Abstract
Hymenocallis henryae is a rare, charismatic spider lily endemic to the Florida panhandle. Currently under review to determine if listing under the Endangered Species Act is warranted, this species has undescribed genetic diversity, information crucial to the listing process. We conducted field observations of 21 historic populations across the species’ geographical range and performed genomic analyses of 279 individuals from 19 extant populations. Most populations had fewer than 40 individuals, while populations with >100 individuals were found exclusively on managed lands. Genetic diversity was uniformly low within populations (HE: 0.074–0.093), with low to moderate inbreeding coefficients (FIS: 0.068–0.431). Genetic differentiation was relatively low among most populations (FST: 0–0.098), although there was statistical support for isolation by distance. In addition, we found high genetic similarity and lack of population structure across the species range. Clonal propagation through fused bulbs is a common reproductive strategy. We confirmed current threats (habitat change, residential development, fire suppression) and identified several coastal populations threatened by sea level rise. It is recommended to continue with in situ protection and management as well as the establishment of ex situ living collections to preserve populations most at risk of extirpation from habitat loss and degradation.
1. Introduction
Conservation biology is a crisis discipline often requiring biologists to make decisions without the comfort of complete scientific certainty [1,2,3]. Conservation biologists aim to maintain biodiversity by recovery of declining or vulnerable species, specifically threatened or endangered species as defined by the Endangered Species Act (ESA). The ESA enforces conservation and protection for species that are within the foreseeable future at risk of extinction. Within the ESA sections, the listing process requires the use of the best available data and an understanding of the current and future threats to species extinction. As main threats such as habitat disturbance and climate change accelerate, the management of threatened or endangered species is becoming increasingly more complex [4]. Mitigation of these threats is one component of conservation management, but a species’ ability to respond to these threats is just as critical.
The patterns and distribution of genetic diversity among populations can determine the ability of a species to respond to changes in environmental conditions and future adaptations [5]. High genetic diversity facilitates adaptive responses of populations to shifting biotic and abiotic conditions [6], while low or loss of genetic diversity has been shown to affect individual fitness and limit adaptive response. Because of this, conservationists have recognized that maintaining genetic diversity as a natural defense against shifting environmental threats is key to the management of populations [7,8]. Rare or endemic species with restricted ranges often have lower genetic diversity compared with their widespread congeners, making them particularly vulnerable to environmental instability [9]. Quantification of levels and patterns of genetic diversity of such species can inform the likelihood of species survival and conservation action decisions.
One rare species in need of understanding its ability to respond to anthropogenic disturbances is Hymenocallis henryae Traub (Amaryllidaceae). Hymenocallis henryae is an at-risk perennial herb that grows from a bulb and produces flowers with six, linear green tepals and a prominent white staminal cup (Figure 1) [10]. This species can reproduce both sexually and asexually [11]; however, the prevalence of clonality within populations is unknown. Two varieties have been described, H. henryae var. henryae and H. henryae var. glaucifolia, differentiated by whether plants grow in loose (var. henryae) or dense (var. glaucifolia) patches, whether glaucescence is slight (var. henryae) or heavy (var. glaucifolia) [12], and whether plants are located on the western side (var. henryae) or on the eastern side (var. glaucifolia) of the Apalachicola River [13,14].

Figure 1.
Reproductive and vegetative characteristics of Hymenocallis henryae: (a) Flowering individual with six narrow pale green perianth segments and six stamens, April 2021, Tyndall Air Force Base. (b) Vegetative plant with three fused basal bulbs with contractile roots, Tate’s Hell State Forest. (c) Vegetative growth with waxy leaves in coastal habitat on Tyndall Air Force Base. (d) Ecotone between a mesic flatwood and a cypress dome swamp; plants were growing adjacent to and in the water.
The New World genus Hymenocallis includes about 50 species, of which 13 are found in Florida and 4 are endemic to the Florida panhandle [15], one of the five richest biodiversity hotspots in North America [16,17,18]. Two of the endemic species, H. godfreyi and H. henryae, are state listed as endangered. Hymenocallis henryae is found in five counties (Figure 2) with 25 documented populations as of 2014 [19,20]. These populations further comprise multiple subpopulations. Populations are predominantly found in mesic flatwoods, coastal wet flatwoods, and forested areas on managed private or public lands [19,21]. The most common habitat for this species is mesic flatwoods, characterized by an open pine canopy with a dense lower understory [20]. In the flatwoods, H. henryae is found in the narrow ecotones between cypress dome swamps and wet prairies with standing water, wet loamy soils, or dry soils [22].
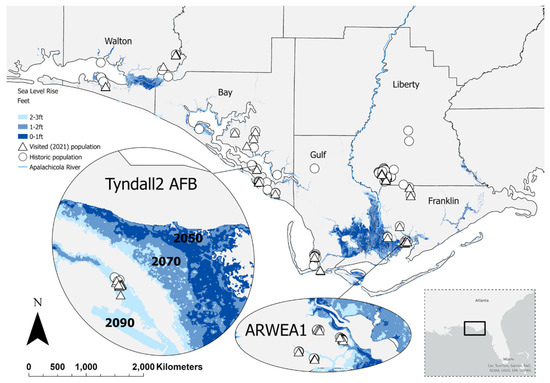
Figure 2.
Predicted sea level rise in the panhandle of Florida ranges from dark blue (0–1 foot, year 2050), blue (1–2 feet, year 2070) to light blue (2–3 ft, year 2090). Hymenocallis henryae occurrences in the panhandle of Florida (Walton, Bay, Gulf, Franklin, Liberty counties) visited in 2021 are indicated by triangles. The location of historic populations is indicated by circles. The Apalachicola River is shown in blue following the Franklin–Liberty county line. Layers provided by the National Oceanic and Atmospheric Administration. See Table 1 for acronyms.
As a species, H. henryae faces multiple threats, including habitat loss and modification, development, overcollection, and coastal flooding or sea level rise [19,23]. The species was petitioned for listing under the Endangered Species Act in 2010 [23,24], and the U.S. Fish and Wildlife Service is preparing a species status review to determine if listing of this species is warranted. To help inform the listing process, we characterized the genetic diversity within and among populations of H. henryae across the species range. Furthermore, we surveyed the historical populations of H. henryae to assess current threats and population size and identified coastal populations at risk of the effects of sea level rise (SLR). Finally, we suggest management practices to conserve the genetic resources of this species.
2. Materials and Methods
2.1. Study Area
The study area represents the geographic extent of H. henryae (Figure 2). This area included publicly managed lands (Apalachicola National Forest [ANF], Tate’s Hell State Forest [THSF], Tyndall Air Force Base [Tyndall], and Eglin Air Force Base [EAFB]), privately managed land (Nokuse Plantation [NP], Apalachicola River Wildlife Environmental Area [ARWEA], St. Joseph’s State Buffer Preserve [SJSBP]), and public unmanaged land (Panama City [PC], and Walton; Table 1). Walton sites represent the location where the type specimens were collected in 1962 [10,25]. Managed lands are subjected to variable prescribed fires of 2–15 years and habitat monitoring (ANF, THSF, NP, ARWEA, SJSBP); however, the wetland habitat of THSF was transformed with the installments of drainage channels. Some managed land, such as Tyndall, were recovering from habitat damage from Hurricane Michael in 2018. This study area in the Florida panhandle falls within the North American coastal plain, a global biodiversity hotspot that contains >1500 endemic plant species and has experienced >70% habitat loss [17,18].

Table 1.
Population locations, abbreviations, number of samples collected, clump counts, observed heterozygosity (HO), expected heterozygosity (HE), and inbreeding coefficient (FIS) for H. henryae populations.
2.2. SLR Projections
We evaluated the impact of sea level rise (SLR) on coastal populations of H. henryae using data from the National Oceanic and Atmospheric Administration [26]. We only included sampled data from the 21 populations (Figure 2, triangle symbols) that were visited during the study because of the census and data consistency. The SLR analysis used three different projections, 0–1 ft (year 2050), 1–2 ft (year 2070), and 2–3 ft (year 2090), using the intermediate SLR projections (ranges from low, intermediate low, intermediate, intermediate high, high) for the southeast coast (Interagency Sea Level Rise tool [26]). Population layers were overlaid with the SLR projection layers and were marked as “affected” if one or more plants or clumps were located in the SLR projection layers.
2.3. Tissue Collection
We visited 21 of 25 (Florida Natural Areas Inventory [FNAI] surveys) historically documented populations with some containing subpopulations (Figure 2) in the spring of 2021, 19 of which were extant. Populations visited were chosen based on accessibility and permit availability and represented the extent of the known species distribution. We collected 279 leaf samples (10–15 cm in length) from the 19 extant populations for genetic diversity analyses. Each plant sampled, which was separated by at least one meter, was georeferenced using a Bad Elf GNSS Surveyor [27]. Voucher photographs of representatives of each population and corresponding permits are deposited at the Willard Sherman Turrell Herbarium, Miami University, Oxford, OH. Samples were stored on ice in the field, then transferred to 4 °C for 1 to 5 days, shipped and transferred to Miami University on ice, and stored at −80 °C prior to DNA extraction.
We assessed clonality by conducting intensive sampling within circular plots of 4 m in diameter in three established plots (two within ANF and one within THSF). We identified a reference clump as the georeferenced central point. One to three leaf samples were collected from all clumps within two meters of the central reference plant within the plot, and the distance (all three plots) and azimuth (one plot) from the reference point were recorded using a compass and tape measure. Up to three leaves per clump were collected. A total of 86 leaf samples were collected across the three plots, with 25–35 samples per plot.
2.4. DNA Isolation and Sequencing
Total genomic DNA was extracted from the 279 collected samples using a protocol similar to that reported by Kim et al. [28]. Briefly, ~125 ng of genomic DNA was digested with PstI and MseI (New England Biolabs [NEB]). After digestion, samples were heat-inactivated for 10 min at 80 °C followed by addition of 2.5 μL of 1 μM P1 Adapter and 0.1 μL of 250 μM MseI adapter. PstI P1 adapters each contained a unique multiplex sequence index (barcode), which is read during the first five nucleotides of the Illumina sequence read. P1 and P2 adapters were added to each sample along with 36 units T4 DNA Ligase (high concentration [HC], Enzymatics, Inc., Beverly, MA, USA), 0.3125 U MseI, 0.25 U PstI in a final reaction volume of 25 μL, which was then incubated at 37 °C for 180 min. Samples were diluted 1:10 in water, and 2.5 μL of this product was used in a PCR amplification with 10x PCR Buffer 1 (Applied Biosystems, Waltham, MA, USA), 0.3 μL of 50 ng/μL MseI +GT primer, 0.05 μL of PstI primer, and 0.2 U AmpliTaq DNA Polymerase (Applied Biosystems). The MseI +2 primer was chosen via a primer test with 8 options, where +GT performed best. Samples were diluted for PCR amplification, and the product was purified and quantified [29]. The library was sequenced at the University of Oregon Genomics and Cell Characterization Core Facility (GC3F) on a NovaSeq 6000 with a SP100 chip, generating 118 bp single-end reads RAD loci were assembled using the STACKS pipeline [30,31,32].
2.5. Genetic Diversity and Clonality Assessment
Prior to conducting analyses, VCFtools [33] was used to separate the clonal and broad (diversity) samples within our variant call format (vcf) files. The diversity samples did not warrant additional filtering because sufficient data were present in all individuals. The majority of analyses were performed using R, version 4.2.1 [34] with the addition of Microsoft Excel [35] to organize data. We calculated our basic population genetic statistics which consisted of the observed heterozygosity (HO), expected heterozygosity (HE), and inbreeding coefficients (FIS) using the ‘basic.stat’ function found in the ‘hierfstat’ package.
Principal component analysis (PCA) with the function ‘dudi.pca’ found in the ‘ade4’ package [36] was used to infer ancestry and genetic structure. Function ‘snmf’ from the ‘LEA’ package [37] was used to develop the ancestry matrix sNMF plots to further investigate the presence of population structure. FST values for all sampled populations (n) were found using the function ‘genetic_diff’ in the package ‘vcfR’ [38]. Pairwise FST was calculated for the diversity survey populations using the functions ‘stamppFst’ and ‘genlight’ found in the package ‘StAMPP’ [39] and ‘vcfR’, respectively. Populations with fewer than 3 samples (Table 1) were removed to avoid overestimation of genetic differentiation in pairwise Fst analysis, leaving 12 populations [40]. These results were visualized in a matrix using the function ‘ggplot’ found in the package ‘ggplot2’. A Mantel test with 999 permutations was used to assess isolation by distance. It was conducted using the genetic distance matrix generated in R using the ‘mantel.randtest’ function and a distance matrix calculated from the center GPS point of each population generated in ArcGIS Pro 3.2.2 [41].
For our clonal assignment analysis, we used ‘vcftools’ to filter out missing data because of the package functions and requirements of ‘poppr’ [42]. With a filter that removed SNPs with 50% or more missing data, clonal plot ANF1 retained 47% of its SNPs, ANF2 44%, and THSF1 27%. We removed the clonal plot THSF1 because of the large amount of missing data. For clonal assignment analyses, we used the function ‘bitwise.dist’ in the ‘poppr’ package to first compute a pairwise genetic distance matrix among individuals within a plot. We assigned clones using a genetic distance threshold by identifying clear gaps in the histogram distribution of individual pairwise genetic distances within a clonal population [43]. A UPGMA tree based on the pairwise distance matrix was constructed using the function ‘upgma’ in the package ‘phangorn’ [44], and the threshold value was mapped onto the UPGMA tree (the corresponding value on the tree was half that as determined by histogram analysis to account for the total branch length between any two individuals). Individuals whose pairwise genetic distance was less than this threshold were identified as clonal ramets; those with greater pairwise genetic distance were considered genets.
3. Results
3.1. Population Assessment
The seven largest populations of the nineteen populations sampled (between 75 and 600+ individuals) were found on managed land with regular prescribed burning regimes (ANF1, ANF2, NP2, THSF1, THSF2, Tyndall2, and Tyndall4; Table 1); many of these populations were found in cypress dome marshes or mesic flatwood marshes (ANF populations, THSF populations, and NP2). The remaining populations contained 39 or fewer individuals (NP1, PC1, PC2, PC3, PC4, SJSBP1, SSBP2, THSF3, Tyandall3, Walton, ARWEA1, ARWEA2). Particularly small populations with fewer than 15 individuals were found in fragmented or developed land [PC1-4], coastal habitats (Tyndall3, ARWEA2), or overgrown, forested habitats (Walton; Table 1). Some subpopulations on private land located in PC were either found with no plants (presumed extirpated) or had few plants due to commercial development, although the PC1 population managed to persist in a highly developed area in an empty lot. A historical coastal population in EAFB was not found and is presumed extirpated. In addition to population size and habitat types, we also observed morphological differences among populations. Almost all populations were flowering in May, except for the Walton population The Walton individuals were smaller in size and found in an overgrown area that lacked the typical hydrology of cypress dome marshes. Plants in this subpopulation were also smaller and less abundant than those found in other populations.
The distribution of SLR flooding is expected to affect a total of six coastal H. henryae populations of the total 21 visited (Figure 2). One coastal EAFB historical population of 20 individuals documented in 1995 was found extirpated and submerged in seawater during our 2021 surveys; our SLR analysis showed that this population was located within the 0–1 ft SLR area (year 2050). At 1–2 ft SLR (year 2070), three populations (ARWEA1, ARWEA2, Tyndall 4) are expected to be affected, and at 2–3 ft SLR (year 2090), two additional populations (Tyndall2, Walton; Figure 2). These impacted populations will be most affected by the encroachment of saltwater on the edges of their habitat, likely changing the current hydrology this species has adapted to.
3.2. Genetic Analysis
The final dataset that we used for downstream analysis included 838 loci and 3905 variable sites that were each present in at least 60% of individuals. The mean genotyped sites per locus was 133.38 ± 0.09 bp). The observed heterozygosity ranged from 0.037 to 0.073, with an average value of 0.058 (Table 1), while the expected heterozygosity (HE) ranged from 0.074 to 0.093, with an average value of 0.088 (Table 1). The observed heterozygosity (HO) in all populations is lower than the expected heterozygosity (HE). This deficiency of HO contributed to inbreeding coefficients (FIS) ranging from 0.068 to 0.431, with an average value of 0.254 (Table 1). We found no statistically significant correlation of population size to either HO or HE (HO: R = 0.1266, p-value = 0.63; HE: R = −0.0033, p-value = 0.99).
Populations displayed pairwise FST values between 0–0.098 (average = 0.020; Table 2) indicating that populations showed little differentiation from one another (FST ≤ 0.05). Although the Pairwise FST range was low, Walton (Avg. FST = 0.055) and PC1 (Avg. FST = 0.063) showed the highest average pairwise FST values among the sampled populations. The rest of the samples showed an average pairwise FST value of less than 0.037. A Mantel test confirmed that genetic distance was significantly correlated with geographic distance (R = 0.5571, p-value = 0.001).

Table 2.
Matrix of pairwise FST values for H. henryae populations with greater than three individuals. The average FST value for each population is indicated. See Table 1 for acronyms.
In PCA analysis, the first three principal components accounted for 5.2% of total variation (Figure 3). The PCA results show one intermixed cluster showing high genetic similarities amongst populations. To further investigate genetic relatedness among populations and confirm these findings, the optimal sNMF model included a single ancestral population (K = 1). This optimal number of ancestral populations (K) was chosen as the lowest cross-entropy value out of 10 K values tested. This finding is consistent with the relatively low genetic differentiation among most individuals as suggested by the PCA analysis. Ancestry matrix and PCA analysis using the 40% and 80% filtered datasets yielded similar results.
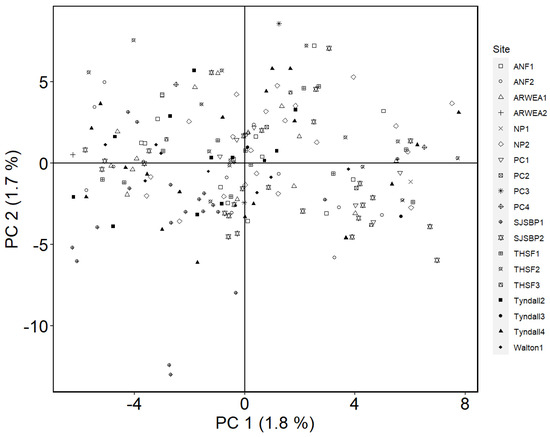
Figure 3.
Principal components analysis for the diversity study. The x-axis 1 represents principal component (PC) one and explains 1.8% of the total variability in the dataset. The second PC explained 1.7% of the variance. The third PC explained 1.7% of the variance, similar to PC2. Each point is an individual plant and symbols indicate different populations (see figure legend). See Table 1 for acronyms.
3.3. Clonal Analysis
We identified an interclonal genetic distance threshold of 0.020 for the plot at ANF1, and 0.015 at ANF2 based on clear gaps in the distribution of pairwise genetic distances (dotted line in Figure 4a,b). We were able to identify clades of individuals on a UPGMA tree with a total genetic distance less than this threshold (Figure 4c,d). Clonal individuals were all found in the same clump (designated a, b, or c in Figure 4); conversely, there were only a few instances where individuals from the same clump were not identified as clones (e.g., 10a and 13b from ANF2). No results support clonal individuals shared between clumps.
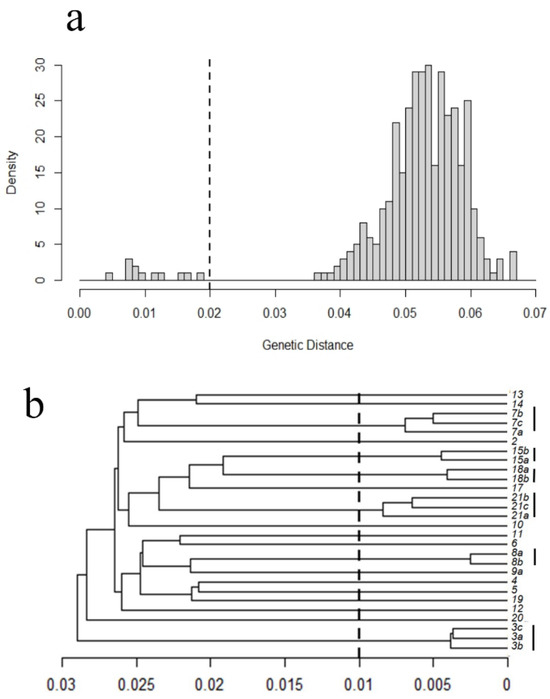
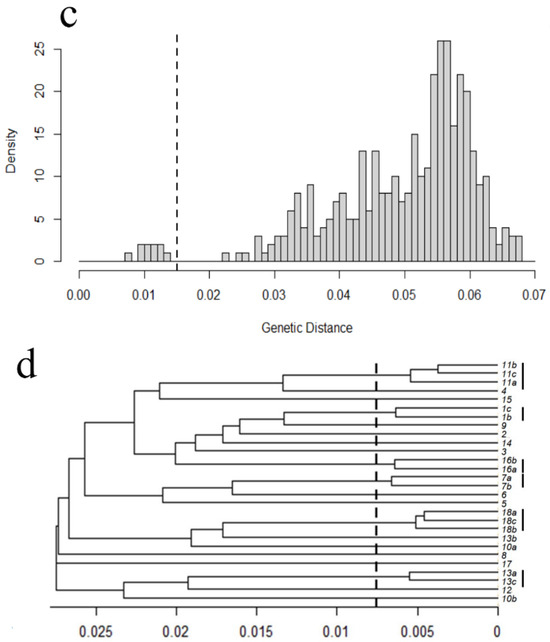
Figure 4.
Clonality assessment for two circular plots, ANF1 (a,b) and ANF2 (c,d): The frequency distribution of pairwise genetic distances among individuals within a test plot (a,c) allows us to determine a genetic divergence threshold (dashed line) that separates putative clones (ramets, to the left of the line) from genetically distinct individuals (genets, to the right of the line). Application of this threshold to UPGMA trees (b,d) that show variation and relatedness among individuals allows us to determine putative clones; those clades that diverge at genetic distances below this threshold are considered clones. In each UPGMA tree, samples taken from the same fused clump of bulbs are indicated by a lowercase letter (e.g., a, b, c after the clump identification number). Clades composed of clonal individuals are indicated by a solid black vertical line to the right of the tree. In almost all cases, clonal individuals belong to the same clump, the exception being 10b and 13b in ANF2 (d). See Table 1 for acronyms.
4. Discussion
Populations at risk from habitat degradation and sea level rise—In 2010, H. henryae was formally petitioned for listing under the ESA based on population declines, limited distribution, habitat loss, and overcollection [23]. Consistent with these threats, we found H. henryae exhibits considerable population size variation across its range, with the majority of observed populations at fewer than 40 individuals. That the largest populations, with hundreds of individuals, are found exclusively on managed lands (ANF, NP, THSF, Tyndall) illustrates the importance of maintaining habitat integrity through prescribed burns, invasive species removal, and hydrological restoration cypress dome swamps and wet prairies for the continued persistence of this species. Indeed, the smallest populations (with fewer than 15 individuals) were all found in degraded habitats, either due to development, overgrowth, or saltwater encroachment as the sea level increases.
According to Sweet et al. [26], the projected sea level rise (SLR) for the next 30 years along the U.S. coast will be 10–12 inches (0.3–0.4 ft per year), on average. Habitats along the coasts will inevitably degrade as saltwater inundates these areas threatening coastal populations of H. henryae. An estimate of a 0–3 ft rise places six coastal populations at risk of saltwater inundation. The impact of this threat is currently unmeasured, yet there is still time to understand the effects of saltwater on this species and if necessary, rescue three at-risk populations before 2070. Because of the lack of population structure, a loss of a few coastal populations using conservative estimates of SLR will likely have little impact on the genetic variation in the species as a whole; however, research on the effects of saltwater intrusion on seed viability and germination would help predict the impact of the future persistence of coastal and coastal-adjacent populations.
Population structure and genetic differentiation—Genetic diversity is a fundamental component of biodiversity conservation and has profound effects on ecological processes including adaptation, community structure, and recovery from disturbance specifically applicable to rare plant populations such as H. henryae [7]. In H. henryae, HE is considerably low (0.037–0.073) similar to other narrowly-endemic or rare species in the region [45,46]. This is lower than the related rare species, H. coronaria (J. LeConte) Kunth, which inhabits river shoals in riparian habitats ranging from Central Alabama through northern Georgia and South Carolina. For H. coronaria, HE ranges between 0.152 and 0.378, as estimated from nuclear microsatellite markers, although this value may be inflated somewhat as H. coronaria is an allotetraploid species [9]. While estimates of genetic diversity using microsatellite markers (H. coronaria) and SNPs (H. henryae) are not directly comparable [47], they do provide some context for understanding the evolutionary potential of H. henryae. The low genetic diversity of H. henryae, combined with its variable and small census size and increased inbreeding coefficients, may lower the species’ capacity to adaptively respond to long-term climatic shifts, while also increasing the risk to demographic and genetic stochasticity [48,49,50].
Furthermore, we found evidence of no population structure across the species range, using both sNMF analysis (K = 1) and genetic PCA. Although the PCA explains only 5.2% of the variation, our results using different filtered datasets are similar. While several studies support the Apalachicola River as a geographic barrier to invertebrates, vertebrates, and plants resulting in genetic discontinuity [51,52], we found no geographically distinct patterns of genetic or morphological variation in H. henryae. This also contrasts findings from H. coronaria, where significant genetic differentiation was found between the western and eastern regions of its extensive range [9,53]. This finding calls into question the taxonomic distinction of H. henryae into two varieties found on either side of the Apalachicola River [18] and suggests H. henryae should be considered a single taxonomic unit without varietal distinctions for listing decisions.
The lack of population structure suggests H. henryae populations share the same ancestral genetic structure, despite their relatively broad geographic distribution across the Florida panhandle. This observation, combined with the low genetic diversity of the species as a whole, is consistent with a past bottleneck followed by population expansion, possibly from wetland refugia established in the last glacial maximum of approximately 9000 ya [9,54]. Retreat into wetland refugia concurrent with population contraction would create a bottleneck effect, reducing the overall diversity of the species; subsequent warming during glacial retreat and expansion of wetland habitat suitable for H. henryae, would promote expansion of the species. The lack of correlation between HE and population size is a characteristic of nonequilibrium population structure and is consistent with past population size changes, including past bottlenecks and subsequent expansion [9,55,56].
Evidence of a single genetic cluster, i.e., lack of population structure, has also been observed in other rare and invasive species [57,58,59,60]. Eserman [58] studied 353 Torreya taxifolia Arn. trees (Taxaceae), a federally endangered conifer endemic to the Florida Panhandle, and demonstrated that genetic diversity is not spatially clustered. However, what differs from our results is that T. taxifolia harbors high genetic diversity. A single genetic cluster and low or no significant genetic differentiation, which is comparable to our results, was reported in the rare snowbed sedge Carex rufina Drejer (Cyperaceae) [60], as well as the invasives Alternanthera philoxeroides (Mart.) Grisb (Amaranthaceae) [59], and Lonicera japonica Thunb. (Caprifoliaceae) [57].
We also found that levels of genetic differentiation between populations were low (FST between 0 and 0.098), with statistical support for isolation by distance across the geographic range of H. henryae spanning more than 240 km. The significance of isolation by distance despite low genetic variance, supports the hypothesis of a range expansion from a single bottlenecked refugia [56]. The alternative hypothesis, that H. henryae represents a panmictic population with few barriers to gene flow between populations is not well supported. The patchy distribution of habitats within the flatwoods restricts gene flow to short distances, spatially limited to within or nearby populations.
The moderate levels of inbreeding found within populations support limited gene flow among populations. While sexual reproduction does occur as evidenced by seed production observed in the field [21], our inbreeding estimates suggest outcrossing is occurring among related individuals within populations, i.e., biparental inbreeding. And while clonal propagation also occurs, it is limited to individuals within a clump connected through the fusion of belowground bulbs; thus, with few exceptions (Figure 4), each clump is a single genet, composed of multiple ramets. While clonal reproduction offers a short-term strategy leading to population growth, and it is a common strategy in members of the Amaryllidaceae (H. coronaria and other related taxa) [9,61,62,63], clonal reproduction in H. henryae is limited to producing vegetative duplicates within a clump. Thus, recruitment of new individuals to populations predictably occurs through locally dispersed seeds, many the products of biparental inbreeding of long-lived individuals (up to 40 years, as seen in H. occidentalis Riddell) [64].
Conservation and Management recommendations—Our results suggest that the majority of small populations of H. henryae are at risk of future decline, due to the combination of low census population size combined with low evolutionary potential associated with overall low levels of genetic diversity and intermediate levels of inbreeding within populations. The overall low genetic diversity of this species specifically makes its long-term survival a concern due to the ongoing changes in land use and climate. Particularly at risk are coastal populations, which are likely to experience habitat encroachment from saltwater inundation due to storm surges from more frequent and powerful hurricanes, as well as climate-change-induced sea level rise. While the loss of a few coastal populations will not necessarily result in a loss of genetic diversity, this is only because of the lack of population structure in the species. Habitat degradation of coastal populations due to the threat of sea level rise, as well as more interior populations due to deficient habitat management, could reduce the redundancy of the species, making it less likely to survive future catastrophic events.
To the extent possible, land managers and decision makers should make use of this genetic study to guide conservation actions. It is essential to continue current habitat maintenance practices performed on managed lands where most of the high census populations are found. For outplanting or reintroduction efforts, germplasm in the form of bulbs and seeds should be collected from across the geographic range of H. henryae and maintained at botanical gardens and other facilities for research, recovery, and public outreach. Although we found no population structure, any efforts to establish new populations (e.g., further inland and less risk to sea level change) or to replace extirpated populations (to maintain redundancy) should be made using germplasm from other populations in the same county, as the importance of local soil microhabitats to seed or bulb growth is unknown. For example, to reintroduce plants to an extirpated population from Eglin AFB, we recommend introducing germplasm from other Walton County populations (Figure 2) rather than Liberty (ANF) or Franklin (Tate’s Hell State Forest) counties. Further research on seed storage, dispersal mechanisms, pollination, and recruitment of H. henryae will aid in determining the ability and methods to safeguard extant populations. As seeds are likely recalcitrant to desiccation and storage methods used in traditional seedbanks [11], cryopreservation of seeds or creating an ex situ collection grown from collected seeds are two options that promote the preservation of at-risk populations and future reintroduction efforts. Therefore, storage experiments should be conducted testing the viability of these seeds by maternal lines under a standard −20 °C (orthodox seeds) and −15 °C (intermediate seeds) after 6 months, 1 year, 5 years, and 10 years [65]. Seeds and plants should be transferred and stored at a seedbank or botanical institution following best practices from the Center for Plant Conservation [65].
Author Contributions
Conceptualization, M.T.V., V.N.-O. and R.C.M.; methodology, M.T.V., V.N.-O. and R.C.M.; validation, M.T.V., V.N.-O. and R.C.M.; formal analysis, M.T.V.; investigation, M.T.V.; resources, V.N.-O. and R.C.M.; data curation, V.N.-O. and R.C.M.; writing—original draft preparation, M.T.V.; writing—review and editing, M.T.V., V.N.-O. and R.C.M.; visualization, M.T.V. and V.N.-O.; supervision, V.N.-O. and R.C.M.; project administration, V.N.-O. and R.C.M.; funding acquisition, M.T.V. and R.C.M. All authors have read and agreed to the published version of the manuscript.
Funding
Financial support was provided under grants USFWS Coordination and Assistance Program Award F20AC10974-00 and USFWS Coastal Program Award F21AC03234-00 to R.C.M., and a Willard Sherman Turrell Herbarium (MU) research award to M.T.V.
Institutional Review Board Statement
The following agencies issued permits for tissue collection: Florida Department of Agriculture and Consumer Services Division of Plant Industry, permit no. #2021-03-002; US Department of Agriculture Forest Service Special Use Permit, permit no. WAK04122020; US Department of Interior Fish and Wildlife Service 10(a)(1)(A) permit to VNO; and Florida Fish And Wildlife Conservation Commission Permit No. SUO-82149, and permission letters from the Nokuse Plantation and Northwest Florida Water Management District.
Data Availability Statement
We are currently in the process of uploading SNP data and vcf files to Dryad (https://doi.org/10.5061/dryad.m63xsj4bb).
Acknowledgments
We thank the following people who helped with field surveys: Jake Rousch, Lydia Ambrose, and Lorainne Ketzler (USFWS PC); Ann Johnson, Jenna Annis, Amy Jenkins, and Camille Eckel (FNAI); Catherine Ricketts (ARWEA); Jason Drake and Brenton Holt (FL FS); Michael R. Jenkins from (FDACS FS); Dylan Shoemaker, Kaylyn Cullen, and Sandra Chafin (SJSBP); Matthew Aresco and Bob Walker (Nokuse Plantation); Tyler McMillan (Northwest Florida Water Management District); Bruce Hagedorn, Melanie Kaeser, and Lisa Keppner (EAFB and TAFB). For lab and technical assistance, we thank William Gregor, Wolfgang Graff, Angelica Vasilatos, Keaka Farleigh, Alfredo Ascanio, Victor Fitzgerald, Ryan Motzko, and Jens Mueller from Miami University, OH, USA. Lauren Eserman-Campbell and Amanda Carmichael (Atlanta Botanical Gardens) provided constructive suggestions for this manuscript. The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the US Fish and Wildlife Service.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Forester, B.R.; Lama, T. The role of genomics in the future of ESA decision-making. In The Codex of the Endangered Species Act: The Next Fifty Years; Baier, L.E., Organ, J.F., Segal, C.E., Eds.; Rowman & Littlefield: Lanham, MD, USA, 2023; Volume 2, pp. 159–186. [Google Scholar] [CrossRef]
- Noss, R.F.; Cartwright, J.M.; Estes, D.; Witsell, T.; Elliott, G.; Adams, D.; Albrecht, M.; Boyles, R.; Comer, P.; Doffitt, C.; et al. Improving species status assessments under the U.S. Endangered Species Act and implications for multispecies conservation challenges worldwide. Conserv. Biol. 2021, 35, 1715–1724. [Google Scholar] [CrossRef]
- Soule, M.E. What is Conservation Biology? A new synthetic discipline addresses the dynamics and problems of perturbed species, communities, and ecosystems. BioScience 1985, 35, 727–734. [Google Scholar] [CrossRef]
- Walther, G.; Post, E.; Convey, P.; Menzel, A.; Parmesank, C.; Beebee, T.J.C.; Fromentin, J.I.O.H.; Bairlein, F. Ecological response to recent climate change. Nature 2002, 416, 389–395. [Google Scholar] [CrossRef]
- Waldvogel, A.M.; Feldmeyer, B.; Rolshausen, G.; Exposito-Alonso, M.; Rellstab, C.; Kofler, R.; Mock, T.; Schmid, K.; Schmitt, I.; Bataillon, T. Evolutionary genomics can improve prediction of species’ responses to climate change. Evol. Lett. 2020, 4, 4–18. [Google Scholar] [CrossRef]
- Soto-Cerda, B.J.; Cloutier, S. Association mapping in plant genomes. In Genetic Diversity in Plants; Çalişkan, M., Ed.; InTech: Rijeka, Croatia, 2012. [Google Scholar] [CrossRef]
- Ottewell, K.M.; Bickerton, D.C.; Byrne, M.; Lowe, A.J. Bridging the gap: A genetic assessment framework for population-level threatened plant conservation prioritization and decision-making. Divers. Distrib. 2016, 22, 174–188. [Google Scholar] [CrossRef]
- Doi, H.; Takahashi, M.; Katano, I. Genetic diversity increases regional variation in phenological dates in response to climate change. Glob. Change Biol. 2010, 16, 373–379. [Google Scholar] [CrossRef]
- Markwith, S.H.; Scanlon, M.J. Multiscale analysis of Hymenocallis coronaria (Amaryllidaceae) genetic diversity, genetic structure, and gene movement under the influence of unidirectional stream flow. Am. J. Bot. 2007, 94, 151–160. [Google Scholar] [CrossRef]
- Smith, G.L.; Flory, W.S. Studies on Hymenocallis henryae (Amaryllidaceae); Springer and New York Botanical Garden Press Stable: Bronx, NY, USA, 1990; Volume 42, pp. 212–220. [Google Scholar] [CrossRef]
- Pacific Bulb Society. (n.d.). How to Grow Bulbs from Seed. Available online: https://www.pacificbulbsociety.org/pbswiki/index.php/HowToGrowBulbsFromSeed (accessed on 21 March 2024).
- Smith, G.L.; Garland, M.A. Nomenclature of Hymenocallis taxa (Amaryllidaceae) in southeastern United States. Taxon 2023, 52, 805–817. [Google Scholar] [CrossRef]
- Florida Natural Areas Inventory (FNAI). Available online: https://www.fnai.org/publications/data-requests (accessed on 10 November 2020).
- Wunderlin, R.P.; Hansen, B.F. Guide to the Vascular Plants of Florida, 3rd ed.; University Press of Florida: Tampa, FL, USA, 2011; Volume 3, p. 800. [Google Scholar]
- Wunderlin, R.P.; Hansen, B.F.; Franck, A.R.; Essig, F.B. Atlas of Florida Plants; Landry, S.M., Campbell, K.N., Eds.; USF Water Institute, Institute for Systematic Botany, University of South Florida: Tampa, FL, USA, 2024. [Google Scholar]
- Volk, M.I.; Hoctor, T.S.; Nettles, B.B.; Hilsenbeck, R.; Putz, F.E.; Oetting, J. Florida Land Use and Land Cover Change in the Past 100 Years. Florida’s Clim. Changes Var. Impacts 2017, 2, 51–82. [Google Scholar] [CrossRef]
- Noss, R.; Platt, W.; Sorrie, B.; Weakley, A.; Means, D.; Costanza, J.; Peet, R. How global biodiversity hotspots may go unrecognized: Lessons from the North American Coastal Plain. Divers. Distrib. 2015, 21, 236–244. [Google Scholar] [CrossRef]
- Blaustein, R.J. Biodiversity Hotspot: The Florida Panhandle. BioScience 2008, 58, 784–790. [Google Scholar] [CrossRef]
- NatureServe Explorer. Available online: https://explorer.natureserve.org/ (accessed on 12 November 2020).
- Florida Natural Areas Inventory. Available online: https://www.fnai.org/PDFs/FieldGuides/Hymenocallis_henryae.pdf (accessed on 10 April 2023).
- Vogel, M.T. Conservation of the Rare Florida Henry’s Spider Lily (Hymenocallis henryae) Using Genomic Analysis. Master’s Thesis, Miami University, Oxford, OH, USA, December 2022. Available online: http://rave.ohiolink.edu/etdc/view?acc_num=miami1668782065354342 (accessed on 12 May 2023).
- Kral, R. A Report on Some Rare, Threatened, or Endangered Forest-Related Vascular Plants of the South; U.S. Dept. of Agriculture Forest Service Technical Publication R8-TP2: Athens, GA, USA, 1983; p. 1305. [Google Scholar]
- Center for Biological Diversity (CBD). Southeast Aquatic Species Petition. In Petition to List 404 Aquatic, Riparian and Wetland Species from the Southeastern United States as Threatened or Endangered under the Endangered Species Act; CBD: Tucson, Arizona, 2010; pp. 614–615. [Google Scholar]
- 16 U.S. Code § 1531—Congressional Findings and Declaration of Purposes and Policy. Available online: https://www.law.cornell.edu/uscode/text/16/1531 (accessed on 20 July 2023).
- Traub, H.P. Specimen: Hymenocallis Henryae—282—MO—(BC:MO-202275/A:3108358) (BC:MO-202276/A:3108359). Tropicos. 1962. Available online: https://www.tropicos.org/specimen/1784859 (accessed on 24 July 2024).
- Sweet, W.V.; Hamlington, B.D.; Kopp, R.E.; Weaver, C.P.; Barnard, P.L.; Bekaert, D.; Brooks, W.; Craghan, M.; Dusek, G.; Frederikse, T.; et al. Global and Regional Sea Level Rise Scenarios for the United States: Updated Mean Projections and Extreme Water Level Probabilities Along U.S. Coastlines. NOAA Technical Report NOS 01. National Oceanic and Atmospheric Administration, National Ocean Service. 2022; p. 111. Available online: https://cdn.oceanservice.noaa.gov/oceanserviceprod/hazards/sealevelrise/noaa-nos-techrpt01-global-regional-SLR-scenarios-US.pdf (accessed on 5 May 2023).
- Bad Elf. GNSS Surveyor BE-3300GPS; Bad Elf.: West Hartford, CT, USA, 2020. [Google Scholar]
- Kim, C.S.; Lee, C.H.; Shin, J.S.; Chung, Y.S.; Hyung, N.I. A simple and rapid method for isolation of high quality genomic DNA from fruit trees and conifers using PVP. Nucleic Acids Res. 1997, 25, 1085–1086. [Google Scholar] [CrossRef]
- Thermo Fisher Scientific. Qubit 4 Fluorometer (Catalog No. Q33238); Thermo Fisher Scientific: Waltham, MA, USA, 2021. Available online: https://www.thermofisher.com/order/catalog/product/Q33238 (accessed on 5 May 2023).
- SBG/dd-RAD-Seq: Floragenex: Your Partner from DNA to Data. Available online: https://www.floragenex.com/sbg-ddrad-seq (accessed on 24 September 2020).
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double Digest RADseq: An Inexpensive Method for De Novo SNP Discovery and Genotyping in Model and Non-Model Species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef]
- Catchen, J.; Hohenlohe, P.; Bassham, S.; Amores, A.; Cresko, W. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 11, 3124–3140. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; McVean, G. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 5 May 2023).
- Microsoft Corporation. Microsoft Excel. 2018. Available online: https://office.microsoft.com/excel (accessed on 5 May 2023).
- Thioulouse, J.; Dray, S.; Dufour, A.; Siberchicot, A.; Jombart, T.; Pavoine, S. Multivariate Analysis of Ecological Data with Ade4; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar] [CrossRef]
- Frichot, E.; Francois, O. LEA: An R package for Landscape and Ecological Association studies. Methods Ecol. Evol. 2015, 6, 925–929. Available online: http://membres-timc.imag.fr/Olivier.Francois/lea.html (accessed on 5 May 2023). [CrossRef]
- Knaus, B.J.; Grunwald, N.J. VCFR: A package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 2017, 17, 44–53. [Google Scholar] [CrossRef]
- Pembleton, L.; Cogan, N.; Forster, J. StAMPP: An R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol. Ecol. Resour. 2013, 13, 946–952. [Google Scholar] [CrossRef]
- Willing, E.-M.; Dreyer, C.; van Oosterhout, C. Estimates of Genetic Differentiation Measured by FST Do Not Necessarily Require Large Sample Sizes When Using Many SNP Markers. PLoS ONE 2012, 7, e42649. [Google Scholar] [CrossRef]
- Esri Inc. ArcGIS Pro (Version 3.0). Esri Inc. 2022. Available online: https://www.esri.com/en-us/arcgis/products/arcgis-pro/overview (accessed on 5 May 2023).
- Kamvar, Z.N.; Brooks, J.C.; Grünwald, N.J. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015, 6, 208. [Google Scholar] [CrossRef]
- Amor, M.D.; Johnson, J.C.; James, E.A. Identification of clonemates and genetic lineages using next-generation sequencing (ddRADseq) guides conservation of a rare species, Bossiaea vombata (Fabaceae). Perspect. Plant Ecol. Evol. Syst. 2020, 45, 125544. [Google Scholar] [CrossRef]
- Schliep; Klaus; Potts, J.A.; Morrison, A.D.; Grimm, W.G. Intertwining phylogenetic trees and networks. Methods Ecol. Evol. 2017, 8, 1212–1220. [Google Scholar] [CrossRef]
- Menges, E.; Dolan, R.; Yahr, R.; Gordon, D. Comparative genetics of seven plants endemic to Florida’s Lake Wales Ridge. Scholarsh. Prof. Work. LAS 2001, 66, 98–114. [Google Scholar]
- Gitzendanner, M.A.; Soltis, P.S. Patterns of genetic variation in rare and widespread plant congeners. Am. Jor. Bot. 2000, 87, 783–792. [Google Scholar] [CrossRef]
- Fischer, M.C.; Rellstab, C.; Leuzinger, M.; Roumet, M.; Gugerli, F.; Shimizu, K.K.; Holderegger, R.; Widmer, A. Estimating genomic diversity and population differentiation—An empirical, of microsatellite and SNP variation in Arabidopsis halleri. BMC Genom. 2017, 18, 69. [Google Scholar] [CrossRef] [PubMed]
- Leimu, R.; Mutikainen, P.; Koricheva, J.; Fischer, M. How general are positive relationships between plant population size, fitness and genetic variation? J. Ecol. 2006, 94, 942–952. [Google Scholar] [CrossRef]
- Honnay, O.; Jacquemyn, H. Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conserv. Biol. 2007, 21, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Willi, Y.; Kristensen, T.N.; Sgrò, C.M.; Weeks, A.R.; Ørsted, M.; Hoffmann, A.A. Conservation genetics as a management tool: The five best-supported paradigms to assist the management of threatened species. Proc. Natl. Acad. Sci. USA 2022, 119, e2105076119. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Morris, A.B.; McLachlan, J.S.; Manos, P.S.; Soltis, P.S. Comparative phylogeography of unglaciated eastern North America. Mol. Ecol. 2006, 14, 671–688. [Google Scholar] [CrossRef]
- Stephens, J.D.; Santos, S.R.; Folkerts, D.R. Genetic differentiation, structure, and a transition zone among populations of the pitcher plant moth Exyra semicrocea: Implications for conservation. PLoS ONE 2011, 6, e22658. [Google Scholar] [CrossRef]
- Meerow, A.W.; Gardner, E.M.; Nakamura, K. Phylogenomics of the Andean tetraploid clade of the American Amaryllidaceae (subfamily Amaryllidoideae): Unlocking a polyploid generic radiation abetted by continental geodynamics. Front. Plant Sci. 2020, 11, 582422. [Google Scholar] [CrossRef]
- Watts, W.A. Late-Quanternary vegetation history at White Pond on the inner coastal plain of South Carolina. Quat. Res. 1980, 13, 187–199. [Google Scholar] [CrossRef]
- Schmidt, K.; Jensen, K. Genetic structure and AFLP variation of remnant populations in the rare plant Pedicularis palustris (Scrophulariaceae) and its relation to population size and reproductive components. Am. J. Bot. 2000, 87, 678–689. [Google Scholar] [CrossRef]
- Tero, N.; Aspi, J.; Siikamäki, P.; Jäkäläniemi, A.; Tuomi, J. Genetic structure and gene flow in a metapopulation of an endangered plant species, Silene tatarica. Mol. Ecol. 2003, 12, 2073–2085. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.F.; Corbett, C.W.; Thixton-Nolan, H.L. A lack of population structure characterizes the invasive Lonicera japonica in West Virginia and across eastern North America. J. Torrey Bot Soc. 2023, 150, 455–466. [Google Scholar] [CrossRef]
- Eserman-Campbell, L. Evaluating the conservation genetics of Florida torreya. Synecology 2022, 4, 8–9. [Google Scholar]
- Ye, W.H.J.; Li, H.L.; Cao, H.L.; Ge, X.J. Genetic uniformity of Alternanthera philoxeroides in South China. Weed Res. 2003, 43, 297–302. [Google Scholar] [CrossRef]
- Westergaard, K.B.; Alsos, I.G.; Engelskjøn, T.; Flatberg, K.I.; Brochmann, C. Trans-Atlantic genetic uniformity in the rare snowbed sedge Carex rufina. Conserv. Genet. 2011, 12, 1367–1371. [Google Scholar] [CrossRef]
- Duchoslav, M.; Staňková, H. Population genetic structure and clonal diversity of Allium oleraceum (Amaryllidaceae), a polyploid geophyte with common asexual but variable sexual reproduction. Folia Geobot. 2015, 50, 123–136. [Google Scholar] [CrossRef]
- Gaikwad, S.P.; Garad, K.U.; Gore, R.D. Crinum solapurense (Amaryllidaceae), a new species from Maharashtra, India. Kew Bull. 2014, 69, 9505. [Google Scholar] [CrossRef]
- Arroyo, J.; Dafni, A. Variations in Habitat, Season, Flower Traits and Pollinators in Dimorphic Narcissus tazetta L. (Amaryllidaceae) in Israel. New Phytol. 1995, 129, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Hymenocallis occidentalis var. Occidentalis—Flora of North America. Available online: http://beta.floranorthamerica.org/Hymenocallis_occidentalis_var._occidentalis (accessed on 5 December 2020).
- Center for Plant Conservation. CPC Best Plant Conservation Practices to Support Species Survival in the Wild; Center for Plant Conservation: Escondido, CA, USA, 2019. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).