Karyological Study of Acanthocephalus lucii (Echinorhynchida): The Occurrence of B Chromosomes in Populations from PCB-Polluted Waters
Abstract
1. Introduction
2. Material and Methods
2.1. Study Sites and Parasite Collection
2.2. Chromosome Analysis
2.3. DNA Extraction, PCR, Cloning and Preparation of 18S rDNA and H3 Histone Probes
2.4. Fluorescence In Situ Hybridization (FISH)
2.5. Microscopy and Image Processing
3. Results
3.1. Karyotype of Acanthocephalus lucii
3.2. Distribution of Ribosomal RNA Genes (rDNA), H3 Histone Genes, and Heterochromatin Blocks
3.3. Meiotic Spermatogenesis and Oogenesis
4. Discussion
4.1. Standard Chromosome Complement
4.2. Supernumerary B Chromosomes and Environmental Pollution?
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Šalgovičová, D.; Zmetáková, Z. Polychorinated Biphenyls in Muscle Tissue of Freshwater Fish in East Slovakia. J. Food Nutr. Res. 2006, 45, 171–178. [Google Scholar]
- Sures, B.; Nachev, M.; Selbach, C.; Marcogliese, D.J. Parasite Responses to Pollution: What We Know and Where We Go in ‘Environmental Parasitology’. Parasite Vector 2017, 10, 65. [Google Scholar] [CrossRef]
- Duarte, G.S.C.; Lehun, A.L.; Leite, L.A.R.; Consolin-Filho, N.; Bellay, S.; Takemoto, R.M. Acanthocephalans Parasites of Two Characiformes Fishes as Bioindicators of Cadmium Contamination in Two Neotropical Rivers in Brazil. Sci. Total Environ. 2020, 738, 140339. [Google Scholar] [CrossRef]
- Nachev, M.; Rozdina, D.; Michler-Kozma, D.N.; Raikova, G.; Sures, B. Metal Accumulation in Ecto- and Endoparasites from the Anadromous Fish, the Pontic Shad (Alosa immaculata). Parasitology 2022, 149, 496–502. [Google Scholar] [CrossRef]
- Mille, T.; Soulier, L.; Caill-Milly, N.; Cresson, P.; Morandeau, G.; Monperrus, M. Differential Micropollutants Bioaccumulation in European Hake and Their Parasites Anisakis sp. Environ. Pollut. 2020, 265, 115021. [Google Scholar] [CrossRef]
- Brázová, T.; Miklisová, D.; Barčák, D.; Uhrovič, D.; Šalamún, P.; Orosová, M.; Oros, M. Hazardous Pollutants in the Environment: Fish Host-Parasite Interactions and Bioaccumulation of Polychlorinated Biphenyls. Environ. Pollut. 2021, 291, 118175. [Google Scholar] [CrossRef]
- Oros, M.; Barčák, D.; Miklisová, D.; Uhrovič, D.; Brázová, T. A Fish-Parasite Sentinel System in an Assessment of the Spatial Distribution of Polychlorinated Biphenyls. Sci. Rep. 2023, 13, 5164. [Google Scholar] [CrossRef]
- Goutte, A.; Molbert, N. Benefits of Parasitism in Polluted Environments: A Review and Perspectives. Front. Ecol. Evol. 2022, 10, 847869. [Google Scholar] [CrossRef]
- Orosová, M.; Marková, A.; Marec, F.; Barčák, D.; Brázová, T.; Oros, M. New Cytogenetic Data on Caryophyllaeus laticeps and Paracaryophyllaeus gotoi, Parasites of Evolutionary Interest. Parasitology 2022, 149, 1094–1105. [Google Scholar] [CrossRef] [PubMed]
- Špakulová, M.; Kráľová-Hromadová, I.; Dudiňák, V.; Reddy, P.V. Karyotype of Acanthocephalus lucii: The First Record of Supernumerary Chromosomes in Thorny-Headed Worms. Parasitol. Res. 2002, 8, 778–780. [Google Scholar] [CrossRef]
- Turčeková, L.; Hanzelová, V.; Špakulová, M. Concentration of Heavy Metals in Perch and Its Endoparasites in the Polluted Water Reservoir in Eastern Slovakia. Helminthologia 2002, 39, 76–80. [Google Scholar]
- Orosová, M.; Marková, A.; Zrzavá, M.; Marec, F.; Oros, M. Chromosome Analysis and the Occurrence of B Chromosomes in Fish Parasite Acanthocephalus anguillae (Palaeacanthocephala: Echinorhynchida). Parasite 2023, 30, 44. [Google Scholar] [CrossRef] [PubMed]
- Kushlan, J. Colonial Waterbirds as Bioindicators of Environmental Change. Colon. Waterbirds 1993, 16, 223–251. [Google Scholar] [CrossRef]
- Binelli, A.; Della Torre, C.; Magni, S.; Parolini, M. Does Zebra mussel (Dreissena polymorpha) Represent the Freshwater Counterpart of Mytilus in Ecotoxicological Studies? A Critical Review. Environ. Pollut. 2015, 196, 386–403. [Google Scholar] [CrossRef]
- Calapoğlu, M.; Sevinç, Z.; Toğay, A.; Kalyoncu, H. Evaluation of Oxidative Stress and Genotoxicity for Environmental Monitoring Using Farmed Rainbow Trout. Fresenius Environ. Bull. 2017, 26, 7105–7113. [Google Scholar]
- Kamollerd, C.; Senaphan, K.; Tengjaroenkul, B.; Monkheang, P.; Neeratanaphan, L. Oxidative Stress and Genetic Differentiation in Experimental Tilapia Fish Exposed to Heavy Metals in a Reservoir Near a Municipal Landfill. Appl. Ecol. Environ. Res. 2019, 17, 12893–12907. [Google Scholar] [CrossRef]
- Neeratanaphan, L.; Kanjanakunti, A.; Intamat, S.; Tengjaroenkul, B. Analysis of Chromosome Abnormalities in the Asian Swamp Eel (Monopterus albus) Affected by Arsenic Contamination Near a Gold Mine Area. Int. J. Environ. Stud. 2020, 77, 815–829. [Google Scholar] [CrossRef]
- Soulivongsa, L.; Tengjaroenkul, B.; Neeratanaphan, L. Effects of Contamination by Heavy Metals and Metalloids on Chromosomes, Serum Biochemistry and Histopathology of the Bonylip Barb Fish Near Sepon Gold-Copper Mine, Lao PDR. Int. J. Environ. Res. Public Health 2020, 17, 9492. [Google Scholar] [CrossRef]
- Šebelová, Š.; Kuperman, B.; Gelnar, M. Abnormalities of the Attachment Clamps of Representatives of the Family Diplozoidae. J. Helminthol. 2002, 76, 249–259. [Google Scholar] [CrossRef]
- Pečínková, M.; Vøllestad, L.A.; Koubková, B.; Huml, J.; Jurajda, P.; Gelnar, M. The Relationship Between Developmental Instability of Gudgeon Gobio gobio and Abundance or Morphology of Its Ectoparasite Paradiplozoon homoion (Monogenea). J. Fish Biol. 2007, 71, 1358–1370. [Google Scholar] [CrossRef]
- Brázová, T.; Orosová, M.; Šalamún, P.; Hanzelová, V. Morphological Abnormalities in Fish Parasites: A Potential Tool for Biomonitoring Natural Contaminants? Parasitol. Res. 2020, 119, 3297–3304. [Google Scholar] [CrossRef]
- Camacho, J.P. B Chromosomes. In The Evolution of the Genome; Gregory, T.R., Ed.; Academic Press: Cambridge, MA, USA, 2005. [Google Scholar] [CrossRef]
- Johnson Pokorná, M.; Reifová, R. Evolution of B Chromosomes: From Dispensable Parasitic Chromosomes to Essential Genomic Players. Front. Genet. 2021, 12, 727570. [Google Scholar] [CrossRef] [PubMed]
- Milani, D.; Ruiz-Ruano, F.J.; Camacho, J.P.M.; Cabral-de-Mello, D.C. Out of Patterns, the Euchromatic B Chromosome of the Grasshopper Abracris flavolineata is not Enriched in High-copy Repeats. Heredity 2021, 127, 475–483. [Google Scholar] [CrossRef]
- Houben, A.; Banaei-Moghaddam, A.M.; Klemme, S.; Timmis, J.N. Evolution and Biology of Supernumerary B Chromosomes. Cell. Mol. Life Sci. 2014, 71, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Dalla Benneta, E.; Antoshechkin, I.; Yang, T.; Nguyen, H.Q.M.; Ferree, P.M.; Akbari, O.S. Genome Elimination Mediated by Gene Expression from a Selfish Chromosome. Sci. Adv. 2020, 6, eaaz980. [Google Scholar] [CrossRef] [PubMed]
- Perrot-Minnot, M.-J.; Cozzarolo, C.-S.; Amin, O.; Barčák, D.; Bauer, A.; García-Varela, M.; Hernández-Orts, J.S.; Le, T.T.Y.; Nachev, M.; Orosová, M.; et al. Hooking the Scientific Community on Thorny-Headed Worms: Interesting and Exciting Facts, Knowledge Gaps and Perspectives for Research Directions on Acanthocephala. Parasite 2023, 30, 23. [Google Scholar] [CrossRef]
- Brázová, T.; Hanzelová, V.; Miklisová, D. Bioaccumulation of Six PCB Indicator Congeners in a Heavily Polluted Water Reservoir in Eastern Slovakia: Tissue-Specific Distribution in Fish and their Parasites. Parasitol. Res. 2012, 111, 779–786. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; Oliveira, S.G.; de Moura, R.C.; Martins, C. Chromosomal Organization of the 18S and 5S rRNAs and Histone H3 Genes in Scarabaeinae coleopterans: Insights into the Evolutionary Dynamics of Multigene Families and Heterochromatin. BMC Genet. 2011, 12, 88. [Google Scholar] [CrossRef]
- Provazníková, I.; Hejníčková, M.; Visser, S.; Dalíková, M.; Paladino, L.Z.C.; Zrzavá, M.; Voleníková, A.; Marec, F.; Nguyen, P. Large-Scale Comparative Analysis of Cytogenetic Markers Across Lepidoptera. Sci. Rep. 2021, 11, 12214. [Google Scholar] [CrossRef]
- Bombarová, M.; Marec, F.; Nguyen, P.; Špakulová, M. Divergent Location of Ribosomal Genes in Chromosomes of Fish Thorny-Headed Worms, Pomphorhynchus laevis and Pomphorhynchus tereticollis (Acanthocephala). Genetica 2007, 131, 141–149. [Google Scholar] [CrossRef]
- Lisitsyna, O. Fauna of Ukraine, Vol 31. Acanthocephala; Naukova Dumka: Kiev, Ukraine, 2019; p. 223. [Google Scholar]
- Orosová, M.; Špakulová, M. Tapeworms Chromosomes: Their Value in Systematics with Instructions for Cytogenetic Study. Folia Parasitol. 2018, 65, 001. [Google Scholar] [CrossRef]
- Dos Santos Guerra, M. Reviewing the Chromosome Nomenclature of Levan et al. Rev. Bras. Genet. 1986, 9, 741–743. [Google Scholar]
- Orosová, M.; Marková, A.; Provazníková, I.; Oros, M.; Radačovská, A.; Čadková, Z.; Marec, F. Molecular Cytogenetic Analysis of a Triploid Population of the Human Broad Tapeworm, Dibothriocephalus latus (Diphyllobothriidea). Parasitology 2021, 148, 787–797. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; Zrzavá, M.; Kubíčková, S.; Rendón, P.; Marec, F. The Role of Satellite DNAs in Genome Architecture and Sex Chromosome Evolution in Crambidae Moths. Front. Genet. 2021, 12, 661417. [Google Scholar] [CrossRef]
- Garey, J.R.; Near, T.J.; Nonnemacher, M.R.; Nadler, S.A. Molecular Evidence for Acanthocephala as a Subtaxon of Rotifera. J. Mol. Evol. 1996, 43, 287–292. [Google Scholar] [CrossRef]
- Hejníčková, M.; Koutecký, P.; Potocký, P.; Provazníková, I.; Voleníková, A.; Dalíková, M.; Visser, S.; Marec, F.; Zrzavá, M. Absence of W Chromosome in Psychidae Moths and Implications for the Theory of Sex Chromosome Evolution in Lepidoptera. Genes 2019, 10, 1016. [Google Scholar] [CrossRef] [PubMed]
- Cabrero, J.; López-León, M.; Teruel, M.; Camacho, J.P.M. Chromosome Mapping of H3 and H4 Histone Gene Clusters in 35 Species of Acridid Grasshoppers. Chromosome Res. 2009, 17, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Orosová, M.; Marec, F.; Oros, M.; Xi, B.W.; Scholz, T. A Chromosome Study and Localization of 18S rDNA in Khawia saurogobii (Cestoda: Caryophyllidea). Parasitol. Res. 2010, 106, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Mutafova, T.; Nedeva, I.; Kanev, I. Chromosomes of Acanthocephalus lucii. J. Helminthol. 1997, 71, 261–262. [Google Scholar] [CrossRef]
- John, B. Chromosomes of Zooparasites I. Acanthocephalus ranae (Acanthocephala: Echinorhynchidae). Chromosoma 1957, 8, 730–738. [Google Scholar] [PubMed]
- Yoshida, K.; Kitano, J. Tempo and Mode in Karyotype Evolution Revealed by a Probabilistic Model Incorporating Both Chromosome Number and Morphology. PLoS Genet. 2021, 17, e1009502. [Google Scholar] [CrossRef]
- Hirai, H.; Taguchi, T.; Saitoh, M.; Kawanaka, M.; Sugiyama, H.; Habe, S. Chromosomal Differentiation of the Schistosoma japonicum Complex. Int. J. Parasitol. 2000, 30, 441–452. [Google Scholar] [CrossRef]
- Anantaraman, S.; Subramoniam, T. Oogenesis in Acanthosentis oligospinus nsp., an Aganthocephalan Parasite of the Fish, Macrones gulio. Proc. Indian Acad. Sci. 1975, 82B, 139–145. [Google Scholar] [CrossRef]
- Atkinson, K.H.; Byram, J.E. The Structure of the Ovarian Ball and Oogenesis in Moniliforrnis dubius (Acanthocephala). J. Morphol. 1976, 148, 391426. [Google Scholar]
- Parenti, U.; Antoniotti, M.L. Oogenesis in Echinorhynchus truttae (Schrank). Nature 1967, 58, 89–91. [Google Scholar]
- Sochorová, J.; Gálvez, F.; Matyášek, R.; Garcia, S.; Kovařík, A. Analyses of the Updated “Animal rDNA Loci Database” with an Emphasis on Its New Features. Int. J. Mol. Sci. 2021, 22, 11403. [Google Scholar] [CrossRef]
- Dubcovsky, J.; Dvořák, J. Ribosomal RNA Multigene Loci: Nomads of the Triticeae genomes. Genetics 1995, 140, 1367–1377. [Google Scholar] [CrossRef]
- Castro, J.; Rodríguez, S.; Pardo, B.G.; Sánchez, L.; Martínez, P. Population Analysis of an Unusual NOR-site Polymorphism in Brown Trout (Salmo trutta L.). Heredity 2001, 86, 291–302. [Google Scholar] [CrossRef]
- Lisitsyna, O.; Barčák, D.; Orosová, M.; Fan, C.K.; Oros, M. Acanthocephalans of Marine and Freshwater Fishes from Taiwan with Description of a New Species. Folia Parasitol. 2023, 70, 021. [Google Scholar] [CrossRef]
- Mason, J.M.; Reddy, H.M.; Capkova Frydrychova, R. Telomere Maintenance in Organisms without Telomerase. In DNA Replication-Current Advances; Seligmann, H., Ed.; InTech: Rijeka, Croatia, 2011. [Google Scholar] [CrossRef]
- Robertson, L.W.; Ludewig, G. Polychlorinated Biphenyl (PCB) Carcinogenicity with Special Emphasis on Airborne PCBs. Gefahrstoffe Reinhalt. Luft 2011, 71, 25–32. [Google Scholar]
- Yang, F.; Wilcox, B.; Jin, S.; Aguirre, A.A.; Rougée, L.; Xu, L.; Lu, Y. Detection and Quantitative Analysis of Polychlorinated Biphenyls in Tilapia from Hawaiian Waters. Chemosphere 2008, 73, 133–137. [Google Scholar] [CrossRef]
- Wójcik, E.; Smalec, E. The Effect of Environmental Factors on Sister Chromatid Exchange Incidence in Domestic Horse (Equus caballus) Chromosomes. Folia Biol. 2013, 61, 199–204. [Google Scholar] [CrossRef]
- Oliveira, N.L.; Cabral-de-Mello, D.C.; Rocha, M.F.; Loreto, V.; Martins, C.; Moura, R.C. Chromosomal Mapping of rDNAs and H3 Histone Sequences in the Grasshopper Rhammatocerus brasiliensis (Acrididae, Gomphocerinae): Extensive Chromosomal Dispersion and Co-localization of 5S rDNA/H3 Histone Clusters in the A Complement and B Chromosome. Mol. Cytogenet. 2011, 4, 24. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.M.Z.d.A.; Pansonato-Alves, J.C.; Utsunomia, R.; Araya-Jaime, C.; Ruiz-Ruano, F.J.; Daniel, S.N.; Hashimoto, D.T.; Oliveira, C.; Camacho, J.P.M.; Porto-Foresti, F.; et al. Delimiting the Origin of a B Chromosome by FISH Mapping, Chromosome Painting and DNA Sequence Analysis in Astyanax paranae (Teleostei, Characiformes). PLoS ONE 2014, 9, e94896. [Google Scholar] [CrossRef] [PubMed]
- Felicetti, D.; Haerter, C.A.G.; Baumgärtner, L.; Paiz, L.M.; Takagui, F.H.; Margarido, V.P.; Blanco, D.R.; Feldberg, E.; da Silva, M.; Lui, R.L. A New Variant B Chromosome in Auchenipteridae: The Role of (GATA)n and (TTAGGG)n Sequences in Understanding the Evolution of Supernumeraries in Trachelyopterus. Cytogenet. Genome Res. 2021, 161, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Stornioli, J.H.F.; Goes, C.A.G.; Calegari, R.M.; dos Santos, R.Z.; Giglio, L.M.; Foresti, F.; Oliveira, C.; Penitente, M.; Porto-Foresti, F.; Utsunomia, R. The B Chromosomes of Prochilodus lineatus (Teleostei, Characiformes) are Highly Enriched in Satellite DNAs. Cells 2021, 10, 1527. [Google Scholar] [CrossRef] [PubMed]
- Dhar, M.K.; Friebe, B.; Koul, A.K.; Gill, B.S. Origin of an Apparent B Chromosome by Mutation, Chromosome Fragmentation and Specific DNA Sequence Amplification. Chromosoma 2002, 111, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Loreto, V.; Cabrero, J.; López-León, M.D.; Camacho, J.P.M.; Souza, M.J. Possible Autosomal Origin of Macro B Chromosomes in Two Grasshopper Species. Chromosome Res. 2008, 16, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Teruel, M.; Cabrero, J.; Perfectti, F.; Camacho, J.P. B Chromosome Ancestry Revealed by Histone Genes in the Migratory Locust. Chromosoma 2010, 119, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Bueno, D.; Palacios-Gimenez, O.M.; Cabral-de-Mello, D.C. Chromosomal Mapping of Repetitive DNAs in the Grasshopper Abracris flavolineata Reveal Possible Ancestry of the B Chromosome and H3 Histone Spreading. PLoS ONE 2013, 8, e66532. [Google Scholar] [CrossRef]
- Volkova, N.V.; Meier, B.; Gonzalez-Huici, V.; Bertolini, S.; Gonzalez, S.; Vohringer, H.; Abascal, F.; Martincorena, I.; Campbell, P.J.; Gartner, A.; et al. Mutational Signatures Are Jointly Shaped by DNA Damage and Repair. Nat. Commun. 2020, 11, 2169–2183. [Google Scholar] [CrossRef]
- So, C.C.; Martin, A. DSB Structure Impacts DNA Recombination Leading to Class Switching and Chromosomal Translocations in Human B cells. PLoS Genet. 2019, 15, e1008101. [Google Scholar] [CrossRef]
- Rassool, F.V.; Tomkinson, A.E. Targeting Abnormal DNA Double Strand Break Repair in Cancer. Cell. Mol. Life Sci. 2010, 67, 3699–3710. [Google Scholar] [CrossRef]
- Baršiené, J. The Chromosome Sets of Trematodes. Parazitologiya 1993, 27, 336–353. [Google Scholar]
- Petkevičiūtė, R.; Baršiené, J. The Comparative Karyological Analysis of Three Species of Trematodes of Genus Notocotylus. Parazitologiya 1988, 22, 21–28. [Google Scholar]
- Baršiené, J.; Kisilienė, V. Karyological Studies of Trematodes within the Genus Echinostoma. Acta Parasitol. 1991, 36, 23–29. [Google Scholar]
- Feldberg, E.; Porto, J.I.R.; Alves-Brinn, M.N.; Mendonça, M.N.C.; Benzaquem, D.C. B Chromosomes in Amazonian Cichlid Species. Genetics 2004, 106, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Pires, L.B.; Sampaio, T.R.; Dias, A.L. Mitotic and Meiotic Behavior of B Chromosomes in Crenicichla lepidota: New Report in the Family Cichlidae. J. Hered. 2015, 106, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Perazzo, G.X.; Noleto, R.B.; Vicari, M.R.; Gava, A.; Cestari, M.M. B Chromosome Polymorphism in South American Cichlid. Neotrop. Biodivers. 2018, 4, 3–9. [Google Scholar] [CrossRef]
- de Campos Júnior, E.O.; Pereira, B.B.; Morelli, S.; Pavanin, E.V.; Pavanin, L.A. Biological Monitoring and B Chromosome Frequency in Bagre (Rhamdia quelen) in Southeast Brazil. Environ. Toxicol. Phar. 2014, 38, 510–517. [Google Scholar] [CrossRef]
- Borisov, Y.M.; Abramov, S.A.; Borisova, M.Y.; Zhigarev, I.A. The Occurrence of Dot-Like Micro B Chromosomes in Korean Field Mice Apodemus peninsulae from the Shore of the Teletskoye Lake (Altai Mountains). Comp. Cytogenet. 2020, 14, 97–105. [Google Scholar] [CrossRef] [PubMed]

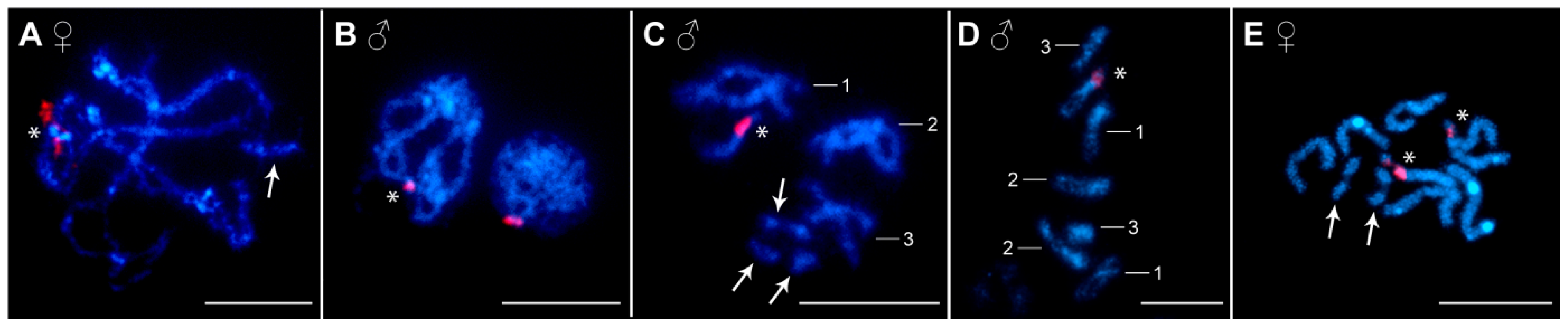
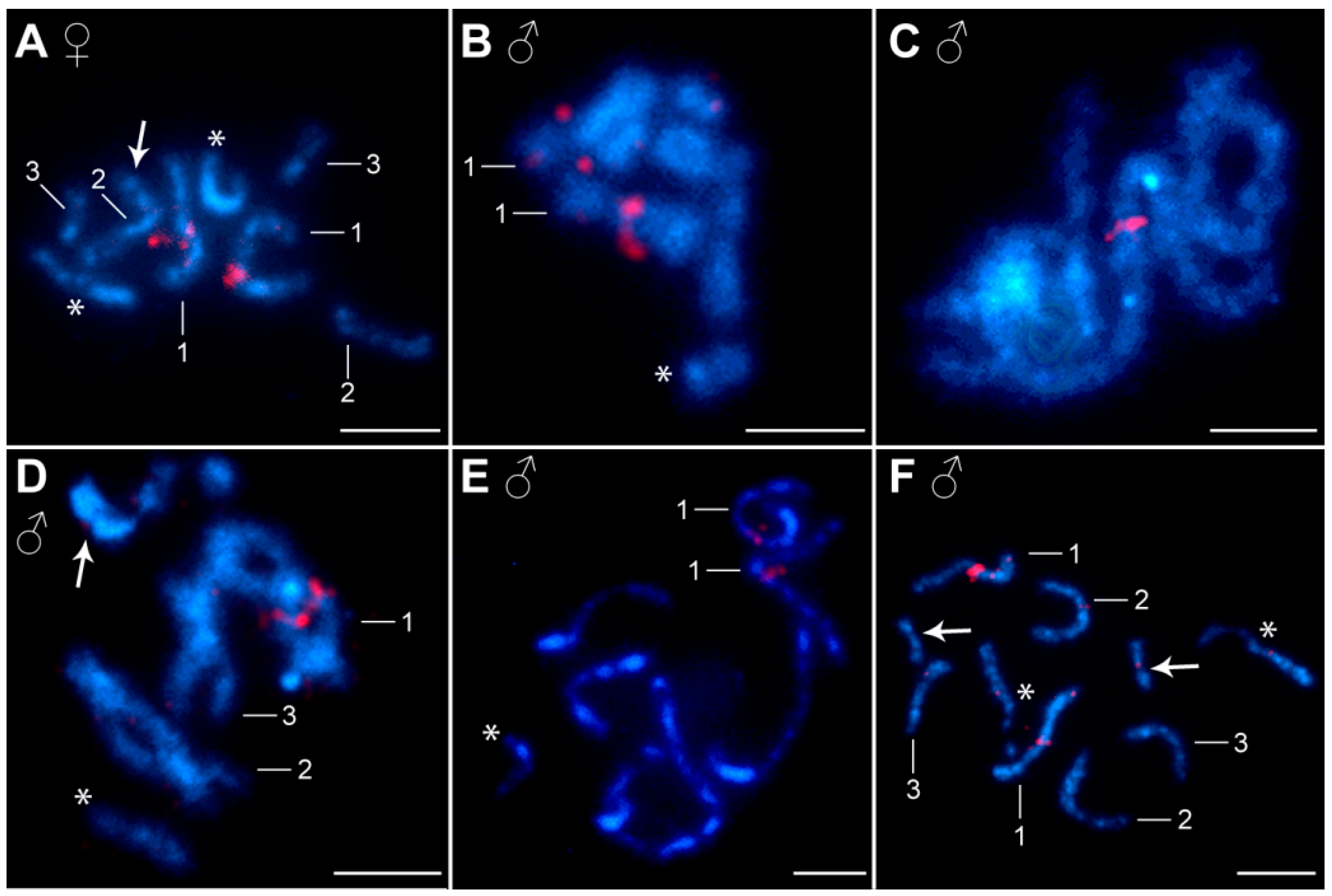
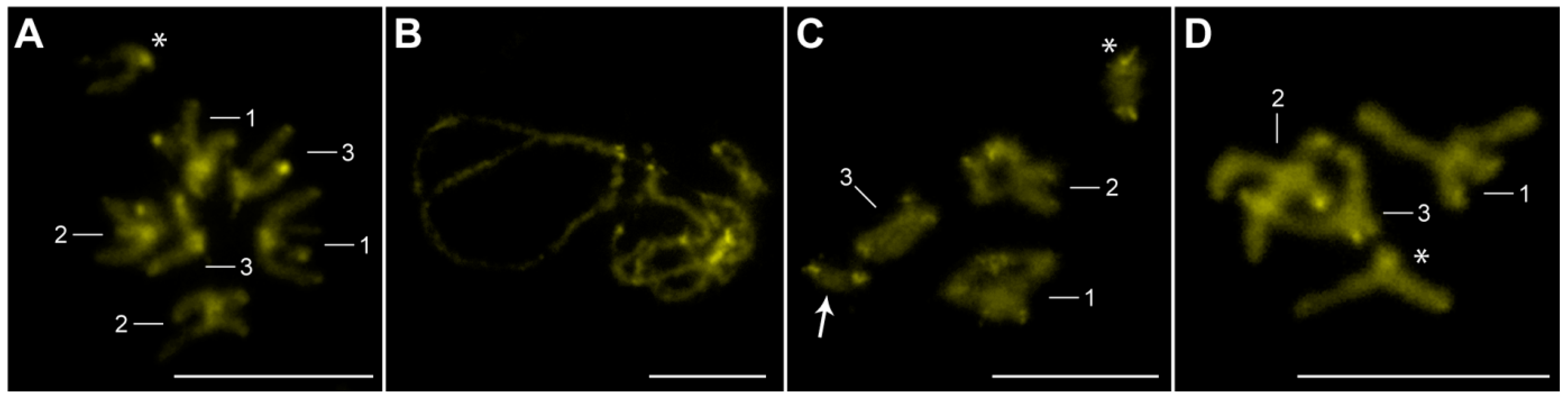
| Chromosome Number | Study Site a | Absolute Length (mean ± SD b) (μm) | Relative Length (mean ± SD) (%) | Centromeric Index (mean ± SD) | Classification c |
|---|---|---|---|---|---|
| 1 | ZŠ | 4.62 ± 0.29 | 32.19 ± 2.02 | 45.79 ± 2.03 | m |
| LB | 5.21 ± 0.27 | 30.87 ± 1.60 | 46.48 ± 1.80 | m | |
| PZ | 5.24 ± 0.22 | 31.64 ± 1.33 | 47.37 ± 1.70 | m | |
| 2 | ZŠ | 4.07 ± 0.32 | 28.33 ± 2.24 | 37.48 ± 1.94 | sm |
| LB | 4.43 ± 0.22 | 26.20 ± 1.31 | 38.09 ± 1.99 | sm | |
| PZ | 4.61 ± 0.24 | 27.82 ± 1.47 | 38.36 ± 1.23 | sm | |
| 3 | ZŠ | 3.19 ± 0.35 | 22.20 ± 2.44 | 35.97 ± 3.08 | sm |
| LB | 3.80 ± 0.26 | 22.50 ± 1.53 | 34.81 ± 2.29 | sm | |
| PZ | 3.61 ± 0.17 | 21.78 ± 0.91 | 28.86 ± 2.22 | sm | |
| X | ZŠ | 2.48 ± 0.24 | 17.27 ± 1.67 | 20.72 ± 1.63 | a |
| LB | 3.45 ± 0.13 | 20.43 ± 0.80 | 23.89 ± 2.61 | a | |
| PZ | 3.11 ± 0.15 | 18.75 ± 0.91 | 15.05 ± 1.68 | a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marková, A.; Orosová, M.; Marec, F.; Barčák, D.; Oros, M. Karyological Study of Acanthocephalus lucii (Echinorhynchida): The Occurrence of B Chromosomes in Populations from PCB-Polluted Waters. Diversity 2024, 16, 140. https://doi.org/10.3390/d16030140
Marková A, Orosová M, Marec F, Barčák D, Oros M. Karyological Study of Acanthocephalus lucii (Echinorhynchida): The Occurrence of B Chromosomes in Populations from PCB-Polluted Waters. Diversity. 2024; 16(3):140. https://doi.org/10.3390/d16030140
Chicago/Turabian StyleMarková, Anna, Martina Orosová, František Marec, Daniel Barčák, and Mikuláš Oros. 2024. "Karyological Study of Acanthocephalus lucii (Echinorhynchida): The Occurrence of B Chromosomes in Populations from PCB-Polluted Waters" Diversity 16, no. 3: 140. https://doi.org/10.3390/d16030140
APA StyleMarková, A., Orosová, M., Marec, F., Barčák, D., & Oros, M. (2024). Karyological Study of Acanthocephalus lucii (Echinorhynchida): The Occurrence of B Chromosomes in Populations from PCB-Polluted Waters. Diversity, 16(3), 140. https://doi.org/10.3390/d16030140







