 2.1–2.3, 4.8–5.3 × 1.8–2.3 | [1] |
| 5.1–8.4 × 1.5–2.5 | [77] | |
| NC-SweB1 | 5 × 1.3 | [50] |
| KBA1 and KBA2 | 5–7 × 1.5–2 | [54] |
| NC-Liverpool | 6 × 2 | [22] |
| BPA1 and BPA2 | 6–8 × 1–2 | [14] |
| JAP1 | 5–6 × 1.5–2 | [46] |
| NC-Sheep+ | 7 × 3 | [49] |
+ Our designate.
2.2.2. Rhoptries
Variable numbers of rhoptries are reported in transverse sections of individual tachyzoites. The numbers of rhoptries are in the 4 to 24 µm range (see Table 3). The variation in the number of rhoptries reported by different researchers may be due in part to the difficulty of distinguishing between rhoptries and dense granules [78,79].

Table 3.
Number of rhoptries present in different Neospora caninum tachyzoites.
| Isolate name | Rhoptry Number. | References |
|---|---|---|
| NC1, NC-2 and NC-3 | 8–12 | [1,80] |
| KBA-1 and KBA-2 | 8–12 | [54] |
| NC-SweB1 | 4–16 | [50] |
| JAP1 | 10–14 | [46] |
| BPA1 and BPA2 | 24 | [14] |
2.2.3. Micronemes
The micronemes in tachyzoites are orientated parallel to the rhoptries on a longitudinal axis in (BPA1, BPA2) or mostly arranged in parallel (NC-Liverpool) [81]. In NC1 micronemes are orientated in either a perpendicular, parallel or random arrangement [13]. The significance of these arrangements is unknown.
2.2.4. Tissue cysts
Variable cyst sizes are reported in tissues (mainly the brain) from different hosts. The largest reported size is 55–107 µm long which was found in the brain of a congenitally infected dog [1] (see Table 4). The tissue cyst thickness might be related to host age [82] or how long the infection has existed in the host [70]. It has been reported that the wall of small sized tissue cysts is thicker than that of larger tissue cysts seen in the spinal cord of a naturally infected calf [83]. The tissue cyst wall produced by NC-Liverpool infection in the Swiss Webster mouse is smooth and slightly thicker (2.4 µm) compared to the NC-5 (1.9 µm) thickness where the wall in outline is irregular [82]. A high density of tissue cysts was observed in a European dog infected by NC-Liverpool compared to the fewer noted in the brain of an American dog infected with NC1 [22,81]. It was reported that NC-Liverpool produced more tissue cysts in mice brain than NC-2 [84]. Tissue cysts might not be seen in infected hosts, even in seropositive animals [20,34,47]. It has also been reported that bradyzoites of NC-2 were able to survive in pepsin-HCl solution (1%) for 30 min in contrast to NC1 which died [85].

Table 4.
Size of tissue cysts of Neospora caninum reported in different infected hosts.
| Source | Tissue cysts size (µm)* | Thickness of the tissue cysts (µm)* | References |
|---|---|---|---|
| Brain of congenitally infected dog | 55 × 25, 65–45, 100 × 77 and 107–75 | 2–3 1.5–3.5 | [1] |
| ND | >1 | [86] | |
| 14–65 long | 0.7–4.5 | [19] | |
| 10–42 × 10–40 | ~ 1 | [18] | |
| Brain of naturally infected dog | 18.1–27.4 × 14.1–23.7 | 0.74–1.12 | [81] |
| Brain of infected Swiss Webster mice | ND | 0.5–4 | [82] |
| Brain of infected bovine foetuses | 6.8–22.7 × 7.4–23.7 | 0.34–1.05, 0.23–0.77, 0.43–1.05 and 0.77–1.22 | [87] |
| 8–10 8–13 | <1 1–2 | [14] | |
| 50 | 2 | [88] | |
| Spinal cord of naturally infected calf | 20–48 14–20 | 1–2 0.53–3.09 and 0.53–1.69 | [83] |
| Brain of naturally infected caprine foetuses | 10–32 15 14–21.8 | 1–2 1 0.51–1.75 | [89] |
| Heart muscle of naturally infected caprine foetus | 10–22 | 1–1.5 | |
| Cerebrum of an aborted goat foetus | 6–20 (mostly 10 µm) | 0.5–1.0 | [90] |
| Brain of gerbil inoculated with oocysts | 24.47–47.07 × 25.33–53.91 | 1.71–2.66 | [57] |
| 28 × 26 and 32 × 27 | >1 | [91] | |
| Brain of mice infected with tachyzoites | 15–107 long | ND | [92] |
| Brain of naturally infected horse | ND | 1.5–3 | [93] |
| ND | 2 | [94] |
*ND, Not determined.
2.2.5. Oocysts
Variable numbers of oocysts and duration of shedding are reported in experiments using naturally or experimentally infected dogs; the number of oocysts produced were between few to millions (see Table 5). The size of oocysts is (9.4–13.4 µm) (Table 5) and the prepatent period (before onset of oocyst shedding) varies extensively between experiments, being typically 5–10 days post infection, but it may be as high as 13 days. Oocyst shedding may continue for a further 10 days [34,95]. Factors that might affect shedding of oocysts by the dog are: the age of host, type of tissue fed to the host and the immune status of the host [10]. For example, higher numbers of oocysts were shed by puppies than adult dogs [35]. The numbers of oocysts shed by dogs fed naturally infected calf tissue were significantly higher than those dogs fed infected mouse carcasses [35]. It was hypothesised that bradyzoites present in tissue cysts in infected mice might be immature, low in number or attenuated by passage in an unnatural intermediate host [35]. Dogs fed skeletal muscle of experimentally infected sheep and goat induced the shedding of oocysts more than other tissues [96].

Table 5.
Sizes and numbers of oocysts shed by dogs.
| Sources | Size of oocysts (µm)* | Oocyst Number* | References |
|---|---|---|---|
| Dog fed infected mice carcasses | 10–11 | few | [34] |
| ND | 4.5 × 106 from dog1 but few from dog 2 | [67] | |
| Dog fed infected mouse brains | 10.6–12.4 × 10.6–12 | ND | [76] |
| Dog fed infected mouse carcasses | ≤11.5 | 100–29,900 | [35] |
| Dog fed experimentally infected calf tissues | 0–503,300 | ||
| Dog fed experimentally infected calf tissues | ND | 792,000 | [95] |
| Dog fed naturally infected bovine placenta | 10–11 | few | [75] |
| Dog fed muscles of experimentally infected sheep | 10.5–12.2, 10.5–11.7, 9.4–13.4 and 10.2–11.4 | 15 × 105 | [28] |
| Dog fed muscles of experimentally infected goat | 8 × 105 | ||
| Dog fed infected guinea pig | 1 × 106–2 × 106 | ||
| Dog fed brain of naturally infected white tailed deer | 10 | 12,300 | [65] |
| Dog fed brain of naturally infected water buffaloes | ND | 43,500–820,655 | [64] |
| Dog fed brain of naturally infected sheep | 6.56–10.84 × 9.73–11.86 | 27,600 | [57] |
| Naturally infected dog | 10.7 | 19–114,000 / g of faeces | [29] |
| 9.89 × 9.95 | ND | [30] | |
| 10 –13 × 10–11 | one million | [26] | |
| 10.4–12.9 × 10.6–11.8 | 400 / g of faeces | [97] | |
| 9.71–10.2 × 9.19–9.76 | 19–143,000 | [24] | |
| Naturally infected fox and coyote | 10–11 | few | [98] |
| Coyote fed experimentally infected calf tissues | 10 | 500 | [68] |
| Dingo fed experimentally infected calf tissues | 10 × 12 | 1,810 | [69] |
*ND, Not determined.
2.3. Pathogenicity
Studies on differences in pathogenicity among N. caninum isolates are common [22,43,87,99,100,101,102]. Since most N. caninum strains are isolated from hosts with clinical signs of disease, little is known about differences among isolates from symptomatic and asymptomatic animals [43] although information in this area is increasing.
Pathogenicity of N. caninum has been assessed in vivo using animal models. The susceptibility of the host plays an important role in parasite infection. Inbred BALB/c mice [103] and immunodeficient mice (nu/nu), INF-γ KO mice and gerbils are all highly susceptible to infection [18,70]. In N. caninum infection, it was demonstrated that the parasite isolate played a significant role in the progression of infection and occurrence of disease [102,104].
2.3.1. Studies with mice
Differences in pathogenicity between two canine isolates (NC1 and NC-2) were first noted in the course and severity of infection in Swiss white mice. Disease caused by NC1 was induced quicker and more severe compared to NC-2 [85]. Similar results were obtained by inoculation of (NC1 and NC-3) tachyzoites into BALB/c mice. NC-3 did not induce clinical signs of neosporosis or brain lesions [105]. NC-SweB1 appeared more pathogenic than NC-3 since severe brain lesions were detected in mice compared to NC3. Inoculation of mice (BALB/c, CBA/ca and ICR) with NC-2 caused a significantly higher level of mortality compared to NC-Liverpool [84].
Marked differences in pathogenicity (virulence) were also demonstrated between NC-Liverpool and NC-SweB1 in a mouse model for central nervous system (CNS) infection [102,106]. NC-Liverpool produced severe clinical signs of neosporosis (brain necrosis and a greater inflammatory response), dis-coordination, paralysis and weight loss in mice compared to NC-SweB1 over the same period.
Infection of BALB/c mice with the bovine isolate NC-Nowra showed it was less pathogenic (70% survival) in mice compared to those infected with NC-Liverpool where all the mice died. Mice infected with NC-Nowra also showed low levels of lesions in their brains (40%) compared to (100%) with NC-Liverpool. Infection of pregnant Qs mice with NC-Nowra did not cause significant amount of foetal loss in mice [59], although it is now recognised that foetal loss in Qs mice is not very reproducible [106]. NC-Liverpool was more virulent in BALB/c mice than NC1 and showed high levels of parasites in the brain [104].
Interferon-gamma gene knock out (KO) mice are susceptible to most intracellular parasites and most N. caninum isolates have the ability to cause death in these mice [18,20,31]. The deer isolate (NC-WTDVA-1, 2 and 3) were mildly virulent to infected KO mice [66].
In a pregnant mouse model (BALB/c), NC-Spain 1H possessed low virulence as determined by transplacental transmission and survival of offspring. The body weight of offspring infected with NC1 was significantly lower in comparison to NC-Spain 1H. The infection with NC-Spain 1H resulted in low levels of transplacental transmission and the offspring survival rate was 100%. The parasite was detected in only one pup brain by PCR. In contrast, NC1 infection resulted in high levels of transplacental transmission (92.8%) and subsequent high neonatal mortality rate over time (76.8%) compared to the NC-Spain 1 H infected group (0.5%) [43]. The transplacental transmission rate in pregnant mice using NC-Nowra was (87%) [59], similar to that obtained with NC-Liverpool (91%) [106].
2.3.2. Studies with gerbils
Inoculation of NC1 into gerbils resulted in either clinical signs of infection (neuromuscular) or the gerbils died [54]. In comparison inoculation of the same dose of tachyzoites of NcIs490 and NcIs580 into gerbils did not cause disease or death [61] suggesting these isolates are attenuated in their ability to cause disease.
The strains NcBrBuf-1, 2, 3, 4 and 5 were not virulent to gerbils [64] but NC1 and NC-Liverpool were virulent [107]. It was reported that gerbils died of acute neosporosis after inoculating them with canine strain NC-6, whereas those inoculated with NC-7 and NC-8 remained asymptomatic [19]. Swiss Webster (SW) out-bred albino mice inoculated with NC-Sp1, NC-6, NC-7 and NC-8 isolates remained asymptomatic [41].
2.3.3. Studies with cattle
Few results are published on the behaviour of N. caninum in cattle that allow comparisons of isolates. Initial studies were focused on correlating foetal loss with infection by N. caninum. Direct foetal inoculation of the BPA1 isolate into pregnant heifers at 118 day’s gestation (dg) resulted in foetal death [32]. Injection of NC-Liverpool tachyzoites into cattle at 70 dg also resulted in foetal death [108].
More recently, researchers have realized that not all isolates of N. caninum may cause fetal death when injected into a pregnant cow. Inoculation of pregnant heifers with an isolate of low virulence (NC-Spain 1H) at 70 dg did not result in foetal death whereas foetal death was detected in heifers inoculated with NC1 as a control. The immune response in heifers inoculated with NC1 was significantly different compared to heifers inoculated with NC-Spain 1H [109]. These studies are of course incredibly important as they provide strong support for the idea of a live vaccine that will prevent abortion in cattle. Vaccination with an isolate that is highly attenuated in its ability to cause fetal death in cattle is a highly attractive quality that is not possessed by most isolates of N. caninum studied to date. Unfortunately the cost of doing cattle experiments means that such experiments are rarely conducted unless industry support is forthcoming.
2.3.4. Studies with sheep
Similar comparative studies with different isolates have yet to be conducted in sheep, however several studies have confirmed that injection of N. caninum into pregnant sheep results in foetal death. Two ewes infected with 1.5 × 108 tachyzoites of NC1 at three months of pregnancy resulted in twin dead lambs, 25 and 26 days post inoculation and foetal lesions were reported in the CNS, skeletal muscles and placenta of the lambs [110]. Eight of 12 infected pregnant ewes with 7 × 105 or 1.7 × 106 tachyzoites of a mixture of NC-Liverpool and NC-2 isolates at 90 dg, regardless of the inoculum dose, resulted in abortion (mean of 138 dg) and the other ewes produced weak lambs or clinically normal lambs [111]. Inoculation of 12 pregnant ewes with 106 tachyzoites of NC-Liverpool at 90 dg resulted in 18 foetuses which all were alive when the dam was killed for necropsy between 115 and 143 dg, except one foetus which was mummified and accompanied by a live sibling at 130 dg. All foetuses showed lesions, mostly in the placenta and CNS [112]. Inoculation of eight pregnant ewes with 106 tachyzoites of NC1 at 90 dg resulted in 11 foetuses; six ewes aborted five single foetuses and one set of twins between 125 and 134 dg and two ewes each gave birth to live lambs at 141 and 146 dg but accompanied by a stillborn sibling [113]. The placental lesions reported by infection with NC1 [113] were more severe than those reported by infection with NC-Liverpool [112]. These observations prompted the unconfirmed suggestion that NC1 appears more virulent than NC-Liverpool in sheep [113]. A subsequent report confirmed that inoculation of pregnant sheep with 105 tachyzoites of NC1 at 90 dg resulted in total foetal loss at 121 to 149 dg [114].
2.4. Antigenicity and Proteins
No significant differences in antigenic reactivity were detected in the immunofluorescent antibody test (IFAT) using sera from infected calves with different bovine isolates (JAP1, JAP-2, JAP-4, JAP-5, BT-2 and BPA1) [47].
Immunoblotting has been used extensively to compare tachyzoites from different isolates of N. caninum and the main conclusion reached by most studies is that no differences were detectable. No major antigenic differences were reported between NC-SweB1 and NC1 isolates, among (NC1, NC-Liverpool, NC-SweB1, BPA1, NC-LivB1and JAP-2), also between NC-Spain 1H and NC1 isolates; [15,31,43]. This may however simply reflect the low resolution of this popular technique [31]. Immunoblot analysis gave similar results when NC1, NC-4 and NC-5 were compared [18].
Two other studies illustrate that no detectable differences exist between isolates in their dominant surface antigens NcSAG1 and NCSRS2. Western blotting with monoclonals 6C11 or 5H5 found no differences in NcSAG1 (NcP29) and NcSRS2 (NcP35) antigens in canine (NC1, NC-2 and NC-Liverpool) and bovine (BPA1 and BPA3) isolates [115]. Similar results for NcSAG1 and NcSRS2 proteins were reported for CN1 (canine) and BPA1 (bovine) isolates by [116].
A small number of immunoblot studies have detected minor antigenic differences between N. caninum isolates. For example, a 28 KDa antigen was uniquely detected in NC-Liverpool but not in NC1 using anti-NC-Liv antiserum. It was suggested that such differences between isolate antigens and antisera raised to them might be due to variations in the stage of adaptation to in vitro culture of the two isolates, or variability in responses by the host used to raise the antisera, rather than to any real differences in their antigenic structure [22]. Others reiterated the idea that these differences might reflect the stage of the parasite during growth [117].
Western blotting of NC-Liverpool and NC-SweB1 revealed the existence of an additional 50 KDa band in NC-Liverpool [102]. Immunoblotting of a buffalo isolate NCBrBuf-4 and NC1 demonstrated the existence of a unique 56.6 KDa band in the buffalo isolate profile [64]. These observations have yet to be further explored and the identification of this mystery protein is currently unknown.
Analysis of the tachyzoite proteome has started to show evidence for differences in the molecular compositon of tachyzoites. Proteome and antigen profiling was used to compare two isolates of N. caninum; KBA-2 [54] from South Korea and JAP1 [46] from Japan. They were similar in 78% (403/516 spots) of the protein spots detected by two-dimensional gels electrophoresis (2-DE) and 80% (73/91 spots) of the antigen spots on 2-DE immunoblotting profiles using rabbit antiserum. Isoelectric focusing (IEF) was performed for a total of 86.1 kV h at pH 4–7 [118]. Comparison of the Korean KBA-2 isolate and VMDL-1 [36] from USA, also showed they were very similar by 2-DE and 2-DE immunoblot analysis. A small number of proteins were observed solely in either VMDL-1 or KBA-2 isolates with different molecular weight and pH 4–7; these included 17 proteins and seven immunoreactive isolate-specific spots were found in KBA-2, and nine proteins and two immunoreactive spots were found only in VMDL-1 [117]. Clearly the proteome offers substantial opportunities for identifying isolate specific molecules that may be linked to pathogenicity.
The kinetics of antibody production in mice infected with N. caninum appear to differ amongst isolates. Mice infected with NC-Liverpool induced an earlier IgG response in the mouse than did NC-SweB1 [102]. In pregnant mice, the level of immunoglobulin IgG1 and IgG2a present in groups infected with NC1 was significantly higher compared to a group infected with NC-Spain 1H and this might be due to differences in the behaviour of the isolate in the host and to the infectious dose given [43].
2.5. DNA Analyses
Information about genetic diversity within N. caninum is increasing and several studies now show there is substantial genetic diversity amongst isolates. Many targets in the genome and several approaches were used to investigate differences among N. caninum isolates.
2.5.1. 18S-like ribosomal DNA (small subunit-rDNA)
Ribosomal DNA has been used extensively in phylogeny because it evolves slowly and has an adequate size and resolution for phylogenetic studies [119]. No significant nucleotide 18S-like rDNA sequence differences were detected among four different bovine (BPA1, BPA2, BPA3 and BPA4) isolates and two canine (NC1 and NC-3) [120], between NC-SweB1 (bovine isolate) and NC1 [121], NC-Liverpool and NC1 [22] and NC-SweB1 and two canine isolates (NC1 and NC-Liverpool) [50]. This can be attributed to the limited amount of nucleotide changes that have occurred during the evolution of N. caninum from its most recent ancestor [120,122].
2.5.2. Internal transcribed spacer sequences (ITS1)
No informative nucleotide sequence differences occur in the ITS1 amongst isolates of N. caninum. Total sequence similarity was identified in the ITS1 region between two canine isolates NC1 and NC-Liverpool [22]. Also no differences were detected between NC-SweB1 and the canine isolates (NC1 and NC-Liverpool) [50,121]. The ITS1 sequence of NC-9 was 99% identical to NC1, NC-Liverpool, BPA1, NC-SweB1, and Nc-NZ1 [20], no differences were detected among NC1, NC-Liverpool, BPA1 and CN1 isolates [9]. However, intra-strain variation at the ITS1 region was reported between NC-Bahia (South American strain) and five other isolates from dogs and cattle of North American (NC-2, NC-Beef and NC-Illinois) and European origin (NC-Liverpool and NcSweB1) [123]. These minor variations in the ITS1 are however insufficiently polymorphic to differentiate between N. caninum isolates. Comparison of five Brazilian buffalo isolates using the ITS1 region with NC1 and NC-Liverpool revealed 2 and 7 bp differences, respectively [64].
2.5.3. RAPD-PCR
Only a limited amount of genetic differences was detected among NC-Liverpool and NC-SweB1 [102]. Differences were also detected among NC-LivB1, NC-Liverpool, NC1 and NC-SweB1 using this method with the B4 primer [38]. Fifty-four polymorphic DNA markers were evaluated using six different isolates by RAPD-PCR, which were capable of differentiating between the individual isolates [15]. Different numbers of bands were also amplified from NC1, NC-Liverpool and NC-SweB1 DNA when two RAPD-PCR primers were evaluated [124]. However, data from this method must be interpreted with caution. Firstly, the validity of this method depends on the purity of template DNA, the absence of extraneous host or other DNA, and on the primers chosen and conditions used for RAPD-PCR. Secondly, the analysis of results can be misleading due to co-migration of heterogeneous fragments [123,125]. As a result of these constraints, this technology has not been used further within the discipline of N. caninum.
2.5.4. Nc5 repeat
Repetitive DNA is common in the genome of N. caninum, and provides a rich source of DNA targets to study. The evidence to date indicates that isolates of N. caninum vary considerably in their repeat content. The diversity present is typically manifest as changes in repeat copy number, as well as changes to the nucleotide sequence of the repeating unit. One of the first repeats to be studied in detail was the Nc5 repeat.
Minor differences were detected between isolates of N. caninum by PCR amplifying the Nc5 repeat sequence. A blast search of sequences deposited in (NCBI database) revealed 95–100% sequence similarity amongst isolates. An alignment of sequences determined from coyote and fox oocyst DNA with published Nc5 sequence of N. caninum [126] revealed 95–99% similarity [98]. Comparison of five Brazilian buffalo isolates with NC1 revealed 6 bp differences [64].
2.5.5. Mini- and microsatellites
Multilocus microsatellite analyses of nine isolates of N. caninum from different hosts and geographical regions of the world revealed distinct genetic profiles amongst them with 12 out of the 13 markers analysed [125]. Additional diverse DNA profiles were reported among Spanish isolates through the study of microsatellites [44]. Nested PCR from 11 microsatellites (targets previously described by [125]) have been developed and applied to clinical samples (brains from aborted bovine fetuses) and new alleles were reported [127].
A much larger study of repetitive sequences investigated 27 different loci and the presence of diversity in 25 cultured isolates of N. caninum and polymorphism was reported within eight loci. These studies extended observations on the types of microsatellites found in the N. caninum genome and also showed that minisatellites were also associated with genetic diversity [128]. A multiplex PCR was subsequently developed using six repetitive sequences (three minisatellites and three microsatellites) (See Figure 2 and Figure 3). This approach has the ability to quickly assess genetic diversity present in any new isolates of N. caninum [128,129]. A variety of different targets can be incorporated into the multiplex, although as shown in Fig. 2 the choice of targets and their corresponding primers can influence the outcome of the PCR.
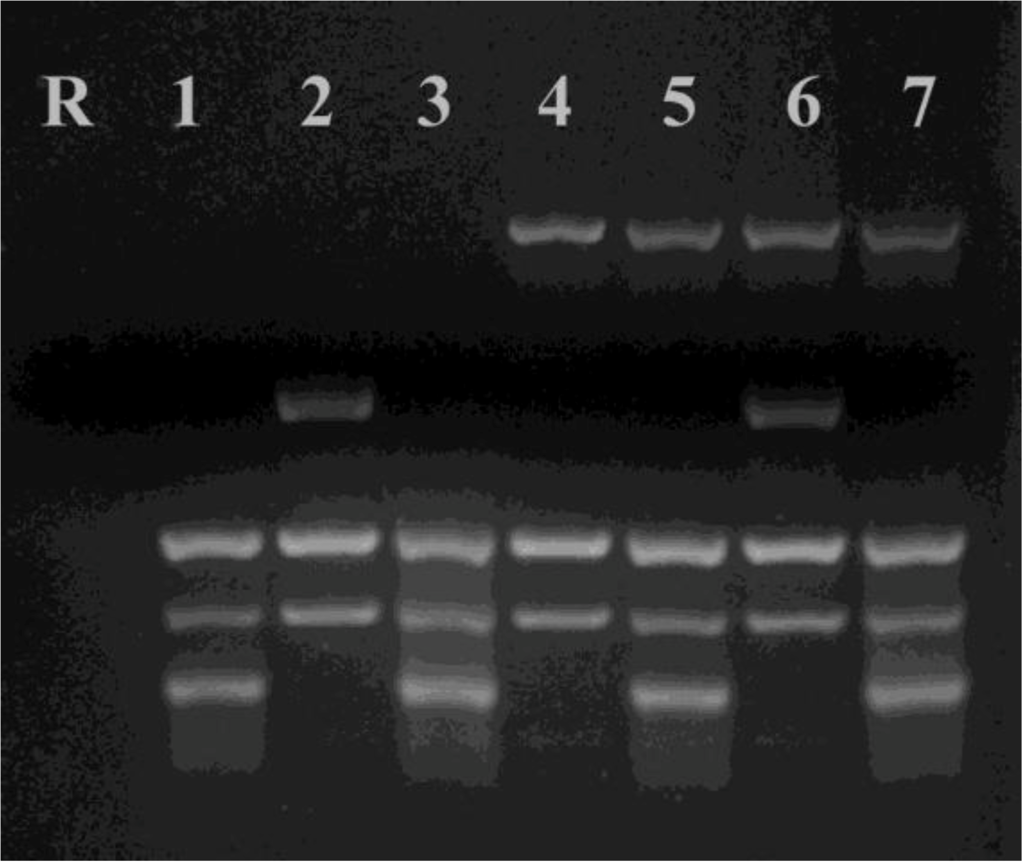
Figure 2.
Selection of DNA targets for inclusion in a multiplex PCR for detection of N. caninum genotypes.The results of using different combinations of targets and primers are shown using Nc-Liverpool DNA. Primers that amplify microsatellite (Tand-3) and minisatellite targets (Tand-12, Tand-13, Cont-13 and Cont-16) were used. R, Control (ddH2O); 1, from bottom band to top, Tand-3, Tand-12 and Tand-13; 2, Tand-12, Tand-13 and Cont-13 (not amplified); 3, Tand-3, Tand-12, Tand-13 and Cont-13 (not amplified); 4, Tand-12, Tand-13 and Cont-16; 5, Tand-3, Tand-12, Tand-13 and Cont-16; 6, Tand-12, Tand-13, Cont-13 and Cont-16; 7, Tand-3, Tand-12, Tand-13, Cont-13 (not amplified) and Cont-16. From the results of these combination experiments we conclude that primers to Tand-3 interfere with amplication of Cont-13. The sizes of the PCR products (bp) for NC-Liverpool [129] are Tand-3, 192; Tand-12, 277; Tand-13, 340; Cont-13, 507; Cont-16, 854. The primers used are described in [129].
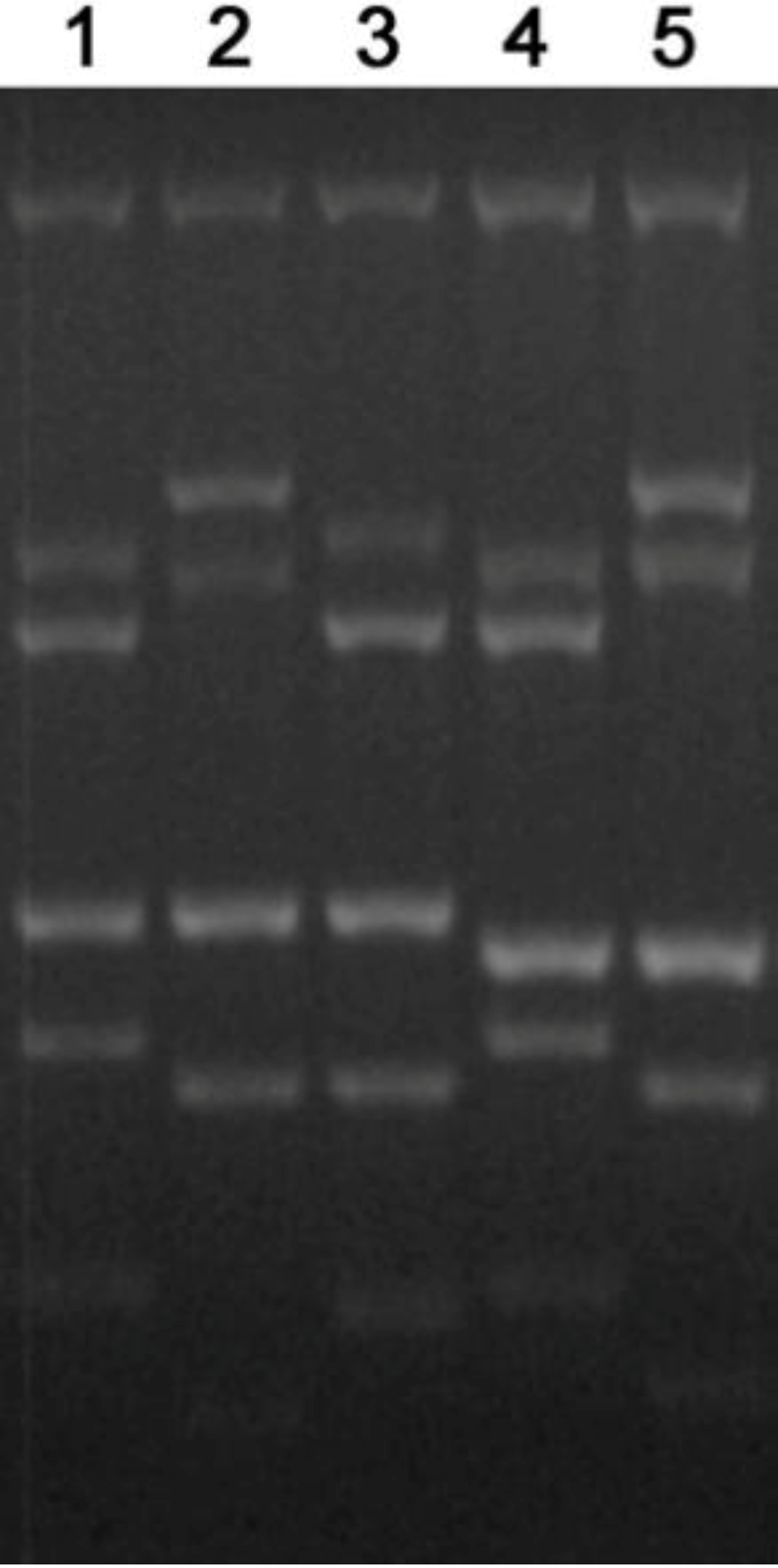
Figure 3.
Multiplex PCR of Neospora caninum DNA was performed with a PCR for the six markers Tand-3, Tand-12, Tand-13, Cont-6, Cont-14 and Cont-16. Lanes 1, BPA6; 2, NC-Beef; 3, JAP1; 4, WA-K9, 5, NC-Bahia. The sizes of the PCR products (bp) for NC-Bahia [129] are Tand-3, 140; Tand-12, 277; Tand-13, 340; Cont-6, 681; Cont-14, 619; Cont-16, 965.
MS10 is an allele that has now been extensively studied because this locus is highly divergent within N. caninum. It was first described by Regidor-Cerrillo et al. [125] who used PCR and sequencing of this locus to study polymorphism amongst nine different isolates of N. caninum. The MS10 locus was subsequently studied within 25 different cultured isolates of N. caninum and was called Tand-3 because different primers were used for PCR [128]. MS10 was also used to genotype a number of different Spanish isolates [43,44]. To date, about 30 different alleles of MS10 have been reported within 42 isolates of N. caninum and 15 clinical samples [30,31,43,44,125,127,128,129].
A nested PCR for MS10 was developed (external primers were designed and based on Regidor-Cerrillo et al. [125] internal primers) and used to detect diversity from oocyst and tissue-derived N. caninum DNA belonging to six isolates [30]. A new nested PCR was designed for MS10 (using different internal and external primers) and used to genotype naturally infected bovine aborted foetuses [127].
Considerable evidence indicates these molecular markers are stable traits. Microsatellite analyses of six loci compared DNA extracted from oocysts from six naturally infected dogs (GER5, 6, 7, 8, 9 and NC-P1) with cell culture tachyzoites obtained from these oocysts after inoculation into γ-interferon knockout mice. No differences were detectable between oocyst and tachyzoite DNA [30]. Such evidence suggests that adaptation of N. caninum tachyzoites to culture is apparently not associated with changes to these repeat populations. In addition, long term in vitro growth of N. caninum also does not appear to result in changes to repeat content [128,129].
2.5.6. Protein-encoding genes
A study of the N. caninum α-tubulin gene showed no variation between NC-Liverpool and NC-SweB1 [130]. No sequence differences were detected in the α- and β-tubulin genes among three canine isolates studied (NC1, NC-Liverpool and WA-K9) [25].
No differences were reported in the (HSP-70) gene from NC1, NC-Liverpool and WA-K9 [25]. No significant differences were reported by comparison of P37, IDA8 and Histone 2A gene sequences derived from NC-Liverpool and NC-SweB1 [131].
No sequence differences were detected in various surface antigen genes between canine and bovine isolates of N. caninum. For example, no variation was detected in comparative analyses of the NcSAG1 and NcSRS2 genes from six isolates of cattle (BPA1 and BPA3) and dogs (NC1, NC-2, CN1 and NC-Liverpool) [116]. Also no differences in bradyzoite stage-specific protein (NcSAG4) genes were reported among four isolates from dogs (NC1 and NC-Liverpool) and cattle (NC-SweB1 and NC-PV1) [132]. No nucleotide differences were detected among four different bovine (NC-SweB1) and canine (NC1, NC-2 and NC-Liverpool) isolates in their dense granule protein genes (NcGRA6 and NcGRA7) [133].
2.6. Growth Rate
Early observations suggested that the NC1 isolate of N. caninum multiplied more quickly in culture than the NC-Liverpool isolate [134] but these observations were subsequently shown to be incorrect. Assessment of relative growth rates in vitro by ³[H] uracil uptake, showed significant differences among six different isolates of N. caninum (including canine and bovine isolates origin). NC-Liverpool multiplied significantly faster compared to other isolates and the growth rate was twice that of NC1 whereas NC-SweB1 was the slowest growing [15].
It was subsequently reported that the NC1 isolate destroyed a cell monolayer more extensively (80%) than the NC-Spain 1H isolate (20%) during the same period which suggests there may be significant differences amongst isolates in their mechanisms of invasion and replication as well as tachyzoite yield from culture. The number of plaque-forming tachyzoites counted in NC1 was significantly higher (36.5%) than that of NC-Spain 1H (17.3%) in a viability assay. The percentage of bradyzoite conversion in NC-Spain 1H was similar to that of NC-Liverpool, but the Spanish isolate produced only intermediate bradyzoites (defined as SAG1 and BAG1-positive) whereas 3.4% of NC-Liverpool contained pure bradyzoites (BAG1) [43].
One study investigated the fitness of three isolates (NC1, NC-Liverpool and NC-SweB1) to proliferate for approximately 250 parasite generations in a cell line in which they had not been cultured before (Marc-145 cell line) [135]. Significant differences in mean fitness were detected among these isolates at the beginning of the experiment whereas no significant differences were reported at the end of the experiment indicating that the ability of these isolates to adapt to growth in a new cell line is similar after 250 generations [135].
In order to illustrate the extremes of diversity in N. caninum growth, we refer readers to the results obtained from the deer isolate. The deer isolate (NC-WTDVA-1) was found to grow very slowly in vitro when compared to other N. caninum isolates. Tachyzoites were first noticed in CV1 cell cultures after 127 days post infection and the average inter-passage interval was more than 100 days [66].
3. Discussion
Neospora caninum is a cyst-forming coccidian parasite which causes neosporosis. This parasite is known to be a major cause of abortion in cattle whilst also causing neuromuscular disorders and death in dogs [3,10,136]. The wide intermediate host range, global distribution and economical impact of N. caninum make it an important infectious agent.
Many strains of N. caninum have been isolated from a variety of hosts such as dog, cow, sheep, deer and water buffalo. Many of these isolations were from animals showing clinical signs of disease, such as an aborted bovine foetuses or a dog with paralysis. Others were from young animals (see Table 1). Some of these isolates show substantial differences in their in vivo pathogenicity in mice, in vitro growth characteristics or DNA sequence at specific loci [15,43,44,59,99,102,129,135,137]. Additional differences occur in the proteome of isolates but this area requires further study to identify and characterise these molecules [118,138]. Nevertheless, a picture is starting to emerge of N. caninum that suggests this species is made up of many diverse heterogeneous populations around the world. This conclusion is strongly supported by the genetic diversity detected among different isolates using mini- and microsatellites [125,129]. Research on Toxoplasma gondii has shown the presence of discrete lineages on a worldwide basis [139] and further research is needed on N. caninum to provide more useful markers for such studies.
The level of genetic diversity detected in N. caninum is a little surprising, since sexual reproduction is considered uncommon in this species [135]. It is also not clear how this surprising level of diversity impacts on the organism from a range of perspectives including pathogenesis.
Different molecular techniques have been used to study N. caninum such as RAPD-PCR [15,102] and also PCR amplification of targets such as ITS1, ss rDNA, Nc5 and the α and β-tubulin genes [22,25,120,123,140,141]. The small number of molecular differences among N. caninum isolates detected using these molecular targets show the limitations of this specific technology and the real value of mini and microsatellite technology. Fingerprinting approaches using other DNA targets for detection of genetic diversity and identification of genetic markers such as mini- and microsatellite sequences have been used successfully to detect differences within individual parasite species [142]. Recently, different mini- and microsatellite DNA markers were used to detect genomic polymorphisms among N. caninum isolates and many alleles were recorded [30,44,125,127,128,129]. Although genetic diversity among N. caninum isolates appears considerable, there is no obvious correlation at this stage between the DNA profile and pathogenicity or geographical locations from which the isolates were derived [44,125,128,129]. The availability of a genome sequence for N. caninum provides many more targets to study and this might further our understanding of the pathogencity of this species.
The N. caninum isolates (such as JAP1, NC-Sheep, NC-Nowra, Nc-Spain 1H and Nc-Goiás) which were derived from asymptomatic calves or sheep appear less virulent compared to those isolates obtained from symptomatic calves (sick calves or aborted foetus) [43,49,58,59,143]. In this context, the definition of virulence is the ability to induce clinical signs of disease in mice; however of importance is that this definition should be extended in the future to include studies in cattle and dogs. The impact these observations on virulence have on the development of vaccines to prevent neosporosis is now clear, since the isolation of naturally attenuated strains of N. caninum has obvious value and may eventually form the basis of effective live vaccines to prevent foetal loss in cattle [144]. The ideal characteristics of a live vaccine would be that it prevents foetal loss in cattle, whilst not persisting in the vaccinated animal nor causing foetal loss. In addition, ideally the strain must not be capable of dissemination to other fauna, nor be able to complete its life cycle in canid hosts. For any population of N. caninum these are difficult criteria to fulfil; however they represent qualities that are well characterised in the cyst-forming coccidia. Indeed they are all measureable and quantifiable. The pathway towards the development of a live vaccine that presents foetal loss in cattle is thus more clear than it was five years ago.
The characterisation of isolate specific antigens is likely to grow in importance, as studies on immunity to N. caninum clarify the role of individual molecules in this process. Initial studies using microarray technology show NC-Liverpool and NC-Nowra differ in their time course and the nature of the host response to infection by these two isolates (Ellis et al., unpublished) suggesting that immunity to N. caninum is complex involving both strain and species-specific components. Opportunities for future marker vaccine development in the N. caninum arena also appear therefore promising and there are many avenues to pursue in such an endeavour.
Acknowledgements
Much of this study was prepared in partial fulfillment of the degree of PhD by SAQ at the University of Technology Sydney.
References and Notes
- Dubey, J.P.; Carpenter, J.L.; Speer, C.A.; Topper, M.J.; Uggla, A. Newly recognized fatal protozoan disease of dogs. J. Amer. Vet. Med. Assn. 1988, 192, 1269–1285. [Google Scholar]
- Dubey, J.P. Review of Neospora caninum and neosporosis in animals. Korean J. Parasitol. 2003, 41, 1–16. [Google Scholar] [CrossRef]
- Reichel, M.P.; Ellis, J.T.; Dubey, J.P. Neosporosis and hammondiosis in dogs. J. Small Anim. Pract. 2007, 48, 308–312. [Google Scholar] [CrossRef]
- Bjerkås, I.; Dubey, J.P. Evidence that Neospora caninum is identical to the Toxoplasma-like parasite of Norwegian dogs. Acta. Vet. Scand. 1991, 32, 407–410. [Google Scholar]
- Bjerkås, I.; Mohn, S.F.; Presthus, J. Unidentified cyst-forming sporozoon causing encephalomyelitis and myositis in dogs. Z. Parasitenkd 1984, 70, 271–274. [Google Scholar] [CrossRef]
- Thilsted, J.P.; Dubey, J.P. Neosporosis-like abortions in a herd of dairy cattle. J. Vet. Diagn. Invest. 1989, 1, 205–209. [Google Scholar] [CrossRef]
- Dubey, J.P. Neosporosis - a newly recognised protozoal infection. Comp. Pathol. Bull. 1992, 24, 4–6. [Google Scholar]
- Dubey, J.P.; Lindsay, D.S. Neosporosis in dogs. Vet. Parasitol. 1990, 36, 147–151. [Google Scholar] [CrossRef]
- Marsh, A.E.; Barr, B.C.; Packham, A.E.; Conrad, P.A. Description of a new Neospora species (Protozoa: Apicomplexa: Sarcocystidae). J. Parasitol. 1998, 84, 983–991. [Google Scholar] [CrossRef]
- Dubey, J.P.; Schares, G.; Ortega-Mora, L.M. Epidemiology and control of neosporosis and Neospora caninum. Clin. Microbiol. Rev. 2007, 20, 323–367. [Google Scholar] [CrossRef]
- Larson, R.L.; Hardin, D.K.; Pierce, V.L. Economic considerations for diagnostic and control options for Neospora caninum-induced abortions in endemically infected herds of beef cattle. J. Amer. Vet. Med. Assn. 2004, 224, 1597–1604. [Google Scholar] [CrossRef]
- Reichel, M.P.; Ellis, J.T. If control of Neospora caninum infection is technically feasible does it make economic sense? Vet. Parasitol. 2006, 142, 23–34. [Google Scholar] [CrossRef]
- Dubey, J.P.; Hattel, A.L.; Lindsay, D.S.; Topper, M.J. Neonatal Neospora caninum infection in dogs: isolation of the causative agent and experimental transmission. J. Amer. Vet. Med. Assn. 1988, 193, 1259–1263. [Google Scholar]
- Conrad, P.A.; Barr, B.C.; Sverlow, K.W.; Anderson, M.; Daft, B.; Kinde, H.; Dubey, J.P.; Munson, L.; Ardans, A. In vitro isolation and characterization of a Neospora sp. from aborted bovine foetuses. Parasitology 1993, 106, 239–249. [Google Scholar] [CrossRef]
- Schock, A.; Innes, E.A.; Yamane, I.; Latham, S.M.; Wastling, J.M. Genetic and biological diversity among isolates of Neospora caninum. Parasitology 2001, 123, 13–23. [Google Scholar]
- Hay, W.H.; Shell, L.G.; Lindsay, D.S.; Dubey, J.P. Diagnosis and treatment of Neospora caninum infection in a dog. J. Amer. Vet. Med. Assn. 1990, 197, 87–89. [Google Scholar]
- Cuddon, P.; Lin, D.S.; Bowman, D.D.; Lindsay, D.S.; Miller, T.K.; Duncan, I.D.; deLahunta, A.; Cummings, J.; Suter, M.; Cooper, B. Neospora caninum infection in English Springer Spaniel littermates. Diagnostic evaluation and organism isolation. J. Vet. Intern. Med. 1992, 6, 325–332. [Google Scholar] [CrossRef]
- Dubey, J.P.; Dorough, K.R.; Jenkins, M.C.; Liddell, S.; Speer, C.A.; Kwok, O.C.; Shen, S.K. Canine neosporosis: clinical signs, diagnosis, treatment and isolation of Neospora caninum in mice and cell culture. Int. J. Parasitol. 1998, 28, 1293–1304. [Google Scholar] [CrossRef]
- Dubey, J.P.; Sreekumar, C.; Knickman, E.; Miska, K.B.; Vianna, M.C.; Kwok, O.C.; Hill, D.E.; Jenkins, M.C.; Lindsay, D.S.; Greene, C.E. Biologic, morphologic, and molecular characterisation of Neospora caninum isolates from littermate dogs. Int. J. Parasitol. 2004, 34, 1157–1167. [Google Scholar] [CrossRef]
- Dubey, J.P.; Vianna, M.C.; Kwok, O.C.; Hill, D.E.; Miska, K.B.; Tuo, W.; Velmurugan, G.V.; Conors, M.; Jenkins, M.C. Neosporosis in Beagle dogs: clinical signs, diagnosis, treatment, isolation and genetic characterization of Neospora caninum. Vet. Parasitol. 2007, 149, 158–166. [Google Scholar] [CrossRef]
- Barber, J.; Trees, A.J.; Owen, M. Isolation of Neospora caninum from a British Dog. Vet. Record 1993, 133, 531–532. [Google Scholar]
- Barber, J.S.; Holmdahl, O.J.M.; Owen, M.R.; Guy, F.; Uggla, A.; Trees, A.J. Characterization of the first European isolate of Neospora caninum (Dubey, Carpenter, Speer, Topper and Uggla). Parasitology 1995, 111, 563–568. [Google Scholar] [CrossRef]
- Gondim, L.F.; Pinheiro, A.M.; Santos, P.O.; Jesus, E.E.; Ribeiro, M.B.; Fernandes, H.S.; Almeida, M.A.; Freire, S.M.; Meyer, R.; McAllister, M.M. Isolation of Neospora caninum from the brain of a naturally infected dog, and production of encysted bradyzoites in gerbils. Vet. Parasitol. 2001, 101, 1–7. [Google Scholar] [CrossRef]
- Basso, W.; Venturini, L.; Venturini, M.C.; Hill, D.E.; Kwok, O.C.; Shen, S.K.; Dubey, J.P. First isolation of Neospora caninum from the feces of a naturally infected dog. J. Parasitol. 2001, 87, 612–618. [Google Scholar] [CrossRef]
- McInnes, L.M.; Irwin, P.; Palmer, D.G.; Ryan, U.M. In vitro isolation and characterisation of the first canine Neospora caninum isolate in Australia. Vet. Parasitol. 2006, 137, 355–363. [Google Scholar] [CrossRef]
- Šlapeta, J.R.; Modrý, D.; Kyselová, I.; Horejš, R.; Lukeš, J.; Koudela, B. Dog shedding oocysts of Neospora caninum: PCR diagnosis and molecular phylogenetic approach. Vet. Parasitol. 2002, 109, 157–167. [Google Scholar] [CrossRef]
- Peters, M.; Wagner, F.; Schares, G. Canine neosporosis: clinical and pathological findings and first isolation of Neospora caninum in Germany. Parasitol. Res. 2000, 86, 1–7. [Google Scholar] [CrossRef]
- Schares, G.; Heydorn, A.O.; Cüppers, A.; Conraths, F.J.; Mehlhorn, H. Hammondia heydorni-like oocysts shed by a naturally infected dog and Neospora caninum NC-1 cannot be distinguished. Parasitol. Res. 2001, 87, 808–816. [Google Scholar] [CrossRef]
- Schares, G.; Pantchev, N.; Barutzki, D.; Heydorn, A.O.; Bauer, C.; Conraths, F.J. Oocysts of Neospora caninum, Hammondia heydorni, Toxoplasma gondii and Hammondia hammondi in faeces collected from dogs in Germany. Int. J. Parasitol. 2005, 35, 1525–1537. [Google Scholar] [CrossRef]
- Basso, W.; Schares, S.; Barwald, A.; Herrmann, D.C.; Conraths, F.J.; Pantchev, N.; Vrhovec, M.G.; Schares, G. Molecular comparison of Neospora caninum oocyst isolates from naturally infected dogs with cell culture-derived tachyzoites of the same isolates using nested polymerase chain reaction to amplify microsatellite markers. Vet. Parasitol. 2009, 160, 43–50. [Google Scholar] [CrossRef]
- Basso, W.; Herrmann, D.C.; Conraths, F.J.; Pantchev, N.; Vrhovec, M.G.; Schares, G. First isolation of Neospora caninum from the faeces of a dog from Portugal. Vet. Parasitol. 2009, 159, 162–166. [Google Scholar] [CrossRef]
- Barr, B.C.; Rowe, J.D.; Sverlow, K.W.; BonDurant, R.H.; Ardans, A.A.; Oliver, M.N.; Conrad, P.A. Experimental reproduction of bovine fetal Neospora infection and death with a bovine Neospora isolate. J. Vet. Diagn. Invest. 1994, 6, 207–215. [Google Scholar] [CrossRef]
- Conrad, P.A.; Barr, B.C.; Anderson, M.L.; Sverlow, K.L. Recombinant neospora antigens and their uses. U.S. Patent 6,716,423, 2004. [Google Scholar]
- McAllister, M.M.; Dubey, J.P.; Lindsay, D.S.; Jolley, W.R.; Wills, R.A.; McGuire, A.M. Dogs are definitive hosts of Neospora caninum. Int. J. Parasitol. 1998, 28, 1473–1478. [Google Scholar] [CrossRef]
- Gondim, L.F.; Gao, L.; McAllister, M.M. Improved production of Neospora caninum oocysts, cyclical oral transmission between dogs and cattle, and in vitro isolation from oocysts. J. Parasitol. 2002, 88, 1159–1163. [Google Scholar] [CrossRef]
- Hyun, C.; Gupta, G.D.; Marsh, A.E. Sequence comparison of Sarcocystis neurona surface antigen from multiple isolates. Vet. Parasitol. 2003, 112, 11–20. [Google Scholar] [CrossRef]
- Davison, H.C.; Trees, A.J.; Guy, F.; Otter, A.; Holt, J.J.; Simpson, V.R.; Jeffrey, M. Isolation of bovine Neospora in Britain. Vet. Record 1997, 141, 607. [Google Scholar]
- Davison, H.C.; Guy, F.; Trees, A.J.; Ryce, C.; Ellis, J.T.; Otter, A.; Jeffrey, M.; Simpson, V.R.; Holt, J.J. In vitro isolation of Neospora caninum from a stillborn calf in the UK. Res. Vet. Sci. 1999, 67, 103–105. [Google Scholar] [CrossRef]
- Trees, A.J.; Williams, D.J.L. Neosporosis in the United Kingdom. Int. J. Parasitol. 2000, 30, 891–893. [Google Scholar]
- Canada, N.; Meireles, C.; Rocha, A.; Sousa, S.; Thompson, G.; Dubey, J.P.; Romand, S.; Thulliez, P.; Correia da Costa, J.M. First Portuguese isolate of Neospora caninum from an aborted fetus from a dairy herd with endemic neosporosis. Vet. Parasitol. 2002, 110, 11–15. [Google Scholar] [CrossRef]
- Canada, N.; Meireles, C.S.; Mezo, M.; González-Warleta, M.; Correia da Costa, J.M.; Sreekumar, C.; Hill, D.E.; Miska, K.B.; Dubey, J.P. First isolation of Neospora caninum from an aborted bovine fetus in Spain. J. Parasitol. 2004, 90, 863–864. [Google Scholar] [CrossRef]
- González-Warleta, M.; Castro-Hermida, J.A.; Carro-Corral, C.; Cortizo-Mella, J.; Mezo, M. Epidemiology of neosporosis in dairy cattle in Galicia (NW Spain). Parasitol. Res. 2008, 102, 243–249. [Google Scholar]
- Rojo-Montejo, S.; Collantes-Fernandez, E.; Regidor-Cerrillo, J.; Alvarez-Garcia, G.; Marugan-Hernandez, V.; Pedraza-Diaz, S.; Blanco-Murcia, J.; Prenafeta, A.; Ortega-Mora, L.M. Isolation and characterization of a bovine isolate of Neospora caninum with low virulence. Vet. Parasitol. 2009, 159, 7–16. [Google Scholar] [CrossRef]
- Regidor-Cerrillo, J.; Gómez-Bautista, M.; Pereira-Bueno, J.; Aduriz, G.; Navarro-Lozano, V.; Risco-Castillo, V.; Férnandez-García, A.; Pedraza-Díaz, S.; Ortega-Mora, L.M. Isolation and genetic characterization of Neospora caninum from asymptomatic calves in Spain. Parasitology 2008, 135, 1651–1659. [Google Scholar] [CrossRef]
- Yamane, I.; Kokuho, T.; Shimura, K.; Eto, M.; Haritani, M.; Ouchi, Y.; Sverlow, K.W.; Conrad, P.A. In vitro isolation of a bovine Neospora in Japan. Vet. Record 1996, 138, 652–652. [Google Scholar]
- Yamane, I.; Kokuho, T.; Shimura, K.; Eto, M.; Shibahara, T.; Haritani, M.; Ouchi, Y.; Sverlow, K.; Conrad, P.A. In vitro isolation and characterisation of a bovine Neospora species in Japan. Res. Vet. Sci. 1997, 63, 77–80. [Google Scholar] [CrossRef]
- Yamane, I.; Shibahara, T.; Kokuho, T.; Shimura, K.; Hamaoka, T.; Haritani, M.; Conrad, P.A.; Park, C.H.; Sawada, M.; Umemura, T. An improved isolation technique for bovine Neospora species. J. Vet. Diagn. Invest. 1998, 10, 364–368. [Google Scholar] [CrossRef]
- Sawada, M.; Kondo, H.; Tomioka, Y.; Park, C.; Morita, T.; Shimada, A.; Umemura, T. Isolation of Neospora caninum from the brain of a naturally infected adult dairy cow. Vet. Parasitol. 2000, 90, 247–252. [Google Scholar] [CrossRef]
- Koyama, T.; Kobayashi, Y.; Omata, Y.; Yamada, M.; Furuoka, H.; Maeda, R.; Matsui, T.; Saito, A.; Mikami, T. Isolation of Neospora caninum from the brain of a pregnant sheep. J. Parasitol. 2001, 87, 1486–1488. [Google Scholar] [CrossRef]
- Stenlund, S.; Bjorkman, C.; Holmdahl, O.J.; Kindahl, H.; Uggla, A. Characterization of a Swedish bovine isolate of Neospora caninum. Parasitol. Res. 1997, 83, 214–219. [Google Scholar] [CrossRef]
- Magnino, S.; Vigo, P.G.; Fabbi, M.; Colombo, M.; Bandi, C.; Genchi, C. Isolation of a bovine Neospora from a newborn calf in Italy. Vet. Record 1999, 144, 456. [Google Scholar]
- Fioretti, D.P.; Rosignoli, L.; Ricci, G.; Moretti, A.; Pasquali, P.; Polidori, G.A. Neospora caninum infection in a clinically healthy calf: Parasitological study and serological follow-up. J. Vet. Med. B–Infect. Dis. Vet. Pub. Health 2000, 47, 47–53. [Google Scholar] [CrossRef]
- Cheah, T.S.; Mattsson, J.G.; Zaini, M.; Sani, R.A.; Jakubek, E.B.; Uggla, A.; Chandrawathani, P. Isolation of Neospora caninum from a calf in Malaysia. Vet. Parasitol. 2004, 126, 263–269. [Google Scholar] [CrossRef]
- Kim, J.H.; Sohn, H.J.; Hwang, W.S.; Hwang, E.K.; Jean, Y.H.; Yamane, I.; Kim, D.Y. In vitro isolation and characterization of bovine Neospora caninum in Korea. Vet. Parasitol. 2000, 90, 147–154. [Google Scholar] [CrossRef]
- Locatelli-Dittrich, R.; Richartz, R.R.T.B.; Joineau, M.E.G.; Pinckney, R.D.; De Sousa, R.S.; Leite, L.C.; Thomaz-Soccol, V. Isolation of Neospora caninum from a blind calf in Parana, southern Brazil. Vet. Record 2003, 153, 366–367. [Google Scholar] [CrossRef]
- Locatelli-Dittrich, R.; Thomaz-Soccol, V.; Richartz, R.R.T.B.; Gasino-Joineau, M.E.; Vander Vinne, R.; Pinckney, R.D. Isolation of Neospora caninum from bovine fetus from a dairy herd in Paraná. Rev. Bras. Parasito. Vet. 2004, 13, 103–109. [Google Scholar]
- Pena, H.F.; Soares, R.M.; Ragozo, A.M.; Monteiro, R.M.; Yai, L.E.; Nishi, S.M.; Gennari, S.M. Isolation and molecular detection of Neospora caninum from naturally infected sheep from Brazil. Vet. Parasitol. 2007, 147, 61–66. [Google Scholar] [CrossRef]
- García-Melo, D.P.; Regidor-Cerrillo, J.; Ortega-Mora, L.M.; Collantes-Fernández, E.; de Oliveira, V.S.F.; de Oliveira, M.A.P.; da Silva, A.C. Isolation and biological characterisation of a new isolate of Neospora caninum from an asymptomatic calf in Brazil. Acta Parasitol. 2009, 54, 180–185. [Google Scholar] [CrossRef]
- Miller, C.M.D.; Quinn, H.E.; Windsor, P.A.; Ellis, J.T. Characterisation of the first Australian isolate of Neospora caninum from cattle. Aust. Vet. J. 2002, 80, 620–625. [Google Scholar] [CrossRef]
- Okeoma, C.M.; Williamson, N.B.; Pomroy, W.E.; Stowell, K.M.; Gillespie, L.M. Isolation and molecular characterisation of Neospora caninum in cattle in New Zealand. NZ. Vet. J. 2004, 52, 364–370. [Google Scholar] [CrossRef]
- Fish, L.; Mazuz, M.; Molad, T.; Savitsky, I.; Shkap, V. Isolation of Neospora caninum from dairy zero grazing cattle in Israel. Vet. Parasitol. 2007, 149, 167–171. [Google Scholar] [CrossRef]
- Pastusiak, K.; Cabaj, W.; Moskwa, B. Isolation, identification and maintenance in cell culture of the first Polish isolate of Neospora caninum. Wiad. Parazytol. 2005, 51, 68. [Google Scholar]
- Goździk, K.; Cabaj, W. Characterization of the first Polish isolate of Neospora caninum from cattle. Acta Parasitol. 2007, 52, 295–297. [Google Scholar] [CrossRef]
- Rodrigues, A.A.R.; Gennari, S.M.; Aguiar, D.M.; Sreekumar, C.; Hill, D.E.; Miska, K.B.; Vianna, M.C.; Dubey, J.P. Shedding of Neospora caninum oocysts by dogs fed tissues from naturally infected water buffaloes (Bubalus bubalis) from Brazil. Vet. Parasitol. 2004, 124, 139–150. [Google Scholar] [CrossRef]
- Gondim, L.F.; McAllister, M.M.; Mateus-Pinilla, N.E.; Pitt, W.C.; Mech, L.D.; Nelson, M.E. Transmission of Neospora caninum between wild and domestic animals. J. Parasitol. 2004, 90, 1361–1365. [Google Scholar] [CrossRef]
- Vianna, M.C.; Sreekumar, C.; Miska, K.B.; Hill, D.E.; Dubey, J.P. Isolation of Neospora caninum from naturally infected white-tailed deer (Odocoileus virginianus). Vet. Parasitol. 2005, 129, 253–257. [Google Scholar] [CrossRef]
- Lindsay, D.S.; Dubey, J.P.; Duncan, R.B. Confirmation that the dog is a definitive host for Neospora caninum. Vet. Parasitol. 1999, 82, 327–333. [Google Scholar] [CrossRef]
- Gondim, L.F.; McAllister, M.M.; Pitt, W.C.; Zemlicka, D.E. Coyotes (Canis latrans) are definitive hosts of Neospora caninum. Int. J. Parasitol. 2004, 34, 159–161. [Google Scholar] [CrossRef]
- King, J.S.; Slapeta, J.; Jenkins, D.J.; Al-Qassab, S.E.; Ellis, J.T.; Windsor, P.A. Australian dingoes are definitive hosts of Neospora caninum. Int. J. Parasitol. 2010.
- Dubey, J.P.; Lindsay, D.S. A review of Neospora caninum and neosporosis. Vet. Parasitol. 1996, 67, 1–59. [Google Scholar] [CrossRef]
- Dubey, J.P. Neosporosis: The first decade of research. Int. J. Parasitol. 1999, 29, 1485–1488. [Google Scholar] [CrossRef]
- Paré, J.; Thurmond, M.C.; Hietala, S.K. Congenital Neospora caninum infection in dairy cattle and associated calfhood mortality. Can. J. Vet. Res. 1996, 60, 133–139. [Google Scholar]
- Wouda, W.; Moen, A.R.; Schukken, Y.H. Abortion risk in progeny of cows after a Neospora caninum epidemic. Theriogenology 1998, 49, 1311–1316. [Google Scholar] [CrossRef]
- Dubey, J.P. Recent advances in Neospora and neosporosis. Vet. Parasitol. 1999, 84, 349–367. [Google Scholar] [CrossRef]
- Dijkstra, T.; Eysker, M.; Schares, G.; Conraths, F.J.; Wouda, W.; Barkema, H.W. Dogs shed Neospora caninum oocysts after ingestion of naturally infected bovine placenta but not after ingestion of colostrum spiked with Neospora caninum tachyzoites. Int. J. Parasitol. 2001, 31, 747–752. [Google Scholar] [CrossRef]
- Lindsay, D.S.; Ritter, D.M.; Brake, D. Oocyst excretion in dogs fed mouse brains containing tissue cysts of a cloned line of Neospora caninum. J. Parasitol. 2001, 87, 909–911. [Google Scholar] [CrossRef]
- Dubey, J.P.; Liddell, S.; Mattson, D.; Speert, C.A.; Howe, D.K.; Jenkins, M.C. Characterization of the Oregon isolate of Neospora hughesi from a horse. J. Parasitol. 2001, 87, 345–353. [Google Scholar] [CrossRef]
- Dubey, J.P.; de Lahunta, A. Neosporosis associated congenital limb deformities in a calf. Appl. Parasitol. 1993, 34, 229–233. [Google Scholar]
- Bjerkås, I.; Jenkins, M.C.; Dubey, J.P. Identification and characterization of Neospora caninum tachyzoite antigens useful for diagnosis of Neosporosis. Clin. Diagn. Lab. Immunol. 1994, 1, 214–221. [Google Scholar]
- Lindsay, D.S.; Speer, C.A.; Toiviokinnucan, M.A.; Dubey, J.P.; Blagburn, B.L. Use of infected cultured cells to compare ultrastructural features of Neospora caninum from dogs and Toxoplasma gondii. Am. J. Vet. Res. 1993, 54, 103–106. [Google Scholar]
- Speer, C.A.; Dubey, J.P. Ultrastructure of tachyzoites, bradyzoites and tissue cysts of Neospora caninum. J. Raptor. Res. 1989, 36, 458–463. [Google Scholar]
- Speer, C.A.; Dubey, J.P.; McAllister, M.M.; Blixt, J.A. Comparative ultrastructure of tachyzoites, bradyzoites, and tissue cysts of Neospora caninum and Toxoplasma gondii. Int. J. Parasitol. 1999, 29, 1509–1519. [Google Scholar] [CrossRef]
- Barr, B.C.; Conrad, P.A.; Dubey, J.P.; Anderson, M.L. Neospora-like encephalomyelitis in a calf: pathology, ultrastructure, and immunoreactivity. J. Vet. Diagn. Invest. 1991, 3, 39–46. [Google Scholar] [CrossRef]
- McGuire, A.M.; McAllister, M.M.; Jolley, W.R.; Anderson-Sprecher, R.C. A protocol for the production of Neospora caninum tissue cysts in mice. J. Parasitol. 1997, 83, 647–651. [Google Scholar] [CrossRef]
- Lindsay, D.S.; Dubey, J.P. Neospora caninum (Protozoa, Apicomplexa) infections in rats. Can. J. Zool. 1990, 68, 1595–1599. [Google Scholar] [CrossRef]
- Dubey, J.P.; Higgins, R.J.; Smith, J.H.; Otoole, T.D. Neospora caninum encephalomyelitis in a British dog. Vet. Record 1990, 126, 193–194. [Google Scholar]
- Barr, B.C.; Anderson, M.L.; Dubey, J.P.; Conrad, P.A. Neospora-like protozoal infections associated with bovine abortions. Vet. Pathol. 1991, 28, 110–116. [Google Scholar]
- Razmi, G.R.; Maleki, M.; Farzaneh, N.; Garoussi, M.T.; Fallah, A.H. First report of Neospora caninum-associated bovine abortion in Mashhad area, Iran. Parasitol. Res. 2007, 100, 755–757. [Google Scholar] [CrossRef]
- Barr, B.C.; Anderson, M.L.; Woods, L.W.; Dubey, J.P.; Conrad, P.A. Neospora-like protozoal infections associated with abortion in goats. J. Vet. Diagn. Invest. 1992, 4, 365–367. [Google Scholar] [CrossRef]
- Dubey, J.P.; Morales, J.A.; Villalobos, P.; Lindsay, D.S.; Blagburn, B.L.; Topper, M.J. Neosporosis-associated abortion in a dairy goat. J. Amer. Vet. Med. Assn. 1996, 208, 263–265. [Google Scholar]
- Basso, W.; Venturini, L.; Venturini, M.C.; Moore, P.; Rambeau, M.; Unzaga, J.M.; Campero, C.; Bacigalupe, D.; Dubey, J.R. Prevalence of Neospora caninum infection in dogs from beef-cattle farms, dairy farms, and from urban areas of Argentina. J. Parasitol. 2001, 87, 906–907. [Google Scholar] [CrossRef]
- McGuire, A.M.; McAllister, M.M.; Jolley, W.R. Separation and cryopreservation of Neospora caninum tissue cysts from murine brain. J. Parasitol. 1997, 83, 319–321. [Google Scholar]
- Lindsay, D.S.; Steinberg, H.; Dubielzig, R.R.; Semrad, S.D.; Konkle, D.M.; Miller, P.E.; Blagburn, B.L. Central nervous system neosporosis in a foal. J. Vet. Diagn. Invest. 1996, 8, 507–510. [Google Scholar] [CrossRef]
- Daft, B.M.; Barr, B.C.; Collins, N.; Sverlow, K. Neospora encephalomyelitis and polyradiculoneuritis in an aged mare with Cushing's disease. Equine Vet. J. 1997, 29, 240–243. [Google Scholar] [CrossRef]
- McCann, C.M.; McAllister, M.M.; Gondim, L.F.; Smith, R.F.; Cripps, P.J.; Kipar, A.; Williams, D.J.; Trees, A.J. Neospora caninum in cattle: experimental infection with oocysts can result in exogenous transplacental infection, but not endogenous transplacental infection in the subsequent pregnancy. Int. J. Parasitol. 2007, 37, 1631–1639. [Google Scholar] [CrossRef]
- Schares, G.; Heydorn, A.O.; Cüppers, A.; Conraths, F.J.; Mehlhorn, H. Cyclic transmission of Neospora caninum: serological findings in dogs shedding oocysts. Parasitol. Res. 2001, 87, 873–877. [Google Scholar] [CrossRef]
- McGarry, J.W.; Stockton, C.M.; Williams, D.J.; Trees, A.J. Protracted shedding of oocysts of Neospora caninum by a naturally infected foxhound. J. Parasitol. 2003, 89, 628–630. [Google Scholar] [CrossRef]
- Wapenaar, W.; Jenkins, M.C.; O'Handley, R.M.; Barkema, H.W. Neospora caninum-like oocysts observed in feces of free-ranging red foxes (Vulpes vulpes) and coyotes (Canis latrans). J. Parasitol. 2006, 92, 1270–1274. [Google Scholar] [CrossRef]
- Lindsay, D.S.; Dubey, J.P. In vitro development of Neospora caninum (Protozoa: Apicomplexa) from dogs. J. Parasitol. 1989, 75, 163–165. [Google Scholar] [CrossRef]
- Thornton, R.N.; Thompson, E.J.; Dubey, J.P. Neospora abortion in New Zealand cattle. NZ. Vet. J. 1991, 39, 129–133. [Google Scholar] [CrossRef]
- Paré, J.; Hietala, S.K.; Thurmond, M.C. An enzyme-linked immunosorbent assay (ELISA) for serological diagnosis of Neospora sp. infection in cattle. J. Vet. Diagn. Invest. 1995, 7, 352–359. [Google Scholar] [CrossRef]
- Atkinson, R.; Harper, P.A.; Ryce, C.; Morrison, D.A.; Ellis, J.T. Comparison of the biological characteristics of two isolates of Neospora caninum. Parasitology 1999, 118, 363–370. [Google Scholar] [CrossRef]
- Lindsay, D.S.; Dubey, J.P. Infections in mice with tachyzoites and bradyzoites of Neospora caninum (Protozoa: Apicomplexa). J. Parasitol. 1990, 76, 410–413. [Google Scholar] [CrossRef]
- Collantes-Fernández, E.; Arnaiz-Seco, I.; Burgos, B.M.; Rodriguez-Bertos, A.; Aduriz, G.; Fernandez-Garcia, A.; Ortega-Mora, L.M. Comparison of Neospora caninum distribution, parasite loads and lesions between epidemic and endemic bovine abortion cases. Vet. Parasitol. 2006, 142, 187–191. [Google Scholar] [CrossRef]
- Lindsay, D.S.; Lenz, S.D.; Cole, R.A.; Dubey, J.P.; Blagburn, B.L. Mouse model for central nervous system Neospora caninum infections. J. Parasitol. 1995, 81, 313–315. [Google Scholar] [CrossRef]
- Quinn, H.E.; Miller, C.M.; Ryce, C.; Windsor, P.A.; Ellis, J.T. Characterization of an outbred pregnant mouse model of Neospora caninum infection. J. Parasitol. 2002, 88, 691–696. [Google Scholar] [CrossRef]
- Dubey, J.P.; Lindsay, D.S. Gerbils (Meriones unguiculatus) are highly susceptible to oral infection with Neospora caninum oocysts. Parasitol. Res. 2000, 86, 165–168. [Google Scholar] [CrossRef]
- Williams, D.J.; Guy, C.S.; Smith, R.F.; Ellis, J.; Bjorkman, C.; Reichel, M.P.; Trees, A.J. Immunization of cattle with live tachyzoites of Neospora caninum confers protection against fetal death. Infec. Immunity 2007, 75, 1343–1348. [Google Scholar] [CrossRef]
- Rojo-Montejo, S.; Collantes-Fernandez, E.; Blanco-Murcia, J.; Rodriguez-Bertos, A.; Risco-Castillo, V.; Ortega-Mora, L.M. Experimental infection with a low virulence isolate of Neospora caninum at 70 days gestation in cattle did not result in foetopathy. Vet. Res. 2009, 40, 49. [Google Scholar] [CrossRef]
- Dubey, J.P.; Lindsay, D.S. Neospora caninum induced abortion in sheep. J. Vet. Diagn. Invest. 1990, 2, 230–233. [Google Scholar] [CrossRef]
- McAllister, M.M.; McGuire, A.M.; Jolley, W.R.; Lindsay, D.S.; Trees, A.J.; Stobart, R.H. Experimental neosporosis in pregnant ewes and their offspring. Vet. Pathol. 1996, 33, 647–655. [Google Scholar]
- Buxton, D.; Maley, S.W.; Thomson, K.M.; Trees, A.J.; Innes, E.A. Experimental infection of non-pregnant and pregnant sheep with Neospora caninum. J. Comp. Pathol. 1997, 117, 1–16. [Google Scholar]
- Buxton, D.; Maley, S.W.; Wright, S.; Thomson, K.M.; Rae, A.G.; Innes, E.A. The pathogenesis of experimental neosporosis in pregnant sheep. J. Comp. Pathol. 1998, 118, 267–279. [Google Scholar] [CrossRef]
- Buxton, D.; Wright, S.; Maley, S.W.; Rae, A.G.; Lundén, A.; Innes, E.A. Immunity to experimental neosporosis in pregnant sheep. Parasite Immunol. 2001, 23, 85–91. [Google Scholar]
- Howe, D.K.; Crawford, A.C.; Lindsay, D.; Sibley, L.D. The p29 and p35 immunodominant antigens of Neospora caninum tachyzoites are homologous to the family of surface antigens of Toxoplasma gondii. Infec. Immunity 1998, 66, 5322–5328. [Google Scholar]
- Marsh, A.E.; Howe, D.K.; Wang, G.; Barr, B.C.; Cannon, N.; Conrad, P.A. Differentiation of Neospora hughesi from Neospora caninum based on their immunodominant surface antigen, SAG1 and SRS2. Int. J. Parasitol. 1999, 29, 1575–1582. [Google Scholar] [CrossRef]
- Shin, Y.S.; Lee, E.G.; Jung, T.S. Exploration of immunoblot profiles of Neospora caninum probed with different bovine immunoglobulin classes. J. Vet. Sci. 2005, 6, 157–160. [Google Scholar]
- Lee, E.G.; Kim, J.H.; Shin, Y.S.; Shin, G.W.; Kim, Y.R.; Palaksha, K.J.; Kim, D.Y.; Yamane, I.; Kim, Y.H.; Kim, G.S.; Suh, M.D.; Jung, T.S. Application of proteomics for comparison of proteome of Neospora caninum and Toxoplasma gondii tachyzoites. J. Chromat. B 2005, 815, 305–314. [Google Scholar] [CrossRef]
- Morrison, D.A.; Ellis, J.T. Effects of nucleotide sequence alignment on phylogeny estimation: a case study of 18S rDNAs of apicomplexa. Mol Biol Evol 1997, 14, 428–441. [Google Scholar] [CrossRef]
- Marsh, A.E.; Barr, B.C.; Sverlow, K.; Ho, M.; Dubey, J.P.; Conrad, P.A. Sequence analysis and comparison of ribosomal DNA from bovine Neospora to similar coccidial parasites. J. Parasitol. 1995, 81, 530–535. [Google Scholar] [CrossRef]
- Holmdahl, J.; Björkman, C.; Stenlund, S.; Uggla, A.; Dubey, J.P. Bovine Neospora and Neospora caninum: One and the same. Parasitol. Today 1997, 13, 40–41. [Google Scholar]
- Johnson, A.M.; Illana, S.; Dubey, J.P.; Dame, J.B. Toxoplasma gondii and Hammondia hammondi: DNA comparison using cloned rRNA gene probes. Exp. Parasitol. 1987, 63, 272–278. [Google Scholar] [CrossRef]
- Gondim, L.F.; Laski, P.; Gao, L.; McAllister, M.M. Variation of the internal transcribed spacer 1 sequence within individual strains and among different strains of Neospora caninum. J. Parasitol. 2004, 90, 119–122. [Google Scholar] [CrossRef]
- Spencer, J.A.; Witherow, A.K.; Blagburn, B.L. A random amplified polymorphic DNA polymerase chain reaction technique that differentiates between Neospora species. J. Parasitol. 2000, 86, 1366–1368. [Google Scholar] [CrossRef]
- Regidor-Cerrillo, J.; Pedraza-Díaz, S.; Gómez-Bautista, M.; Ortega-Mora, L.M. Multilocus microsatellite analysis reveals extensive genetic diversity in Neospora caninum. J. Parasitol. 2006, 92, 517–524. [Google Scholar] [CrossRef]
- Kaufmann, H.; Yamage, M.; Roditi, I.; Dobbelaere, D.; Dubey, J.P.; Holmdahl, O.J.; Trees, A.; Gottstein, B. Discrimination of Neospora caninum from Toxoplasma gondii and other apicomplexan parasites by hybridization and PCR. Mol. Cell. Probe. 1996, 10, 289–297. [Google Scholar]
- Pedraza-Diaz, S.; Marugan-Hernandez, V.; Collantes-Fernandez, E.; Regidor-Cerrillo, J.; Rojo-Montejo, S.; Gomez-Bautista, M.; Ortega-Mora, L.M. Microsatellite markers for the molecular characterization of Neospora caninum: application to clinical samples. Vet. Parasitol. 2009, 166, 38–46. [Google Scholar] [CrossRef]
- Al-Qassab, S.; Reichel, M.P.; Ivens, A.; Ellis, J.T. Genetic diversity amongst isolates of Neospora caninum, and the development of a multiplex assay for the detection of distinct strains. Mol. Cell. Probe. 2009, 23, 132–139. [Google Scholar] [CrossRef]
- Al-Qassab, S.; Reichel, M.P.; Ellis, J. A second generation multiplex PCR for typing strains of Neospora caninum using six DNA targets. Mol. Cell. Probe. 2009, 24, 20–26. [Google Scholar] [CrossRef]
- Siverajah, S.; Ryce, C.; Morrison, D.A.; Ellis, J.T. Characterization of an alpha tubulin gene sequence from Neospora caninum and Hammondia heydorni, and their comparison to homologous genes from Apicomplexa. Parasitology 2003, 126, 561–569. [Google Scholar]
- Ellis, J.T.; Ryce, C.; Atkinson, R.; Balu, S.; Holmdahl, O.J.M. Neospora caninum: gene discovery through analysis of expressed sequence taqs. Int. J. Parasitol. 2000, 30, 909–913. [Google Scholar]
- Fernández-García, A.; Risco-Castillo, V.; Zaballos, A.; Alvarez-García, G.; Ortega-Mora, L.M. Identification and molecular cloning of the Neospora caninum SAG4 gene specifically expressed at bradyzoite stage. Mol. Biochem. Parasitol. 2006, 146, 89–97. [Google Scholar] [CrossRef]
- Walsh, C.P.; Vemulapalli, R.; Sriranganathan, N.; Zajac, A.M.; Jenkins, M.C.; Lindsay, D.S. Molecular comparison of the dense granule proteins GRA6 and GRA7 of Neospora hughesi and Neospora caninum. Int. J. Parasitol. 2001, 31, 253–258. [Google Scholar] [CrossRef]
- Innes, E.A.; Panton, W.R.M.; Marks, J.; Trees, A.J.; Holmdahl, J.; Buxton, D. Interferon-gamma inhibits the intracellular multiplication of Neospora caninum, as shown by incorporation of ³[H] uracil. J. Comp. Pathol. 1995, 113, 95–100. [Google Scholar] [CrossRef]
- Pérez-Zaballos, F.J.; Ortega-Mora, L.M.; Álvarez-García, G.; Collantes-Fernández, E.; Navarro-Lozano, V.; García-Villada, L.; Costas, E. Adaptation of Neospora caninum isolates to cell-culture changes: An argument in favor of its clonal population structure. J. Parasitol. 2005, 91, 507–510. [Google Scholar] [CrossRef]
- Dubey, J.P.; Barr, B.C.; Barta, J.R.; Bjerkås, I.; Björkman, C.; Blagburn, B.L.; Bowman, D.D.; Buxton, D.; Ellis, J.T.; Gottstein, B.; Hemphill, A.; Hill, D.E.; Howe, D.K.; Jenkins, M.C.; Kobayashi, Y.; Koudela, B.; Marsh, A.E.; Mattsson, J.G.; McAllister, M.M.; Modrý, D.; Omata, Y.; Sibley, L.D.; Speer, C.A.; Trees, A.J.; Uggla, A.; Upton, S.J.; Williams, D.J.L.; Lindsay, D.S. Redescription of Neospora caninum and its differentiation from related coccidia. Int. J. Parasitol. 2002, 32, 929–946. [Google Scholar] [CrossRef]
- Collantes-Fernández, E.; Lopez-Perez, I.; Álvarez-García, G.; Ortega-Mora, L.M. Temporal distribution and parasite load kinetics in blood and tissues during Neospora caninum infection in mice. Infec. Immunity 2006, 74, 2491–2494. [Google Scholar] [CrossRef]
- Shin, Y.S.; Shin, G.W.; Kim, Y.R.; Lee, E.Y.; Yang, H.H.; Palaksha, K.J.; Youn, H.J.; Kim, J.H.; Kim, D.Y.; Marsh, A.E.; Lakritz, J.; Jung, T.S. Comparison of proteome and antigenic proteome between two Neospora caninum isolates. Vet. Parasitol. 2005, 134, 41–52. [Google Scholar] [CrossRef]
- Lehmann, T.; Marcet, P.L.; Graham, D.H.; Dahl, E.R.; Dubey, J.P. Globalization and the population structure of Toxoplasma gondii. Proc.Nat.Acad.Sci.USA 2006, 103, 11423–11428. [Google Scholar]
- Müller, N.; Sager, H.; Hemphill, A.; Mehlhorn, H.; Heydorn, A.O.; Gottstein, B. Comparative molecular investigation of Nc5-PCR amplicons from Neospora caninum NC-1 and Hammondia heydorni-Berlin-1996. Parasitol. Res. 2001, 87, 883–885. [Google Scholar] [CrossRef]
- Yamage, M.; Flechtner, O.; Gottstein, B. Neospora caninum: specific oligonucleotide primers for the detection of brain "cyst" DNA of experimentally infected nude mice by the polymerase chain reaction (PCR). J. Parasitol. 1996, 82, 272–279. [Google Scholar] [CrossRef]
- Gasser, R.B. Molecular tools--advances, opportunities and prospects. Vet. Parasitol. 2006, 136, 69–89. [Google Scholar] [CrossRef]
- Shibahara, T.; Kokuho, T.; Eto, M.; Haritani, M.; Hamaoka, T.; Shimura, K.; Nakamura, K.; Yokomizo, Y.; Yamane, I. Pathological and immunological findings of athymic nude and congenic wild type BALB/c mice experimentally infected with Neospora caninum. Vet. Pathol. 1999, 36, 321–327. [Google Scholar]
- Reichel, M.P.; Ellis, J.T. Neospora caninum—how close are we to development of an efficacious vaccine that prevents abortion in cattle? Int. J. Parasitol. 2009, 39, 1173–1187. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).