New Species of Bacidia s.l. from the Azores and the Resurrection of Genus Woessia
Abstract
1. Introduction
2. Materials and Methods
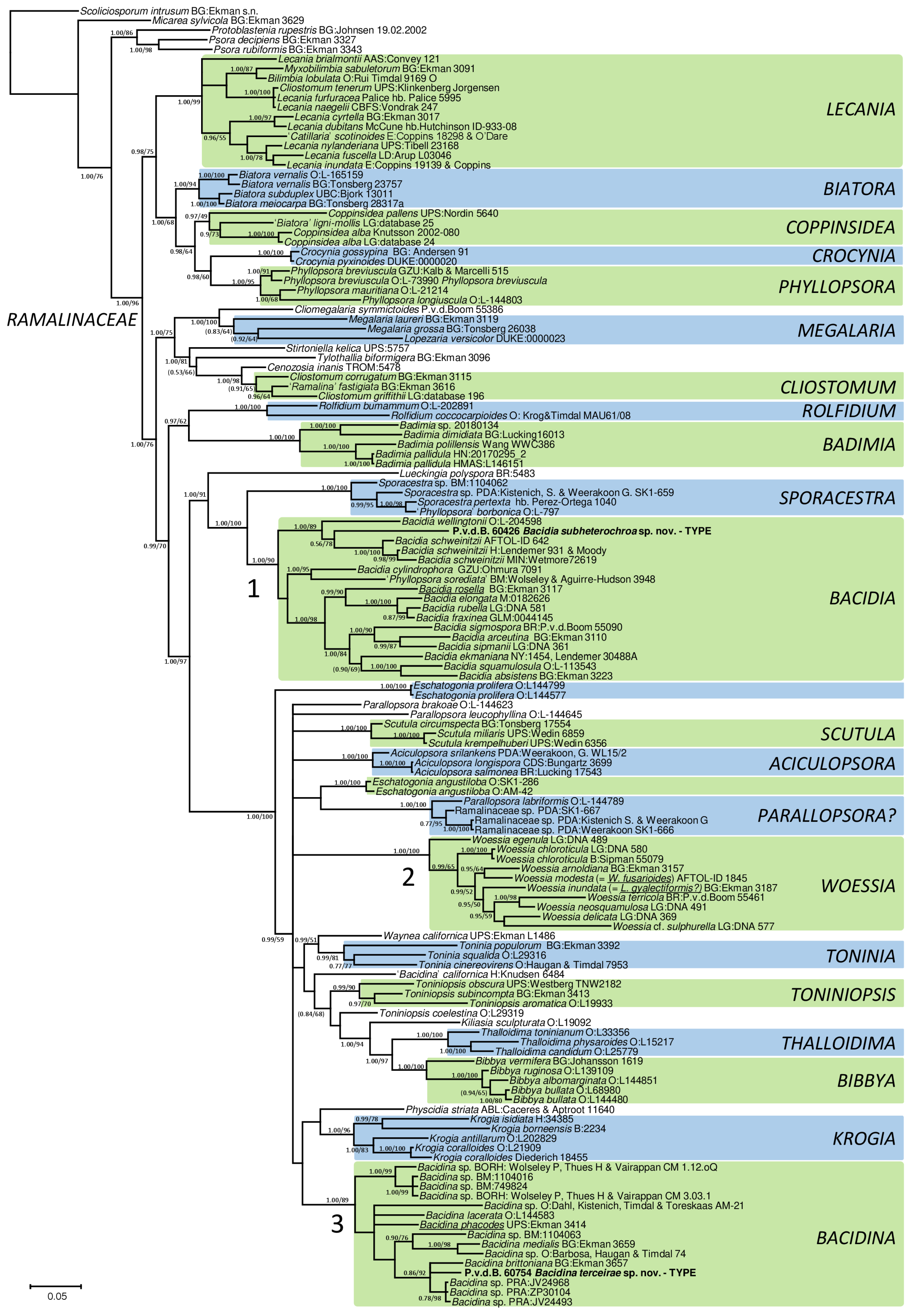
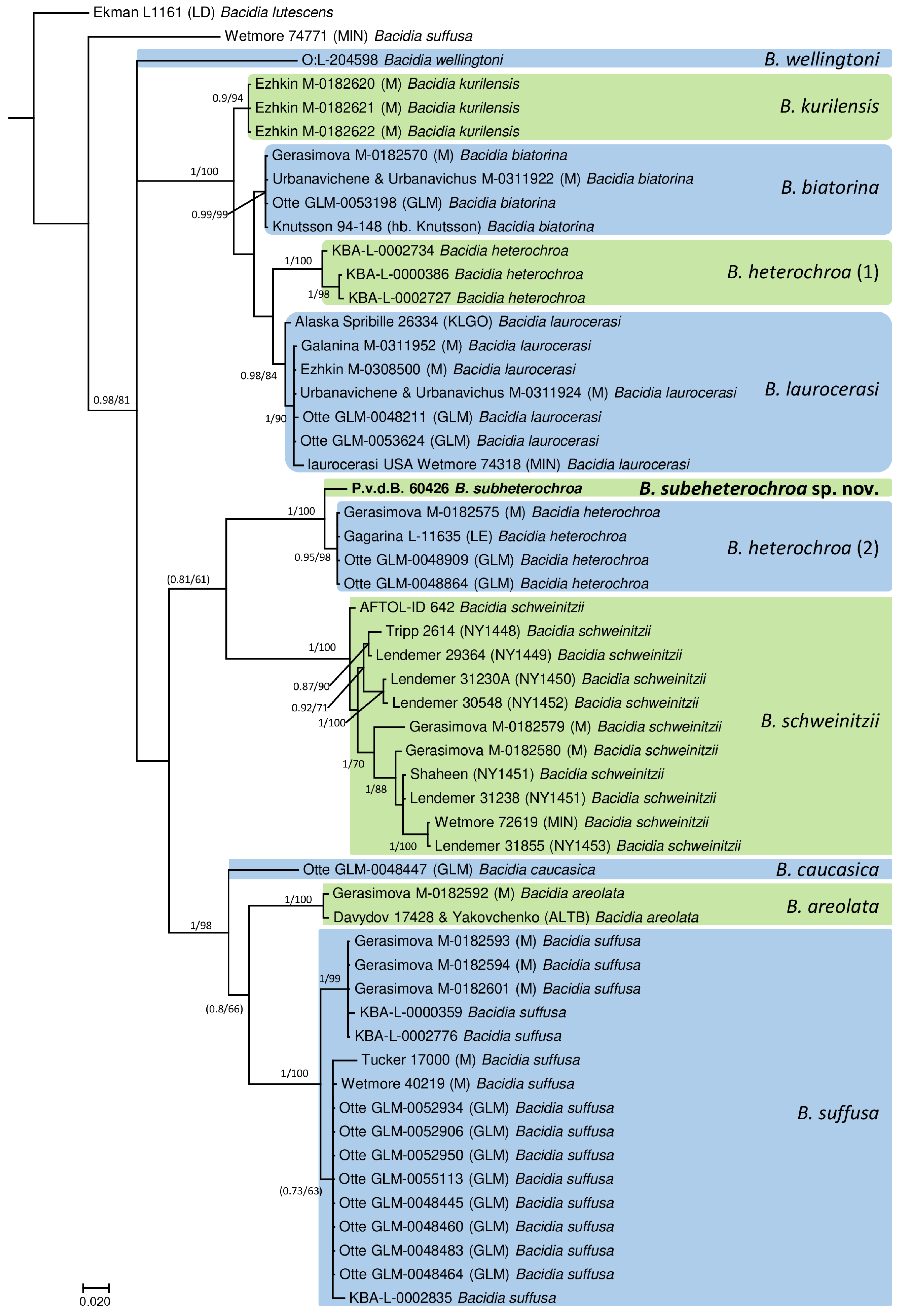
3. Results
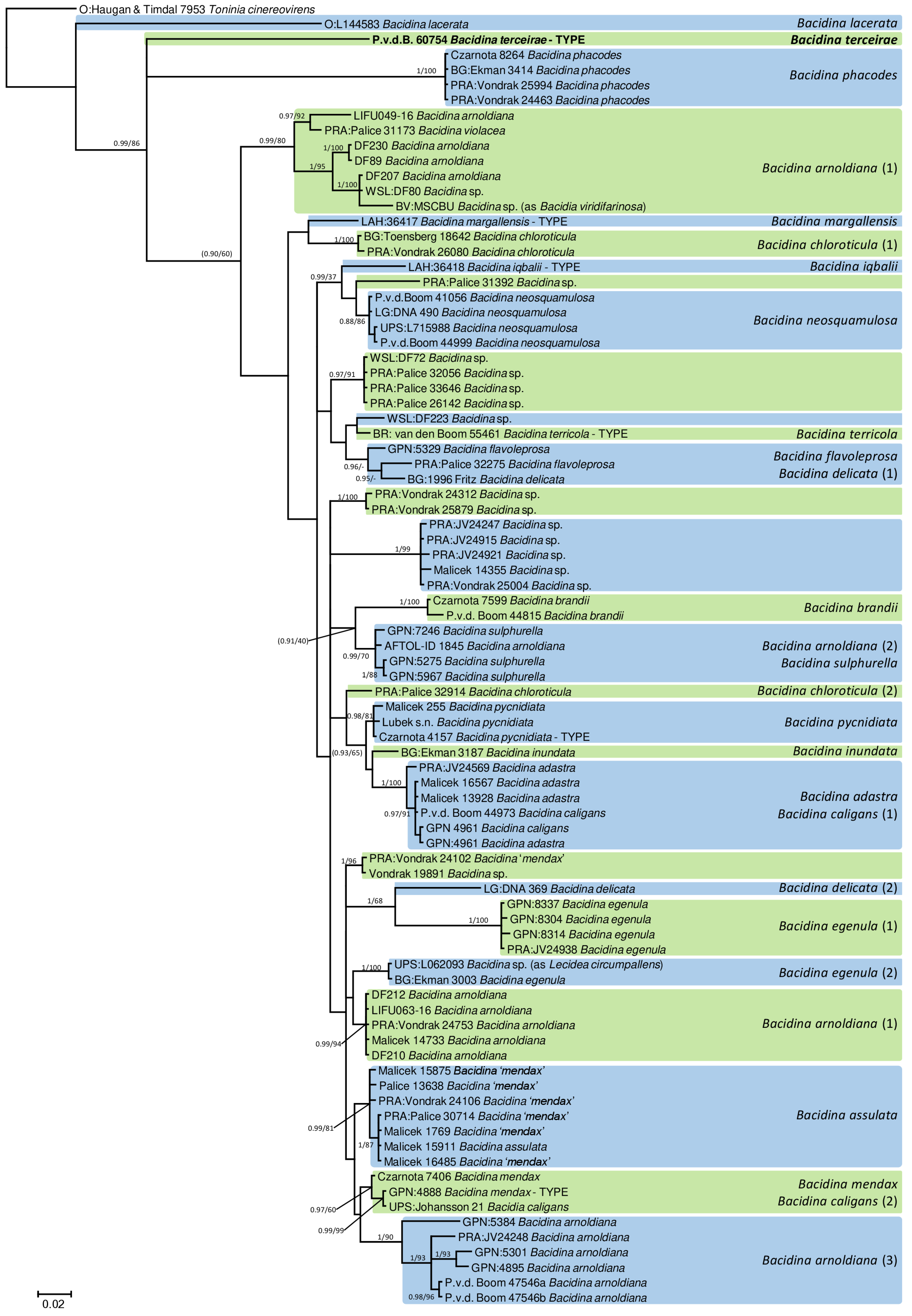
3.1. Taxonomy
3.2. New Combinations into Woessia:
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aptroot, A.; Rodrigues, A.F.; Schumm, F.; Câmara, S.; Gabriel, R. Lista dos líquenes e fungos liquenícolas (Fungi). List of lichens and lichenicolous fungi (Fungi). In Listagem dos Organismos Terrestres e Marinhos dos Açores. A list of the Terrestrial and Marine Biota from the Azores; Botges, P.A.V., Costa, A., Cunha, R., Gabriel, R., Gonçalves, V., Martins, A.F., Melo, I., Parente, M., Raposeiro, P., Rodrigues, P., et al., Eds.; Princípia: Cascais, Portugal, 2010; Volume 532, pp. 59–79. [Google Scholar]
- van den Boom, P.P.G. Lichens and lichenicolous fungi of the Azores (Portugal), collected on São Miguel and Terceira with the descriptions of seven new species. Acta Bot. Hung. 2016, 58, 199–222. [Google Scholar] [CrossRef]
- Etayo, J. Lichenicolous fungi of the Azores islands. I. Terceira. Ascomycete. Org 2018, 10, 107–116. [Google Scholar]
- Diederich, P.; Millanes, A.M.; Etayo, J.; van den Boom, P.P.G.; Wedin, M. Finding the needle in the haystack: A revision of Crittendenia, a surprisingly diverse lichenicolous genus of Agaricostilbomycetes, Pucciniomycotina. Bryologist 2022, 125, 248–293. [Google Scholar] [CrossRef]
- Breuss, O. New and remarkable lichen records from the Azores (São Miguel Island). Herzogia 2018, 31, 430–435. [Google Scholar]
- Breuss, O. Neue und bemerkenswerte Flechtenfunde von den Azoren (Insel São Miguel) II. Osterr. Z. Für Pilzkd. 2018, 27, 31–36. [Google Scholar]
- van den Boom, P.P.G.; Alvarado, P. Lichens and lichenicolous fungi of Faial (Azores, Portugal) with descriptions of three new species. Herzogia 2019, 32, 421–437. [Google Scholar] [CrossRef]
- van den Boom, P.P.G.; Magain, N. Three new lichen species from Macaronesia belonging in Ramalinaceae, with the description of a new genus. Plant Fungal Syst. 2020, 65, 167–175. [Google Scholar] [CrossRef]
- Clerc, P. Synopsis of Usnea (lichenized Ascomycetes) from the Azores with additional information on species in Macaronesia. Lichenologist 2006, 38, 191–212. [Google Scholar] [CrossRef]
- Moberg, R.; Purvis, W. Studies on the lichens of the Azores. Part 4. The genus Heterodermia. In Lichen Studies Dedicated to Rolf Santesson. 32. Symbolae Botanicae Upsalienses; Tibell, L., Hedberg, I., Eds.; Acta Universitatis Upsaliensis: Uppsala, Sweden, 1997; pp. 187–194. [Google Scholar]
- van den Boom, P.P.G. Notes on the genus Anisomeridium (lichenized Ascomycotina) from Madeira and the Azores (Macaronesia). Phytotaxa 2015, 205, 65–70. [Google Scholar] [CrossRef]
- van den Boom, P.P.G.; Lücking, R.; Sipman, H.J.M. Notes on Graphidaceae in Macaronesia, with descriptions of four new species. Diversity 2023, 15, 817. [Google Scholar] [CrossRef]
- Vězda, A. Bacidina genus novum familiae Lecideaceae (Ascomycetes lichenisati). Folia Geobot. Phytotaxon 1991, 25, 431–432. [Google Scholar] [CrossRef]
- Vězda, A. Foliicole Flechten aus der Kolchis (West-Transkaukasien, UdSSR). Folia Geobot. Phytotaxon 1983, 18, 45–70. [Google Scholar] [CrossRef]
- Wirth, V. Phytosoziologie, Okologie und Systematik bei Flechten. Berichte Dtsch. Bot. Ges. 1983, 96, 103–115. [Google Scholar] [CrossRef]
- Ekman, S. Proposal to conserve the name Bacidina against Lichingoldia and Woessia (lichenized Ascomycotina). Taxon 1996, 45, 687–688. [Google Scholar] [CrossRef]
- Ekman, S. Four new and two resurrected species of Bacidina from Sweden, with notes and a preliminary key to the known Scandinavian species. Nord. J. Bot. 2023, 2023, e03846. [Google Scholar] [CrossRef]
- Hauck, M.; Wirth, V. New combinations in Bacidina. Herzogia 2010, 23, 15–17. [Google Scholar]
- McNeill, J.; Barrie, F.R.; Burdet, H.M.; Demoulin, V.; Hawksworth, D.L.; Marhold, K.; Nicolson, D.H.; Prado, J.; Silva, P.C.; Skog, J.E.; et al. International Code of Botanical Nomenclature (Vienna Code); Lubrecht & Cramer Ltd.: Port Jervis, NY, USA, 2006; Volume 146, pp. 1–568. [Google Scholar]
- Kistenich, S.; Timdal, E.; Bendiksby, M.; Ekman, S. Molecular systematics and character evolution in the lichen family Ramalinaceae (Ascomycota: Lecanorales). Taxon 2018, 67, 871–904. [Google Scholar] [CrossRef]
- Sérusiaux, E.; van den Boom, P.P.G.; Brand, M.A.; Coppins, B.J.; Magain, N. Lecania falcata, a new species from Spain, the Canary Islands and the Azores, close to Lecania chlorotiza. Lichenologist 2012, 44, 577–590. [Google Scholar] [CrossRef]
- Miadlikowska, J.; Kauff, F.; Högnabba, F.; Oliver, J.C.; Molnár, K.; Fraker, E.; Gaya, E.; Hafellner, J.; Hofstetter, V.; Gueidan, C.; et al. A multigene phylogenetic synthesis for the class Lecanoromycetes (Ascomycota): 1307 fungi representing 1139 infrageneric taxa, 317 genera and 66 families. Mol. Phylogenetics Evol. 2014, 79, 132–168. [Google Scholar] [CrossRef]
- Kistenich, S.; Rikkinen, J.K.; Thüs, H.; Vairappan, C.S.; Wolseley, P.A.; Timdal, E. Three new species of Krogia (Ramalinaceae, lichenised Ascomycota) from the Paleotropics. MycoKeys 2018, 40, 69–88. [Google Scholar] [CrossRef]
- Gabriel, R. Base de Dados da Biodiversidade dos Açores. Líquenes. Universidade dos Açores. Available online: http://www.azoresbioportal.angra.uac.pt/listagens.php?sstr=1&lang=pt2008 (accessed on 11 October 2020).
- Schumm, F. Flechten Madeiras, der Kanaren und Azoren; Book on Demand: Wangen, Germany, 2008; p. 294. [Google Scholar]
- Schumm, F.; Aptroot, A. Flechten Madeiras, der Kanaren und Azoren—Band 2 Ergänzungsband; Book on Demand: Wangen, Germany, 2013; p. 457. [Google Scholar]
- Pino-Bodas, R.; Ahti, T.; Stenroos, S. Cladoniaceae of the Azores. Herzogia 2017, 30, 445–462. [Google Scholar] [CrossRef]
- Guzow-Krzemińska, B.; Sérusiaux, E.; van den Boom, P.P.G.; Brand, A.M.; Launis, A.; Łubek, A.; Kukwa, M. Understanding the evolution of phenotypical characters in the Micarea prasina group (Pilocarpaceae) and descriptions of six new species within the group. MycoKeys 2019, 57, 1–30. [Google Scholar] [CrossRef] [PubMed]
- van den Boom, P.P.G. Lichens and lichenicolous fungi of the Azores (Pico, São Jorge), additional records and four new species. Acta Bot. Hung. 2020, 62, 417–434. [Google Scholar] [CrossRef]
- van den Boom, P.P.G. Lichen and lichenicolous fungus records from Santa Maria, Azores archipelago. Botanica 2021, 27, 34–43. [Google Scholar] [CrossRef]
- van den Boom, P.P.G. Foliicolous lichens and their lichenicolous fungi in Macaronesia and Atlantic Europe. Bibl. Lichenol. 2021, 111, 197. [Google Scholar]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Mullis, K.; Faloona, F.A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987, 155, 335–350. [Google Scholar]
- White, T.J.; Bruns, T.D.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J., White, T.J., Eds.; Academic Press: London, UK, 1990; 482p. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for Basidiomycetes —Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Cubeta, M.A.; Echandi, E.; Abernethy, T.; Vilgalys, R. Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology 1991, 81, 1395–1400. [Google Scholar] [CrossRef]
- Zoller, S.; Scheidegger, C.; Sperisen, C. PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 1999, 31, 511–516. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Arita, M.; Karsch-Mizrachi, I.; Cochrane, G. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2021, 49, D121–D124. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Poelt, J. Five additional genera of conidial lichen-forming fungi from Europe. Plant Syst. Evol. 1986, 154, 195–211. [Google Scholar] [CrossRef]
- Ekman, S.; Svensson, M.; Westberg, M.; Zamora, J.C. Additions to the lichen flora of Fennoscandia III. Graph. Scr. 2019, 31, 34–46. [Google Scholar]
- Ekman, S. The corticolous and lignicolous species of Bacidia and Bacidina in North America. Opera Bot. 1996, 127, 1–148. [Google Scholar]
- Gerasimova, J.V.; Otte, V.; Urbanavichene, I.N.; Urbanavichus, G.P.; Beck, A. High diversity of Bacidia (Ramalinaceae, Lecanorales) species in the Caucasus as revealed by molecular and morphological analyses. Lichenologist 2023, 55, 275–296. [Google Scholar] [CrossRef]
- Vondrák, J.; Svoboda, S.; Malíček, J.; Palice, Z.; Kocourková, J.; Knudsen, K.; Mayrhofer, H.; Thüs, H.; Schultz, M.; Košnar, J.; et al. From Cinderella to Princess: An exceptional hotspot of lichen diversity in a long-inhabited central-European landscape. Preslia 2022, 94, 143–181. [Google Scholar] [CrossRef]
- Smith, C.W.; Aptroot, A.; Coppins, B.J.; Fletcher, A.; Gilbert, O.L.; James, P.W.; Wolseley, P.A. (Eds.) The Lichens of Great Britain and Ireland; British Lichen Society: London, UK, 2009; p. 1046. [Google Scholar]
- Kistenich, S.; Bendiksby, M.; Vairappan, C.S.; Weerakoon, G.; Wijesundara, S.; Wolseley, P.A.; Timdal, E. A regional study of the genus Phyllopsora (Ramalinaceae) in Asia and Melanesia. MycoKeys 2019, 53, 23–72. [Google Scholar] [CrossRef] [PubMed]
- Gerasimova, J.V.; Ezhkin, A.K.; Davydov, E.A.; Beck, A. Multilocus-phylogeny of the lichen-forming genus Bacidia s. str. (Ramalinaceae, Lecanorales) with special emphasis on the Russian Far East. Lichenologist 2021, 53, 441–455. [Google Scholar] [CrossRef]
- Czarnota, P.; Guzow-Krzeminska, B. ITS rDNA data confirm a delimitation of Bacidina arnoldiana and B. sulphurella and support a description of a new species within the genus Bacidina. Lichenologist 2012, 44, 743–755. [Google Scholar] [CrossRef]
- Kondratyuk, S.Y.; Lőkös, L.; Farkas, E.; Jang, S.-H.; Liu, D.; Halda, J.; Persson, P.-E.; Hansson, M.; Kärnefelt, I.; Thell, A.; et al. Three new genera of the Ramalinaceae (lichen-forming Ascomycota) and the phenomenon of presence of ‘extraneous mycobiont DNA’ in lichen associations. Acta Bot. Hung. 2019, 61, 275–323. [Google Scholar] [CrossRef]


| Species | Specimen Voucher | mtSSU | LSU | RPB1 |
|---|---|---|---|---|
| Aciculopsora longispora | CDS:Bungartz 3699 | MN334224 | ||
| Aciculopsora salmonea | BR:Lucking 17543 | MG925842 | MG926137 | |
| Aciculopsora srilankens | PDA:Weerakoon, G. WL15/2 | MK400212 | ||
| Bacidia absistens | BG:Ekman 3223, SE-93 | MG925845 | MG926139 | |
| Bacidia arceutina | BG:Ekman 3110, SE-23 | MG925846 | MG926041 | MG926140 |
| Bacidia cylindrophora | GZU:Ohmura 7091 | MG925908 | ||
| Bacidia ekmaniana | NY:1454, Lendemer 30488A | KX151746 | ||
| Bacidia elongata | M:0182626 | MW506352 | MW493330 | |
| Bacidia fraxinea | GLM:0044145 | MW506373 | MW489428 | MW540441 |
| Bacidia rosella | BG:Ekman 3117 | AY300877 | AY300829 | AY756412 |
| Bacidia rubella | LG:DNA 581 | JQ796831 | ||
| Bacidia schweinitzii | AFTOL-ID 642 | DQ972998 | DQ782911 | DQ782830 |
| Bacidia schweinitzii | H:Lendemer 931 & Moody, AFTOL-ID 4969 | KJ766354 | KJ766527 | |
| Bacidia schweinitzii | MIN:Wetmore72619, SE-111 | MG926045 | MG926146 | |
| Bacidia sigmospora | BR:P.v.d.Boom 55090 | MW622007 | ||
| Bacidia sipmanii | LG:DNA 361 | JQ796832 | JQ796844 | |
| Bacidia sp. (as Phyllopsora sorediata) | BM:Wolseley & Aguirre-Hudson 3948, 1007 | MG925905 | MG926094 | MG926196 |
| Bacidia squamulosula | O:L-113543, SE-314 | MG925856 | MG926051 | MG926152 |
| Bacidia subheterochroa sp. nov. | P.v.d.B. 60426 | PQ615684 | PQ615682 | |
| Bacidia wellingtonii | O:L-204598 | MG925853 | ||
| Bacidina brittoniana | BG:Ekman 3657 SE-224 | MG926050 | MG926151 | |
| Bacidina californica | H:Knudsen 6484 AFTOL-ID 4892 | KJ766356 | KJ766529 | |
| Bacidina lacerata | O:L144583, Timdal 10213 519 | MG925896 | ||
| Bacidina medialis | BG:Ekman 3659 SE-225 | MG925850 | MG926044 | MG926145 |
| Bacidina phacodes | UPS:Ekman 3414 SE-78 | MG926049 | ||
| Bacidina sp. | O:Dahl, Kistenich, Timdal & Toreskaas AM-21 | MW307634 | ||
| Bacidina sp. | O: Barbosa, Haugan & Timdal 74 | W307633 | ||
| Bacidina sp. | PRA:JV24968 | OK465496 | ||
| Bacidina sp. | PRA:ZP30104 | OK465497 | ||
| Bacidina sp. | PRA:JV24493 | OK465495 | ||
| Bacidina sp. (as undetermined Ramalinaceae) | BM:1104063 | MK400200 | ||
| Bacidina sp. (as undetermined Ramalinaceae) | BORH: Wolseley P, Thues H & Vairappan CM 1.12.oQ | MK400201 | ||
| Bacidina sp. (as undetermined Ramalinaceae) | BM:1104016 | MK400192 | ||
| Bacidina sp. (as undetermined Ramalinaceae) | BM:749824 | MK400193 | ||
| Bacidina sp. (as undetermined Ramalinaceae) | BORH: Wolseley P, Thues H & Vairappan CM 3.03.1 | MK400206 | ||
| Bacidina terceirae sp. nov. | P.v.d.B. 60754 | PQ615685 | PQ615683 | |
| Badimia dimidiata | BG:Lucking 16013 | AY567774 | MG926052 | |
| Badimia pallidula | HN:20170295_2 | MW349653 | MT791315 | |
| Badimia pallidula | HMAS:L146151, Wang 20192889 | MT791324 | MT791317 | |
| Badimia polillensis | Wang WWC386 | MT791325 | MT791318 | |
| Badimia sp. | 20180134 | MT791323 | MT791316 | |
| Biatora meiocarpa | BG:Tonsberg 28317a BG | AM292710 | ||
| Biatora subduplex | UBC:Bjork 13011 AFTOL-ID 4912 | KJ766360 | KJ766533 | KJ766838 |
| Biatora vernalis | O:L-165159, J.T. Klepsland JK09-L616 | KF360418 | KF360446 | |
| Biatora vernalis | BG:Tonsberg 23757 | DQ838753 | DQ838752 | |
| Biatora vetera | Knutsson 2002-080 | AY567771 | ||
| Bibbya albomarginata | O:L144851, Timdal 10481 5468 | MG925927 | MG926115 | MG926212 |
| Bibbya bullata | O:L68980, Elix & Streimann 40393 SE-251 | MG925929 | MG926116 | |
| Bibbya bullata | O:L144480, Bannister s.n., 4823 | MG925928 | ||
| Bibbya ruginosa | O:L139109, Timdal 10087 5469 | MG925937 | MG926121 | MG926218 |
| Bibbya vermifera | BG:Johansson 1619 SE-109 | MG925852 | MG926047 | MG926148 |
| Bilimbia lobulata | O:Rui Timdal 9169 O | AM292712 | MG926062 | MG926161 |
| Cenozosia inanis | TROM:5478 | MG925866 | MG926066 | |
| Cliomegalaria symmictoides | P.v.d.Boom 55386 | MW622006 | MW621867 | |
| Cliostomum corrugatum | BG:Ekman 3115 | AY567722 | MG926067 | MG926166 |
| Cliostomum griffithii | LG:database 196 | GU138667 | ||
| Cliostomum sp. (as Ramalina fastigiata) | BG:Ekman 3616 | AY756375 | AY756360 | AY756422 |
| Cliostomum tenerum | UPS:Klinkenberg Jorgensen | AM292722 | ||
| Coppinsidea alba (as Biatora veteranorum) | LG:database 24 | GU138664 | ||
| Coppinsidea pallens | UPS:Nordin 5640 | AM292709 | ||
| Coppinsidea sp. (as Biatora ligni-mollis) | LG:database 25 | GU138665 | ||
| Crocynia gossypina | BG:Andersen 91 | AY567766 | ||
| Crocynia pyxinoides | DUKE:0000020, AFTOL-ID 111 | AY584615 | AY584653 | |
| Eschatogonia angustiloba | O:SK1-286 | MW307646 | ||
| Eschatogonia angustiloba | O:AM-42 | MW307643 | ||
| Eschatogonia prolifera | O:L144799, Timdal 10429 447 | MG925871 | MG926170 | |
| Eschatogonia prolifera | O:L144577, Timdal 10207 446 | MG925870 | MG926070 | MG926169 |
| Killiasia sculpturata | O:L19092, Haugan & Timdal YAK17,30, 5470 | MG925938 | MG926122 | MG926219 |
| Krogia antillarum | O:L202829, AM39 | MH174274 | ||
| Krogia borneensis | B:2234 | MH174277 | ||
| Krogia coralloides | O:L21909, Krog & Timdal MAU51,83, SE-387 | MG925875 | MG926072 | MG926173 |
| Krogia coralloides | Diederich 18455 | MH174278 | ||
| Krogia isidiata | H:34385 | MH174279 | ||
| Lecania brialmontii | AAS:Convey 121 | AM292726 | MG926112 | MG926209 |
| Lecania cyrtella | BG:Ekman 3017 | AY300891 | AY300840 | MG926175 |
| Lecania dubitans | McCune herb. Hutchinson ID-933-08 | AM292732 | ||
| Lecania furfuracea | Palice herb. Palice 5995 | AM292734 | ||
| Lecania fuscella | LD:Arup L03046 | AM292735 | ||
| Lecania inundata | E:Coppins 19139 & Coppins | AM292740 | ||
| Lecania naegelii | CBFS:Vondrak 247 | AM292741 | ||
| Lecania nylanderiana | UPS:Tibell 23168 | AM292742 | ||
| Lecania sp. (as Catillaria scotinoides) | E:Coppins 18298 & O’Dare | AM292720 | ||
| Lopezaria versicolor | DUKE:0000023, AFTOL-ID 108 | AY584622 | AY584651 | |
| Lueckingia polyspora | BR:5483 | MG925882 | MG926082 | |
| Megalaria grossa | BG:Tonsberg 26038 | AY762095 | AY756356 | AY756419 |
| Megalaria laureri | BG:Ekman 3119, SE-125 | MG925884 | MG926182 | |
| Micarea sylvicola | BG:Ekman 3629 | AY567769 | AY756331 | AY756392 |
| Myxobilimbia sabuletorum | BG:Ekman 3091 | AY567721 | AY756346 | AY756413 |
| Parallopsora brakoae | O:L-144623, Timdal 10253 | MG925891 | ||
| Parallopsora labriformis | O:L-144789, Timdal 10419 | MG925895 | MG926188 | |
| Parallopsora leucophyllina | O:L-144645, 464 | MG925897 | MG926189 | |
| Parallopsora sp. (as undetermined Ramalinaceae) | PDA: SK1-667 | MK400228 | ||
| Parallopsora sp. (as undetermined Ramalinaceae) | PDA:Kistenich S. & Weerakoon G, Ne141 | MK400210 | ||
| Parallopsora sp. (as undetermined Ramalinaceae) | PDA:Weerakoon SK1-666 | MK400225 | ||
| Phyllopsora breviuscula | GZU:Kalb & Marcelli 515 1305 | MG925893 | MG926088 | MG926186 |
| Phyllopsora breviuscula | O:L-73990, 528 | MG925892 | MG926087 | MG926185 |
| Phyllopsora longiuscula | O:L-144803, 454 | MG925899 | MG926090 | MG926191 |
| Phyllopsora mauritiana | O:L-21214, SE-386 | MG925900 | MG926091 | MG926192 |
| Physcidia striata | ABL:Caceres & Aptroot 11640 | MG925910 | MG926098 | |
| Protoblastenia rupestris | BG:Johnsen 19.02.2002 | AY756358 | ||
| Psora decipiens | BG:Ekman 3327 | AY567772 | MG926102 | AY756396 |
| Psora rubiformis | BG:Ekman 3343 | AY756374 | AY756359 | |
| Rolfidium bumammum | O:L-202891, 4821 | MG925930 | MG926117 | MG926213 |
| Rolfidium coccocarpioides | O: Krog&Timdal MAU61/08 | AY762096 | AY756361 | |
| Scoliciosporum intrusum | BG:Ekman s.n. | AY567767 | AY756329 | AY756391 |
| Scutula circumspecta | BG:Tonsberg 17554 SE-228 | MG925848 | MG926143 | |
| Scutula krempelhuberi | UPS:Wedin 6356 | AY567789 | ||
| Scutula miliaris | UPS:Wedin 6859 | AY567790 | ||
| Sporacestra pertexta | hb. Perez-Ortega 1040 | MG925903 | MG926093 | MG926194 |
| Sporacestra sp. (as Phyllopsora borbonica) | O:L-797 | MG925890 | MG926086 | MG926184 |
| Sporacestra sp. (as undetermined Ramalinaceae) | BM:1104062 | MK400191 | ||
| Sporacestra sp. (as undetermined Ramalinaceae) | PDA:Kistenich, S. & Weerakoon G. SK1-659 | MK400223 | ||
| Stirtoniella kelica | UPS:5757 | MG925923 | ||
| Thalloidima candidum | O:L25779, Bratli & Timdal 8733 SE-59 | MG925932 | MG926118 | MG926215 |
| Thalloidima physaroides | O:L15217, SE-250 | MG925936 | MG926120 | MG926217 |
| Thalloidima toninianum | O:L33356, SE-72 | MG926125 | MG926222 | |
| Toninia cinereovirens | O:Haugan & Timdal 7953 | AY567724 | AY756365 | AY756429 |
| Toninia populorum | BG:Ekman 3392 SE-274 | MG925843 | MG926039 | MG926138 |
| Toninia squalida | O:L29316, Haugan 4970 SE-70 | MG925940 | MG926123 | MG926220 |
| Toniniopsis aromatica | O:L19933, Haugan & Timdal 4819 SE-58 | MG925926 | MG926113 | MG926210 |
| Toniniopsis coelestina | O:L29319, Haugan 5985 SE-61 | MG925933 | MG926119 | |
| Toniniopsis obscura | UPS:Westberg TNW2182, SE-306 | MG925943 | MG926126 | MG926223 |
| Toniniopsis subincompta | BG:Ekman 3413 SE-100 | MG925851 | MG926046 | MG926147 |
| Tylothallia biformigera | BG:Ekman 3096, SE-28 | MG925946 | MG926129 | MG926226 |
| Waynea californica | UPS:Ekman L1486, SE-214 | MG925947 | MG926130 | |
| Woessia arnoldiana | BG:Ekman 3157 SE-118 | MG925854 | MG926048 | MG926149 |
| Woessia cf. sulphurella | LG:DNA 577 | JQ796839 | ||
| Woessia chloroticula | LG:DNA 580 | JQ796833 | ||
| Woessia chloroticula | B:Sipman 55079 AFTOL-ID 4990 | KJ766357 | KJ766530 | |
| Woessia delicata | LG:DNA 369 | JQ796834 | JQ796845 | |
| Woessia egenula | LG:DNA 489 | JQ796836 | JQ796846 | |
| Woessia inundata | BG:Ekman 3187 SE-87 | MG925855 | MG926150 | |
| Woessia modesta | DUKE:AFTOL-ID 1845 | DQ986810 | DQ986798 | KJ766904 |
| Woessia neosquamulosa | LG:DNA 491 | JQ796838 | JQ796848 | |
| Woessia terricola | BR:P.v.d.Boom 55461 | MW622008 |
| Species | Herbarium | ITS |
|---|---|---|
| Bacidia areolata | Davydov 17428 & Yakovchenko (ALTB) | MW491455 |
| Bacidia areolata | Gerasimova M-0182592 (M) | MH048614 |
| Bacidia biatorina | Gerasimova M-0182570 (M) | MW523509 |
| Bacidia biatorina | Knutsson 94-148 (hb. Knutsson) | AF282079 |
| Bacidia biatorina | Otte GLM-0053198 (GLM) | MW523511 |
| Bacidia biatorina | Urbanavichene & Urbanavichus M-0311922 (M) | MW523510 |
| Bacidia caucasica | Otte GLM-0048447 (GLM) | MW523553 |
| Bacidia heterochroa (1) | KBA-L-0000386 | ON352606 |
| Bacidia heterochroa (1) | KBA-L-0002727 | ON352612 |
| Bacidia heterochroa (1) | KBA-L-0002734 | ON352613 |
| Bacidia heterochroa (2) | Gagarina L-11635 (LE) | MW523516 |
| Bacidia heterochroa (2) | Gerasimova M-0182575 (M) | MW523515 |
| Bacidia heterochroa (2) | Otte GLM-0048864 (GLM) | MW523518 |
| Bacidia heterochroa (2) | Otte GLM-0048909 (GLM) | MW523517 |
| Bacidia kurilensis | Ezhkin M-0182620 (M) | MH048610 |
| Bacidia kurilensis | Ezhkin M-0182621 (M) | MH048611 |
| Bacidia kurilensis | Ezhkin M-0182622 (M) | MH048612 |
| Bacidia laurocerasi | Alaska Spribille 26334 (KLGO) | MN483106 |
| Bacidia laurocerasi | Ezhkin M-0308500 (M) | MW491460 |
| Bacidia laurocerasi | Galanina M-0311952 (M) | MH048609 |
| Bacidia laurocerasi | Otte GLM-0048211 (GLM) | MW523513 |
| Bacidia laurocerasi | Otte GLM-0053624 (GLM) | MW523514 |
| Bacidia laurocerasi | Urbanavichene & Urbanavichus M-0311924 (M) | MW523512 |
| Bacidia laurocerasi | USA Wetmore 74318 (MIN) | AF282078 |
| Bacidia lutescens | Ekman L1161 (LD) | AF282082 |
| Bacidia schweinitzii | AFTOL-ID 642 | DQ782850 |
| Bacidia schweinitzii | Gerasimova M-0182579 (M) | MW491454 |
| Bacidia schweinitzii | Gerasimova M-0182580 (M) | MH048613 |
| Bacidia schweinitzii | Lendemer 29364 (NY1449) | KX151763 |
| Bacidia schweinitzii | Lendemer 30548 (NY1452) | KX151761 |
| Bacidia schweinitzii | Lendemer 31230A (NY1450) | KX151766 |
| Bacidia schweinitzii | Lendemer 31238 (NY1451) | KX151764 |
| Bacidia schweinitzii | Lendemer 31855 (NY1453) | KX151765 |
| Bacidia schweinitzii | Shaheen (NY1451) | MG461696 |
| Bacidia schweinitzii | Tripp 2614 (NY1448) | KX151762 |
| Bacidia schweinitzii | Wetmore 72619 (MIN) | AF282080 |
| Bacidia subheterochroa sp. nov. | P.v.d.B. 60426 | PQ615680 |
| Bacidia suffusa | Gerasimova M-0182593 (M) | MH048616 |
| Bacidia suffusa | Gerasimova M-0182594 (M) | MH048617 |
| Bacidia suffusa | Gerasimova M-0182601 (M) | MH048615 |
| Bacidia suffusa | KBA-L-0000359 | ON352605 |
| Bacidia suffusa | KBA-L-0002776 | ON352614 |
| Bacidia suffusa | KBA-L-0002835 | ON352616 |
| Bacidia suffusa | Otte GLM-0048445 (GLM) | MW523551 |
| Bacidia suffusa | Otte GLM-0048460 (GLM) | MW523552 |
| Bacidia suffusa | Otte GLM-0048464 (GLM) | MW523555 |
| Bacidia suffusa | Otte GLM-0048483 (GLM) | MW523554 |
| Bacidia suffusa | Otte GLM-0052906 (GLM) | MW523548 |
| Bacidia suffusa | Otte GLM-0052934 (GLM) | MW523547 |
| Bacidia suffusa | Otte GLM-0052950 (GLM) | MW523549 |
| Bacidia suffusa | Otte GLM-0055113 (GLM) | MW523550 |
| Bacidia suffusa | Tucker 17000 (M) | MH048618 |
| Bacidia suffusa | Wetmore 40219 (M) | MH048619 |
| Bacidia suffusa | Wetmore 74771 (MIN) | AF282091 |
| Bacidia wellingtoni | O:L-204598 | MG925953 |
| Species | Herbarium | ITS |
|---|---|---|
| Bacidina adastra | PRA:JV24569 | OK332938 |
| Bacidina adastra | Malicek 16567 | PQ524005 |
| Bacidina adastra | Malicek 13928 | OQ717318 |
| Bacidina adastra | GPN:4961 | MG461694 |
| Bacidina arnoldiana | GPN:5301 | JN972439 |
| Bacidina arnoldiana | GPN:5384 | JN972441 |
| Bacidina arnoldiana | GPN:4895 | JN972440 |
| Bacidina arnoldiana | AFTOL-ID 1845 | HQ650650 |
| Bacidina arnoldiana | DF230 | KX098347 |
| Bacidina arnoldiana | DF207 | KX098343 |
| Bacidina arnoldiana | LIFU049-16 | KX132958 |
| Bacidina arnoldiana | DF89 | KX098348 |
| Bacidina arnoldiana | DF212 | KX098346 |
| Bacidina arnoldiana | LIFU063-16 | KX132972 |
| Bacidina arnoldiana | PRA:Vondrak 24753 | OQ717717 |
| Bacidina arnoldiana | Malicek 14733 | OP730571 |
| Bacidina arnoldiana | DF210 | KX098344 |
| Bacidina arnoldiana | PRA:JV24248 | OK332939 |
| Bacidina arnoldiana | P.v.d. Boom 47546a | KX239021 |
| Bacidina arnoldiana | P.v.d. Boom 47546b | KX239020 |
| Bacidina assulata | Malicek 15911 | PP768145 |
| Bacidina brandii | Czarnota 7599 | KX239029 |
| Bacidina brandii | P.v.d. Boom 44815 | KX239025 |
| Bacidina caligans | UPS:Johansson 21 | AF282096 |
| Bacidina caligans | GPN 4961 | JN972442 |
| Bacidina caligans | P.v.d. Boom 44973 | KX239024 |
| Bacidina chloroticula | PRA:Palice 32914 | OQ717320 |
| Bacidina chloroticula | BG:Toensberg 18642 | AF282098 |
| Bacidina chloroticula | PRA:Vondrak 26080 | OQ717319 |
| Bacidina delicata | BG:1996 Fritz | AF282097 |
| Bacidina delicata | LG:DNA 369 | JQ796854 |
| Bacidina egenula | GPN:8337 | KY379234 |
| Bacidina egenula | GPN:8304 | KY379235 |
| Bacidina egenula | GPN:8314 | KY379233 |
| Bacidina egenula | PRA:JV24938 | OL396592 |
| Bacidina egenula | BG:Ekman 3003 | AF282095 |
| Bacidina flavoleprosa | PRA:Palice 32275 | OQ717718 |
| Bacidina flavoleprosa | GPN:5329 | JN972443 |
| Bacidina inundata | BG:Ekman 3187 | AF282094 |
| Bacidina iqbalii | LAH:36418-TYPE | MT952885 |
| Bacidina lacerata | O:L144583 | MG925993 |
| Bacidina margallensis | LAH:36417 | NR_182351 |
| Bacidina mendax | GPN:4888-TYPE | JN972444 |
| Bacidina mendax | Malicek 15875 | OQ918722 |
| Bacidina mendax | PRA:Palice 30714 | OQ717321 |
| Bacidina mendax | Malicek 1769 | KX239016 |
| Bacidina mendax | Palice 13638 | KX239028 |
| Bacidina mendax | PRA:Vondrak 24102 | OQ717722 |
| Bacidina mendax | PRA:Vondrak 24106 | OQ717721 |
| Bacidina mendax | Malicek 16485 | PQ524006 |
| Bacidina mendax | Czarnota 7406 | KX239023 |
| Bacidina neosquamulosa | UPS:L715988 | OP256854 |
| Bacidina neosquamulosa | P.v.d.Boom 41056 | KX239026 |
| Bacidina neosquamulosa | LG:DNA 490 | JQ796855 |
| Bacidina neosquamulosa | P.v.d.Boom44999 | KX239018 |
| Bacidina phacodes | Czarnota 8264 | KX239036 |
| Bacidina phacodes | BG:Ekman 3414 | AF282100 |
| Bacidina phacodes | PRA:Vondrak 25994 | OQ717322 |
| Bacidina phacodes | PRA:Vondrak 24463 | OQ717723 |
| Bacidina pycnidiata | Malicek 255 | KX239034 |
| Bacidina pycnidiata | Lubek s.n. | KX239032 |
| Bacidina pycnidiata | Czarnota 4157-TYPE | KX239033 |
| Bacidina sp. | WSL:DF223 | KX098339 |
| Bacidina sp. | WSL:DF80 | KX098341 |
| Bacidina sp. | WSL:DF72 | KX098340 |
| Bacidina sp. | PRA:Vondrak 24312 | OQ717719 |
| Bacidina sp. | PRA:Vondrak 25879 | OQ717720 |
| Bacidina sp. | PRA:JV24247 | OK332883 |
| Bacidina sp. | PRA:JV24915 | OK332884 |
| Bacidina sp. | PRA:JV24921 | OK332885 |
| Bacidina sp. | PRA:Palice 32056 | OQ717726 |
| Bacidina sp. | PRA:Palice 33646 | OQ717327 |
| Bacidina sp. | PRA:Palice 26142 | OQ717326 |
| Bacidina sp. | PRA:Palice 31392 | OQ727426 |
| Bacidina sp. | Malicek 14355 | PP410490 |
| Bacidina sp. | PRA:Vondrak 25004 | OQ717324 |
| Bacidina sp. | Vondrak 19891 | MT803542 |
| Bacidina sp. (as Bacidia viridifarinosa) | BV:MSCBU | PP889388 |
| Bacidina sp. (as Lecidea circumpallens) | UPS:L062093 | OP256856 |
| Bacidina sulphurella | GPN:5275 | JN972445 |
| Bacidina sulphurella | GPN:5967 | JN972446 |
| Bacidina sulphurella | GPN:7246 | JN972447 |
| Bacidina terceirae | P.v.d.B. 60754 | PQ615681 |
| Bacidina terricola | BR: van den Boom 55461 | NR_172202 |
| Bacidina violacea | PRA:Palice 31173 | OQ717328 |
| Toninia cinereovirens | O:Haugan & Timdal 7953 | AF282104 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boom, P.P.G.v.d.; Alvarado, P. New Species of Bacidia s.l. from the Azores and the Resurrection of Genus Woessia. Diversity 2025, 17, 187. https://doi.org/10.3390/d17030187
Boom PPGvd, Alvarado P. New Species of Bacidia s.l. from the Azores and the Resurrection of Genus Woessia. Diversity. 2025; 17(3):187. https://doi.org/10.3390/d17030187
Chicago/Turabian StyleBoom, P. P. G. van den, and P. Alvarado. 2025. "New Species of Bacidia s.l. from the Azores and the Resurrection of Genus Woessia" Diversity 17, no. 3: 187. https://doi.org/10.3390/d17030187
APA StyleBoom, P. P. G. v. d., & Alvarado, P. (2025). New Species of Bacidia s.l. from the Azores and the Resurrection of Genus Woessia. Diversity, 17(3), 187. https://doi.org/10.3390/d17030187







