Abstract
Commiphora wightii is an endangered, endemic species found in the Thar Desert of Rajasthan, India and adjoining areas of Pakistan. The populations of this plant are rapidly dwindling due to overexploitation for their medicinally important resin. Analysis of nucleotide sequences of the internal transcribed spacer of rDNAs revealed low genetic diversity (π = 0.03905; θw = 0.05418) and high population structure (ΦST = 0.206). Parsimony based assessment and Bayesian analyses were conducted on the dataset. Mantel’s test showed a statistically significant positive correlation between genetic and geographic distance (r2 = 0.3647; p = 0.023). Anthropogenic overexploitation of C. wightii for its natural resources has resulted in population fragmentation. Initiatives should be taken immediately to preserve the diversity of this important plant species.
1. Introduction
Commiphora wightii (Arn.) Bhandari, of the family Burseraceae [1], is a small tree that has a thick main stem with crooked knotty branches. The plant is primarily found in the arid to semi arid regions of the Thar Desert (in the state of Rajasthan, India) and adjoining parts of Pakistan. It grows on plains and hilly areas with preference for dry rocky terrains. The plant is reported to be apomictic [2]. Mature plants (5–7 yrs old) produce a medicinally important resin which is called ‘guggulu’ in Sanskrit. Guggulu is mentioned in the classical medical treatises Charaka Samhita (1000 BC) and Shushruta Samhita (600 BC); in Western texts it is known as ‘guggul’ [3]. The major bioactive components of guggul resin are Z-guggulsterone and E-guggulsterone [4] that are used for the treatment of hypercholesterolemia and cardiovascular diseases [5,6]. Recent studies have shown that guggulsterones also possess anti-cancer activity [7,8,9]. Due to its medicinal importance the resin has high commercial value. Those seeking to obtain guggulu, resort to inflicting severe and often fatal wounds to the main stem of the plant for quick extraction of the resin. Due to the presence of resin, the main stem burns well and hence it is also used as fuel wood. These anthropogenic activities have resulted in a drastic decrease in the number of populations. In response to these situations the Government of India has placed C. wightii in the RET (Rare, Endangered and Threatened) category. If such exploitation continues without sustainable and appropriate management of the genetic resources, the diversity and natural abundance of this species will be eroded further [10]. Conserving and managing a species without regard to its population structure may fail to conserve the full spectrum of functions provided by that species, thus it becomes critical to understand the nature of its population structure.
The nuclear ribosomal transcription unit (NRTU) is comprised of 18S, 5.8S and 28S genes, two internal transcribed spacers (ITS-1 and ITS-2), and an intergenic spacer (IGS). After transcription, the NRTU is processed to produce mature rRNAs that are key components of cytoplasmic ribosomes. NRTU are found in hundreds to thousands of tandem copies and usually several NRTU clusters are present within plant genomes. The conserved regions (18S and 28S genes) of NRTU are used to infer phylogenetic relationships at higher taxonomy levels, whereas the more rapidly evolving segments (ITS and IGS) are used for studies at the genic or population levels [11,12]. For over a decade, sequences of internal transcribed spacers (ITS) of NRTUs have been widely used to infer phylogenetic relationships, genetic diversity and to unravel evolution in a wide range of complexes in plants [12,13,14,15,16,17]. Although NRTUs are found in thousands of copies within a genome, intra-genomic diversity is generally low [14]. This homogeneity among NRTUs is attributed to concerted evolution [14,18], a process that acts through gene conversion and unequal crossing over. Despite the fact that homogenization is a norm among NRTUs in a genome, extensive intra-individual and intra-specific variation has been observed in various plant species [19,20]. Evidence is accumulating that suggests that intra-individual variation of nuclear ribosomal ITS regions should not be considered as exceptional [21]. Because of the influence of concerted evolution, the occurrence of ancestral polymorphisms is not the most likely ultimate cause for intra-genomic variability in this marker. Instead, a more frequent origin is the merging of different ITS copies within the same genome as a consequence of gene flow. Once the two copies meet, the fate of the polymorphism depends on genetic, reproductive and population-level factors: specifically, the number and location of ribosomal loci (on the same or different chromosomes), the occurrence of polyploidy and/or apomixis [15,19,22], and the relative abundance of different ITS copies in the breeding populations [21].
Molecular phylogenetic analysis using NRTU sequence data of Mexican Bursera spp. [23,24,25] revealed Commiphora spp. and Bursera spp. to be closely related and resolved them as monophyletic sister groups, with considerable differentiation between them. C. wightii was not included in the study. Weeks et al. [26] included C. wightii in their attempt to reassess biogeographical studies of 48 species belonging to Burseraceae of Gondwanan origin, but the sample was acquired from the arid lands greenhouse, Arizona, USA. Moreover, these studies did not capture genetic variation among Commiphora spp. Our earlier studies to genetically dissect C. wightii populations by RAPD markers revealed low diversity and high population differentiation [27]. An important prerequisite for development of an effective conservation strategy is the proper evaluation of the distribution and level of genetic variation [28]. Information on population structure and genetic variation of C. wightii is lacking, which is surprising, considering the endangered status of the plant. The principal objectives of our present study was to utilize the nucleotide data of the ITS1-5.8S-ITS2 sequences to (a) determine the genetic variability within C. wightii (b) evaluate the degree of differentiation among C. wightii populations and its relationship with geographical distribution, and (c) offer genetic information that can be used in development of a C. wightii conservation strategy.
2. Material and Methods
C. wightii plants only produce leaves during the short rainy season (July-October). Fresh young leaves showing no signs of necrotic lesions were sampled from 32 plants growing naturally, belonging to seven populations (Table 1), collected from the state of Rajasthan, India (Figure 1) during September 2007. Voucher specimens were deposited at the herbarium of SKN University College at Jobner, Rajasthan. For each specimen total genomic DNA was isolated from 0.4 g of leaves following a modified CTAB DNA extraction procedure [29] developed in this laboratory for plants having significant quantities of secondary metabolites. The DNA samples were quantified on a Fluorometer (VersaFluor, Bio-Rad Lab., Hercules, CA, USA) with Hoechst 33258 and using Calf thymus DNA as a standards. The quality of DNA was checked both spectrophotometrically and by agarose (0.8%) gel electrophoresis as well.

Table 1.
Location of the sampled population of C. wightii.
| Population | Terrain | Population Code | Sample size | Latitude | Longitude | Elevation (ft) |
| Mangliawas | Hilly | M | 7 | 26.283 | 74.500 | 1,433 |
| Ajmer | Hilly | A | 4 | 26.450 | 74.633 | 1,548 |
| Kishangarh | Hilly | KG | 4 | 26.566 | 74.866 | 1,420 |
| Jobner | Plains | J | 8 | 26.966 | 75.383 | 1,243 |
| Bobas | Plains | B | 3 | 26.905 | 75.498 | 1,220 |
| Hirnoda | Plains | H | 3 | 26.885 | 75.333 | 1,116 |
| Galta Hills | Hilly | GH | 3 | 26.916 | 75.857 | 1,698 |
The internal transcribed spacers (ITS1 and ITS2) and the 5.8S-coding region were amplified by primers ITS4 and ITS5 [30]. Reaction conditions were according to Haque et al. [29]. Amplified products were resolved by electrophoresis on 1.4% agarose gels prepared with 1× TAE (Tris-acetate-EDTA) buffer system, run at 5 V/cm, and stained with ethidium bromide solution (0.5 µg L-1). A 100 bp DNA ladder (Hyperladder IV, Bioline Ltd., London, UK) was used to estimate the molecular weights of the amplified products. The stained gels were viewed briefly and bands were excised and eluted from the gel using the MinElute gel extraction kit (Qiagen). The samples were directly sequenced on an automated sequencer ABI-3730xl at a commercial facility (Macrogen Inc., Seoul, South Korea). To obtain proper and reliable sequences, all the amplified samples were sequenced in both directions with two flanking (ITS5 and ITS4) and two internal (ITS2 and ITS3) primers which provided four overlapping sequences, two for the sense strand and two for the antisense strand for each sample selected for the study (Supplementary Data 1).
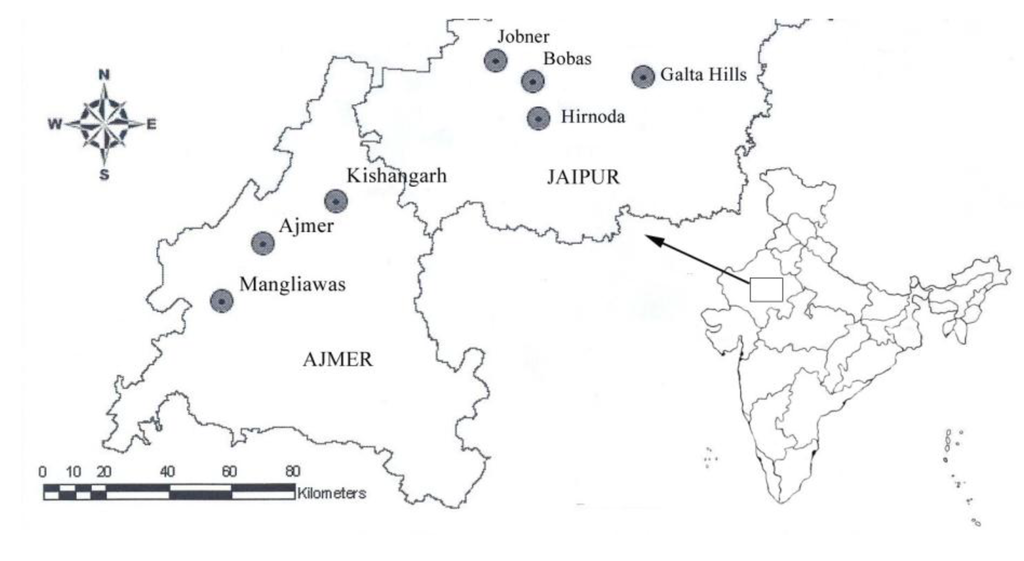
Figure 1.
Location of the seven populations of C. wightii included in the study from the state of Rajasthan, India.
2.1. Data Analysis
Complimentary strands of the ITS region were assembled using Sequencher 4.8 (Gene Codes Corporation, MI, USA) and checked for homology using the NCBI BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/). Nucleotide multiple alignments were performed with ClustalX [31] using the default parameters (Supplementary data 2). Nucleotide polymorphism, as measured by θw [32] and diversity, as measured by π [33], were calculated using DnaSP v4.5 [34].
Analysis of molecular variance (AMOVA) was performed using GenAlEx 6.1 [35] to assess genotypic variations across all the populations studied. This analysis, apart from partitioning of total genetic variation into within-group and among-group variation components, also provided a measure of intergroup genetic distance as the proportion of the total variation residing between populations. The program was also used for a subsequent Mantel’s test [36] to evaluate the relationship between genetic and geographic distance. The significance of both the analysis was tested using 999 random permutations.
Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 [37] and a Maximum Parsimony (MP) tree was constructed using the Close-Neighbour-Interchange algorithm. A bootstrap consensus tree was inferred from 1000 replicates, and branches corresponding to partitions reproduced in less than 50% of bootstrap replicates were collapsed. To check for congruency, a Bayesian analysis using Metropolis-coupled Markov chain Monte Carlo (MCMCMC) was also implemented on the ITS dataset using MrBAYES software [38]. Prior to phylogenetic estimation, we used the program jModelTest 0.1.1 [39] to select which of the 88 models of DNA substitution better fits the data. The Bayesian analysis consisted of four chains with random starting trees and the GTR+G model of nucleotide substitution. The four chains, were run for 3 × 106 generations and trees were sampled every 100th generation. After plotting likelihood values for the four analyses we ascertained that the Markov chains reached stationarity approximately after 100,000 generations. We discarded the first 7,500 trees as burn-in and the remaining trees were used to construct a 50% majority rule consensus tree on which posterior probability scores were indicated.
3. Results and Discussion
ITS regions from 32 C. wightii individuals were sequenced and analyzed. The absence of other variable regions in the nuclear DNA of plants that could provide useful markers at both intra-family [13] and intra-genomic level differentiation [21], makes ITS ostensibly the best molecule for phylogenetic studies. The present study focuses on the insights of inter-individual variability to address questions related to diversity.
The amplified region of ITS1-5.8S rDNA-ITS2 in C. wightii varied between 711 to 727 bp. Similarity search of the sequences at the NCBI database showed ~95% similarity with other Commiphora spp., the sequences were deposited in the GenBank Database (Accession numbers- EU419958 to EU419989). The average GC content of the sequences was 64.6%, the substitution probabilities are given in Table 2 and the overall transition/transversion bias (R) was 1.319. Multiple sequence alignment of all the 32 ITS sequences resulted in a matrix of 757 characters (Supplementary data 2) out of which 70 were parsimony informative.

Table 2.
Maximum composite likelihood estimate of the pattern of nucleotide substitution. Each entry shows the probability of substitution from one base (row) to another base (column) instantaneously. Only entries within a row need to be compared. Rates of different transitional substitutions are shown in bold and those of transversional substitutions are shown in italics.
| A | T | C | G | |
| A | - | 4.84 | 8.22 | 22.43 |
| T | 3.94 | - | 10.13 | 8.57 |
| C | 3.94 | 5.96 | - | 8.57 |
| G | 10.33 | 4.84 | 4.22 | - |
The overall estimate for nucleotide polymorphism is presented in Table 3. A five-fold level of polymorphism was observed between the populations, with the highest in Galta Hills (π = 0.05493 ± 0.01707, θw = 0.05307 ± 0.03221) and the lowest in Hirnoda (π = 0.01107 ± 0.00374, θw = 0.01107 ± 0.00709). Variability within NRTU families usually depends upon number of gene copies, rates of mutation, concerted evolution, number and chromosomal location of NRTU clusters, and proportion of sexual and asexual reproduction [40]. Polymorphism may arise when concerted evolution is not fast enough to homogenise repeats in face of high rates of mutation [41] or by loss of sexual recombination [19]. Also, concerted evolution is retarded in agamospermous plants [18]. As C. wightii is reported to be apomictic, lack of sexual reproduction could be an important contributor to polymorphism. Also, concerted evolution is retarded in agamospermous plants. In a simulation model, Adolfsson and Bengtsson [42] concluded that the spread of apomixis in a population is not necessarily associated with a substantial decrease in genetic variability and most of it is retained. These cumulative factors should increase the polymorphism among C. wightii populations, but the converse was observed. Our earlier studies revealed that sexuality in C. wightii is not uncommon [27], and the plant does revert to sexual mode of reproduction during its life cycle. Sexual reproduction may lower ITS sequence polymorphism by accelerating concerted evolution and homogenizing the ITS repeats.

Table 3.
Nucleotide diversity of the ITS1- 5.8S- ITS2 fragment for the seven populations. π and θw refer to nucleotide diversity according to Nei and Li [43] and Watterson’s parameter [32] respectively. Figures in parenthesis denote standard deviation.
| Population | π | θw |
|---|---|---|
| Mangliawas | 0.03473 (0.00908) | 0.03446 (0.01590) |
| Ajmer | 0.03078 (0.01080) | 0.03148 (0.01742) |
| Kishangarh | 0.03650 (0.00843) | 0.03424 (0.01890) |
| Jobner | 0.04445 (0.00550) | 0.04966 (0.02180) |
| Bobas | 0.03661 (0.01121) | 0.03614 (0.02210) |
| Hirnoda | 0.01107 (0.00374) | 0.01107 (0.00709) |
| Galta Hills | 0.05493 (0.01707) | 0.05307 (0.03221) |
| All Samples | 0.03905 (0.00307) | 0.05418 (0.01685) |
AMOVA was used to partition the genetic diversity and test whether there is any hierarchy of ITS sequence variation among individuals (Table 4). The genetic differentiation between the populations is high (ΦST = 0.206) (Nei [33] classified GST > 0.15 as high, ΦST and GST both denote fixation index and are comparable).

Table 4.
Hierarchical analysis of molecular variance (AMOVA) within/among C. wightii populations. d.f.: degrees of freedom; SSD: sum of squared deviations; ΦST: fixation index; p-value: the probability of having a more extreme variance component than the observed values by chance alone.
| Source of variation | d.f. | SSD | Estimated variance | Total variance (%) | ΦST | p-value |
|---|---|---|---|---|---|---|
| Among population | 6 | 301.118 | 6.046 | 21 | 0.206 | 0.01 |
| Within population | 25 | 508.796 | 23.359 | 79 | ||
| Total | 31 | 885.094 | 29.405 |
The constructed maximum parsimony tree separated the samples into five major clusters (data not shown). The majority rule consensus tree prepared by Bayesian analyses of the ITS sequence data also showed that the populations were grouped according to their geographic location (Figure 2). Apart from Bobas, samples from a particular population did not separate into a single cluster and were found to be mixed. Samples collected from Ajmer, Kishangarh and Mangliawas were distributed in two clusters, so were the Hirnoda, Jobner and Galta Hills samples. It seems that genetic exchange might be taking place between samples from nearby areas (i.e., districts), while long distance transfer is being restricted; this may be responsible for high population structure and differentiation.
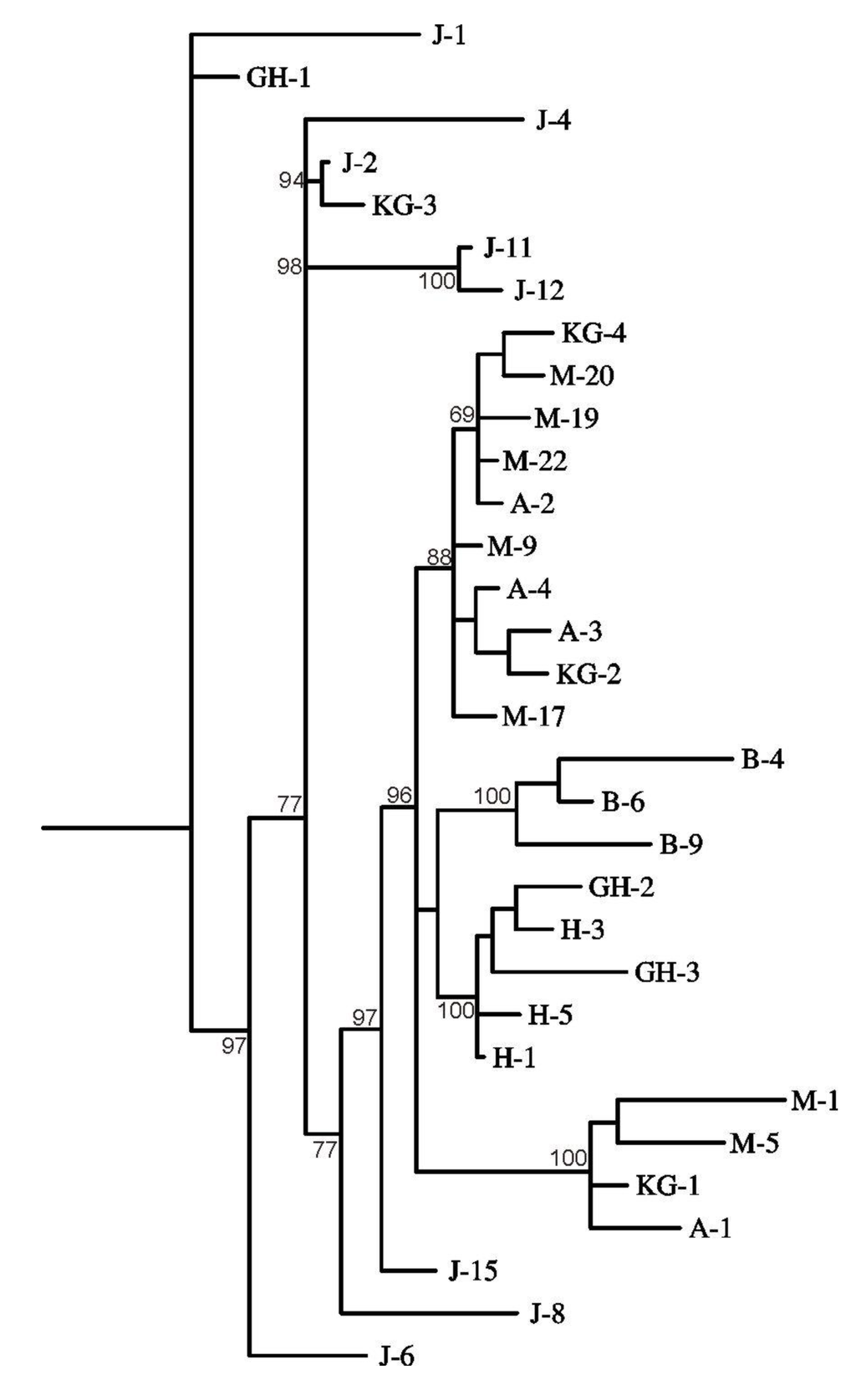
Figure 2.
Relationship between the taxa studied, inferred using the maximum likelihood method (MrBayes). Numbers at the nodes are Bayesian posterior probabilities.
4. Conclusions
It appears that C. wightii plants have evolved under reproductive isolation, probably due to population fragmentation and habitat loss. The reproductive isolation, coupled with the reported apomictic behaviour may have a detrimental effect on genetic variation. Mantel’s test showed an average correlation (r2 = 0.3647) between genetic and geographic distance that was statistically significant (p = 0.023). These samples appeared to be the remnants of a previously large population that was present in the area. Since population differentiation is high, the population continuum has been disrupted, possibly due to over-exploitation, unsustainable utilization and other anthropogenic activities. Educating the villagers and promoting scientific gum-resin tapping practices seem to be the best conservation strategies, along with habitat restoration. Besides, initiative should be taken on surveying and mapping the distribution of the plant for future germplasm collection and maintenance.
Supplementary Data 1.
Screenshot showing complete sequence assembly of ITS region obtained with four primers.

Supplementary Data 2.
Complete alignment matrix of the ITS sequences from C. wightii.
Acknowledgements
We thank the late Dr. M L Sharma and Dr. M Krishnamohan for helping us to locate and identify C. wightii plants. The authors gratefully acknowledge BTISNet SubDIC (BT/BI/065/2004) for providing facilities for bioinformatic analyses and the Government of Jharkhand, Dept. of Agriculture for providing infrastructure development fund. This study was financially supported by the Department of Biotechnology, Govt. of India (NBDB Grant 102/IFD/SAN/1192/2004-05).
References and Notes
- APG, II. An update of the Angiosperm Phylogenetic Group classification for the orders and families of flowering plants: APG II. Bot. J. Lin. Soc. 2003, 141, 399–436. [Google Scholar] [CrossRef]
- Gupta, P.; Shivanna, K.R.; Mohan Ram, H.Y. Apomixis and polyembryony in the guggul plant, Commiphora wightii. Ann. Bot. 1996, 78, 67–72. [Google Scholar] [CrossRef]
- Murray, M.T. The Healing Power of Herbs: The Enlightened Person’s Guide to the Wonders of Medicinal Plants; Prima Publishers: Roseville, CA, USA, 1995. [Google Scholar]
- El Ashry, E.S.; Rashed, N.; Salama, O.M.; Saleh, A. Components, therapeutic value and uses of myrrh. Pharmazie 2003, 58, 163–168. [Google Scholar]
- Singh, R.B.; Niaz, M.A.; Ghosh, G. Hypolipidemic and antioxidant effects of Commiphora mukul as an adjunct to dietary therapy in patients with hypercholesterolemia. Cardiovasc. Drugs Ther. 1994, 8, 659–664. [Google Scholar] [CrossRef]
- Deng, R. Therapeutic effects of guggul and its constituent guggulsterone: Cardiovascular benefits. Cardiovasc. Drug Rev. 2007, 25, 375–390. [Google Scholar]
- Sarfaraz, S.; Siddiqui, I.A.; Syed, D.N.; Afaq, F.; Mukhtar, H. Guggulsterone modulates MAPK and NF-kappaB pathways and inhibits skin tumorigenesis in SENCAR mice. Carcinogenesis 2008, 29, 2011–2018. [Google Scholar] [CrossRef]
- Singh, S.V.; Choi, S.; Zeng, Y.; Hahm, E.R.; Xiao, D. Guggulsterone-induced apoptosis in human prostate cancer cells is caused by reactive oxygen intermediate dependent activation of c-Jun NH2-terminal kinase. Cancer Res. 2007, 67, 7439–7449. [Google Scholar] [CrossRef]
- Xiao, D.; Singh, S.V. Z-Guggulsterone, a constituent of Ayurvedic medicinal plant Commiphora mukul, inhibits angiogenesis in vitro and in vivo. Mol. Cancer Ther. 2008, 7, 171–180. [Google Scholar] [CrossRef]
- Khan, T.I.; Dular, A.K.; Deepika, M.S. Biodiversity conservation in the Thar Desert; with emphasis on endemic and medicinal plants. Environmentalist 2003, 23, 137–144. [Google Scholar] [CrossRef]
- Soltis, D.; Soltis, P. Choosing an approach and an appropriate gene for phylogenetic analysis. In Molecular Systematics of Plants II. DNA Sequencing; Soltis, D., Soltis, P., Doyle, J., Eds.; Kluwer Academic Publishers: Boston, MA, USA, 1998; pp. 1–42. [Google Scholar]
- Alvarez, I.; Wendel, J.F. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 2003, 29, 417–434. [Google Scholar] [CrossRef]
- Baldwin, B.G.; Markos, S. Phylogenetic utility of the external transcribed spacer (ETS) of 18S-26S rDNA: Congruence of ETS and ITS trees of Calycadenia (Compositae). Mol. Phylogenet. Evol. 1998, 10, 449–463. [Google Scholar] [CrossRef]
- Baldwin, B.; Sanderson, M.; Porter, J.; Wojciechowski, M.; Campbell, C.; Donoghue, M. The ITS region of nuclear ribosomal DNA- a valuable source of evidence on angiosperm phylogeny. Ann. Mo. Bot. Gard. 1995, 82, 247–277. [Google Scholar] [CrossRef]
- Hershkovitz, M.A.; Zimmer, E.A.; Hahn, W.J. Ribosomal DNA sequences and angiosperm systematics. In Molecular Systematics and Plant Evolution; Hollingsworth, P.M., Bateman, R.M., Gornall, R.J., Eds.; Taylor & Francis: London, UK, 1999; pp. 268–326. [Google Scholar]
- Kelch, D.G.; Baldwin, B.G. Phylogeny and ecological radiation of New World thistles (Cirsium, Cardueae-Compositae) based on ITS and ETS rDNA sequence data. Mol. Ecol. 2003, 12, 141–151. [Google Scholar] [CrossRef]
- Lee, J.Y.; Mummenho, V.K.; Bowman, J.L. Allopolyploidization and evolution of species with reduced floral structures in Lepidium L. (Brassicaceae). Proc. Natl. Acad. Sci. USA 2002, 99, 16835–16840. [Google Scholar] [CrossRef]
- Ainouche, M.L.; Bayer, R.J. On the origins of the tetraploid Bromus species (section Bromus, Poaceae): insights from the internal transcribed spacer sequences of nuclear ribosomal DNA. Genome 1997, 40, 730–743. [Google Scholar] [CrossRef]
- Campbell, C.S.; Wojciechowski, M.F.; Baldwin, B.G.; Alice, L.A.; Donoghue, M.J. Persistent nuclear ribosomal DNA sequence polymorphism in the Amelanchier agamic complex (Rosaceae). Mol. Biol. Evol. 1997, 14, 81–90. [Google Scholar]
- Hughes, C.E.; Bailey, C.D.; Harris, S.A. Divergent and reticulate species relationships in Leucaena (Fabaceae) inferred from multiple data sources: Insights into polyploid origins and nrDNA polymorphism. Am. J. Bot. 2002, 89, 1057–1073. [Google Scholar] [CrossRef]
- Feliner, G.N.; Larena, B.G.; Aguilar, J.F. Fine scale geographic structure, intra-individual polymorphism and recombination in nuclear ribosomal internal transcribed spacers in Armeria (Plumbaginaceae). Ann. Bot. 2004, 93, 189–200. [Google Scholar] [CrossRef]
- Buckler, E.S.; Ippolito, A.; Holtsford, T.P. The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics 1997, 145, 821–832. [Google Scholar]
- Becerra, J.X. Insects on plants: macroevolutionary chemical trends in host use. Science 1999, 276, 253–256. [Google Scholar] [CrossRef]
- Becerra, J.X.; Venable, D.L. Nuclear ribosomal DNA phylogeny and its implications for evolutionary trends in Mexican Bursera (Burseraceae). Am. J. Bot. 1999, 86, 1047–1057. [Google Scholar] [CrossRef]
- Becerra, J.X. Evolution of Mexican Bursera (Burseraceae) inferred from ITS, ETS, and 5S nuclear ribosomal DNA sequences. Mol. Phylogenet. Evol. 2003, 26, 300–309. [Google Scholar] [CrossRef]
- Weeks, A.; Daly, D.C.; Simpson, B.B. The phylogenetic history and biogeography of the frankincense and myrrh family (Burseraceae) based on nuclear and chloroplast sequence data. Mol. Phylogenet. Evol. 2005, 35, 85–101. [Google Scholar] [CrossRef]
- Haque, I.; Bandopadhyay, R.; Mukhopadhyay, K. Population genetic structure of the endangered and endemic medicinal plant Commiphora wightii. Mol. Biol. Rep. 2009. [Google Scholar] [CrossRef]
- Milligan, B.G.; Leebens-Mack, J.; Strand, A.E. Conservation genetics: beyond the maintenance of marker diversity. Mol. Ecol. 1994, 3, 423–435. [Google Scholar] [CrossRef]
- Haque, I.; Bandopadhyay, R.; Mukhopadhyay, K. An optimised protocol for fast genomic DNA isolation from high secondary metabolites and gum containing plants. Asian J. Plant Sci. 2008, 7, 304–308. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplifcation and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M., Gelfand, D., Sninsky, J., White, T., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, G.D. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 24, 4876–4882. [Google Scholar]
- Watterson, G.A. On the number of segregating sites in genetical models without recombination. Theor. Pop. Biol. 1975, 7, 256–276. [Google Scholar]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar]
- Rozas, J.; Rozas, R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 1999, 15, 174–175. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel, population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MrBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Posada, D. Selection of Models of DNA Evolution with jModelTest. In Bioinformatics for DNA Sequence Analysis; Posada, D., Ed.; SpringerLink: Secaucus, NJ, USA, 2009; Volume 537, pp. 93–112. [Google Scholar]
- Dover, G.A.; Linares, A.R.; Bowen, T.; Hancock, J.M. Detection and quantification of concerted evolution and molecular drive. Methods Enzymol. 1993, 224, 525–541. [Google Scholar] [CrossRef]
- Appels, R.; Honeycutt, R.L. rDNA: Evolution over a billion years. In DNA systematics; Dutton, S., Ed.; CRC Press: Boca Raton, FL, USA, 1986; pp. 81–135. [Google Scholar]
- Adolfsson, S.; Bengtsson, B.O. The spread of apomixis and its effect on resident genetic variation. J. Evol. Biol. 2007, 20, 1933–1940. [Google Scholar] [CrossRef]
- Nei, M.; Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).