Abstract
We investigated the evolutionary relationships between the taxa in the butterfly genus Iolana Bethune-Baker, 1914 and others in the subtribe Scolitantidina using information from nine DNA markers (COI-COII, ND1, ITS2, 28S, CAD, EF-1α, wg, and H3). We show that the genus Iolana originated about 10 mya in Central Asia and gradually expanded to the west to reach Europe about 5 mya. We then compared our inferred phylogeny with that of the Iolana larval host plants in the genus Colutea, reconstructed using three DNA markers (ITS, matK, and rpl32). Although the host plant phylogeny was weakly resolved, the close spatiotemporal correlation between Iolana butterflies and their larval hosts suggests that they may have co-evolved. Based on the molecular results and the morphology of male and female genitalia, we confirm nine species in the genus Iolana, which are distributed in allopatry from Europe and North Africa to Central Asia. We synonymize I. andreasi andreasi Sheljuzhko, 1919 (=I. andreasi khayyami Bernardi, 1964 syn. nov.) and I. iolas wullschlegeli Oberthür, 1914 (=I. iolas protogenes Fruhstorfer, 1917 syn. nov.).
1. Introduction
Iolana butterflies are undoubtedly among the largest and most charismatic species of lycaenid blues in Eurasia, but they are also among the most threatened. The larvae of Iolana are specialists on deciduous shrubby and sub-shrubby plants in the genus Colutea (Fabaceae), whose inflated pods they burrow to feed on the seeds [1]. Their life cycles are also facultatively associated with ants [2]. Iolana iolas has been listed as endangered in the Swiss Red List [3] and is classified among the species with the highest conservation priority [4], while in Romania, it is regarded as critically endangered [5]. In Catalonia, the taxon debilitata is listed as vulnerable [6], and it seems to be on the brink of extinction in the Maghreb ([7] and online updates, www.lepidofrance.fr/les-papillons-de-jour-du-maroc).
The genus Iolana was erected by Bethune-Baker [8], primarily based on genitalia characteristics, to accommodate four species: Lycaena iolas Ochsenheimer, 1816 (type species); L. gigantea Grum-Grshimailo, 1885; L. coeligena Oberthür, 1876 (now in genus Caerulea Forster, 1938); and L. astraea Freyer, [1851] (now in genus Glaucopsyche Scudder, 1872). While the original concept of the genus clearly did not stand the test of time, modern molecular studies (e.g., [9]) have proven Iolana to indeed be a well-supported monophyletic group with new taxa discovered and described as recently as 2004 (Iolana kermani Dumont, 2004). The genus, which is widely distributed from North Africa and Southern Europe to Central Asia and the Himalayas, is best known for its type species Iolana iolas, which has been long considered the only species in Europe and much of the Palearctic region. However, over time, studies have highlighted the possibility of the presence of multiple species under Iolana [10,11,12,13]. In 1972, Bernardi introduced a “superspecies” concept, where he listed several “semi-species” (gilgitica, gigantea, andreasi, alfierii, debilitata, and iolas) under superspecies Iolana iolas, some with multiple subspecies [11]. Later, in a detailed revision of the genus, Dumont [14] reiterated the distinctiveness of the genitalia in Iolana and recognized nine species, an arrangement largely confirmed by a recent molecular study that investigated the phylogeny of the subtribe Scolitantidina [15]. The latter study, however, did not include molecular data for Iolana arjanica Rose, 1979 and I. gilgitica (Tytler, 1926), nor for any representatives from genera Palaeophilotes Forster, 1938; Sinia Forster, 1940; and Subsolanoides Koiwaya, 1989. Two subsequent large-scale genomic studies [16,17] made available numerous sequences from some of these missing genera with suggestions on their taxonomic statuses, phylogenetic positions, and divergence times. Here, we compile a new molecular dataset for all species in Iolana as well as representatives from all genera in the subtribe Scolitantidina (equivalent to the Glaucopsyche section sensu Eliot [18]) with the aim of confirming or clarifying the statuses of these genera. We also analyze and discuss the geographic evolutionary history of the genus Iolana with respect to geography and the diversification of its Colutea host plants.
2. Materials and Methods
DNA sequencing and analysis. Specimens used in this study were selected mainly from the collections of RV (Institut de Biologia Evolutiva CSIC-UPF, Barcelona, Spain), WtH (Mömlingen, Germany), and AN (Tehran, Iran). Legs from selected specimens were submitted to the Center for Biodiversity Genomics in Guelph, ON, Canada, where samples were barcoded using mini-primers (LepF/mLepR and mLepF/LepR) [19]. Another part of the sequences (COI, ND1, ITS2, and CAD) were generated at Institut de Biologia Evolutiva (CSIC-UPF), Barcelona, Spain, using primers and protocols included in Supplementary Information Table S1. Additional molecular data for COI-COII, 28S, EF-1α, wg, and H3 genes were downloaded from Genbank (Supplementary Information Table S2). Since the species-level taxonomy at the sub-tribe level was not the primary focus of this study, in order to improve the phylogenetic resolution at the backbone, we substituted missing CAD sequences with those of closely related species in two cases: Philotiella leona OK744181 for P. speciosa, and Pseudophilotes vicrama OK743243 for P. baton. We used Polyommatus icarus to root the tree. A total of 171 new sequences were submitted to GenBank (for accessions, see Supplementary Table S2). A combined dataset was assembled using MEGA 11.0.8 [20]. Alignment of sequences was carried out using MUSCLE modules implemented in AliView 1.28 [21] and double-checked visually. After final alignment, the dataset contained a total of 8613 base pairs (bps). The partitioned Nexus file was analyzed using the IQtree web server (https://iqtree.cibiv.univie.ac.at) [22]. Excluding short and incomplete sequences, a Median-Joining Network of full-length (658 bps) COI barcode sequences of Iolana was constructed in PopART [23].
Eight calibration points for Scolitantidina were adopted from two recent comprehensive time-calibrated backbone trees provided by Wiemers et al. [24] and Kawahara et al. [16] (Table 1). Estimated node ages were added as MRCA priors in BEAUTi 2.7.5 [25] under normal distributions. The analysis was allowed to run in BEAST 2.7.3 [26] for 20 million generations and was repeated multiple times to check for convergence and stationarity, and the results were tested using TRACER 1.7.1 [27]. The resulting consensus tree was viewed in FigTree 1.4.4. [28] and edited using the open-source software GIMP 2.10.32 (gimp.org).

Table 1.
Calibration points used in our molecular clock analysis. Values represent millions of years.
We used the R package BioGeoBEARS [29] to reconstruct the biogeographic history of Iolana. The model with the highest likelihood was DIVALIKE+x, which, in addition to the likelihood implementation of the processes assumed by DIVA, also considers geographic distances between the different areas and reflects them as pie charts with all possible ranges for each node and their respective probabilities.
Host plant phylogeny. Host plant data for Iolana species were taken from the comprehensive review by Dumont [14]. Some authors have split Colutea species into more localized species, but this should not substantially affect our interpretations since we cover these taxa as subspecific entities in our analysis. Four genes (ITS1-ITS2, matK, and rpl32) for all available “Coluteid” species were selected from previous studies [30,31,32,33,34,35,36] and downloaded from GenBank (Supplementary Information Table S3). Alignment was carried out in AliView 1.28 [21], and a Maximum Likelihood tree for the combined dataset (3248 base pairs in length) was inferred using IQtree web server (https://iqtree.cibiv.univie.ac.at) [22]. A partitioned Bayesian analysis in MRBAYES 3.2.6 [37] produced a similar topology (Supplementary Figure S1).
Genitalia dissections. The abdomens of the butterflies were detached from the thorax using forceps and macerated for 20 min at 90 °C in 10% potassium hydroxide. Genitalia dissections were performed under a Leica MZ8 stereomicroscope in a small Petri dish (ø = 50 mm). The cleaning procedure was carried out in several washes with ethanol at increasing concentrations from 10% to 70%. The abdomen was dissected by tearing one of the pleurae along its entire length with two-pointed forceps. Then, the male genitalia were dissected by separating the valvae from their articulations or by removing the tegumen. The phallus was extirpated together with the vesica. Female genitalia were cleaned and arranged in order to show the lamella antevaginalis. For photography, the genitalia were immersed in a thin layer of 70% ethanol, pressed under a coverslip, and visualized with a Leica Z16 macroscope equipped with a CF500 camera and LAS 5.0 (Leica®) image capture software. We preserved dissections in vials containing EtOH 70% at room temperature in an air-conditioned room (22–26 °C). The photographs obtained were processed with Photoshop (R) to show the valvae of the males and the lamella antevaginalis of the females, separating them from the background and enhancing the images. Genitalia terminology follows Higgins [38].
3. Results
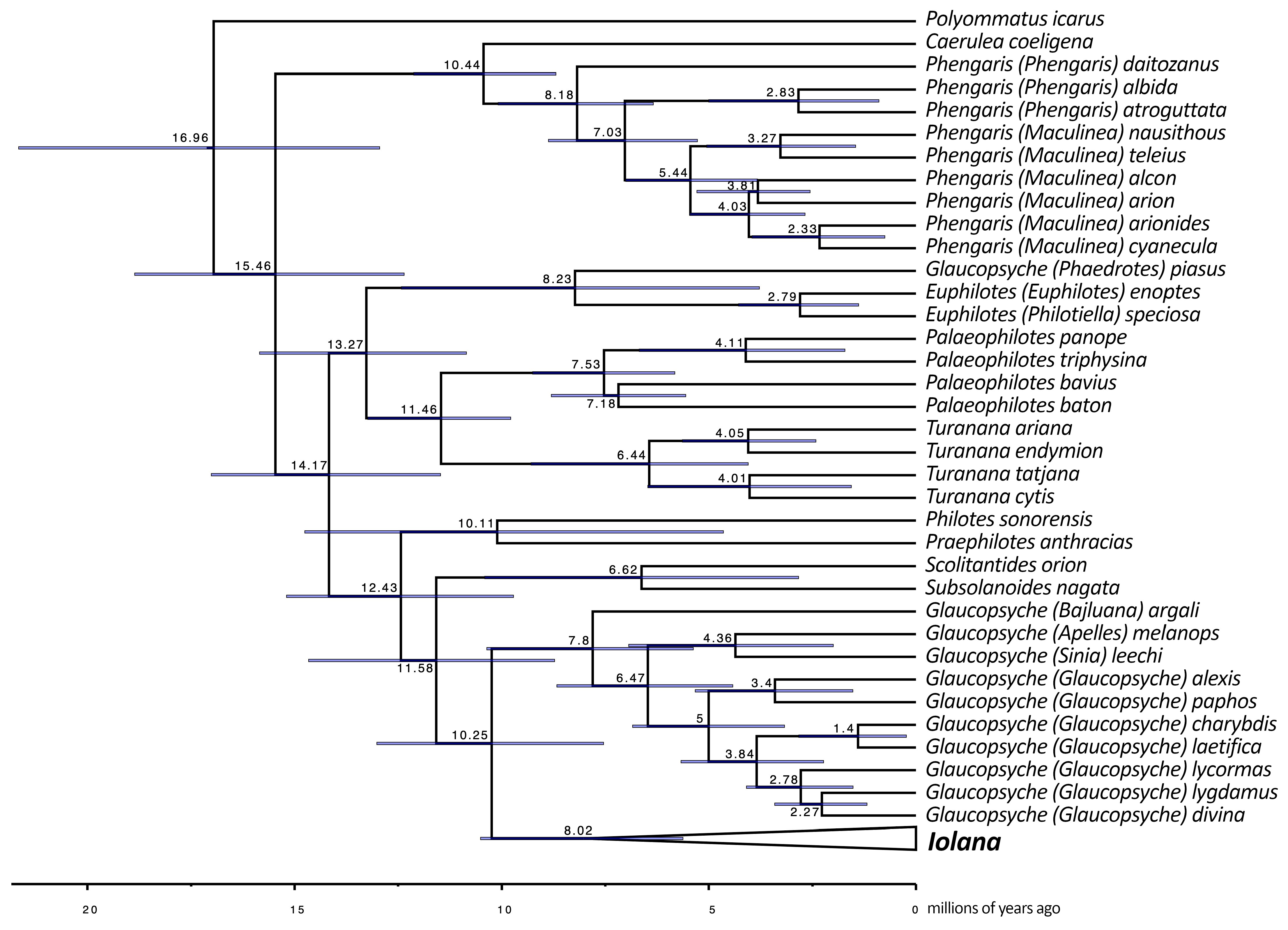
Butterfly phylogeny. Our ML and Bayesian analyses produced well-supported topologies (Figure 1; see Supplementary Figures S2 and S3 for node support values). We noticed that the COI sequence of specimen SCAUB:BN006, identified as Sinocupido lokiangensis in GenBank [16], showed the same haplotype as that of another record (OQ311407), identified as Palaeophilotes triphysina [17]. An examination of the original descriptions, types, and voucher photos for both of these records made it clear that the voucher specimen for Sinocupido lokiangensis is misidentified and it belongs to Palaeophilotes triphysina. Thus, in our dataset, we merged the available sequences for these two under the latter species.
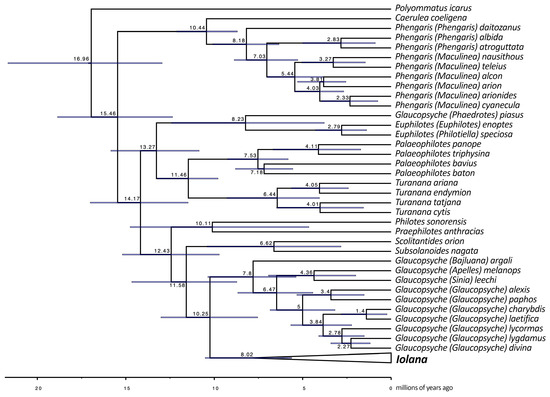
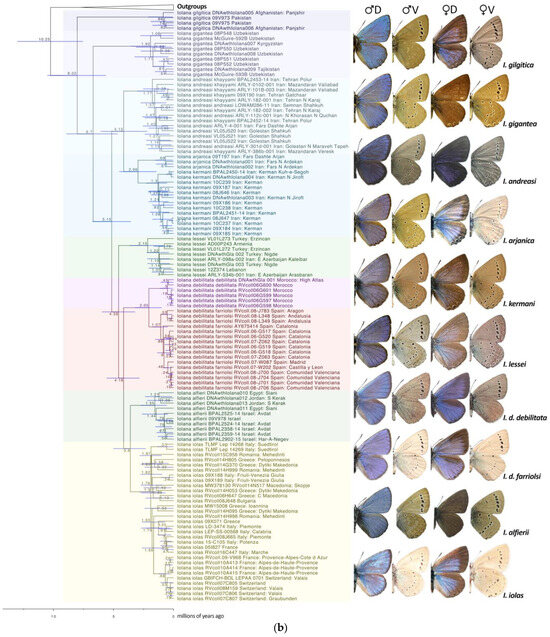
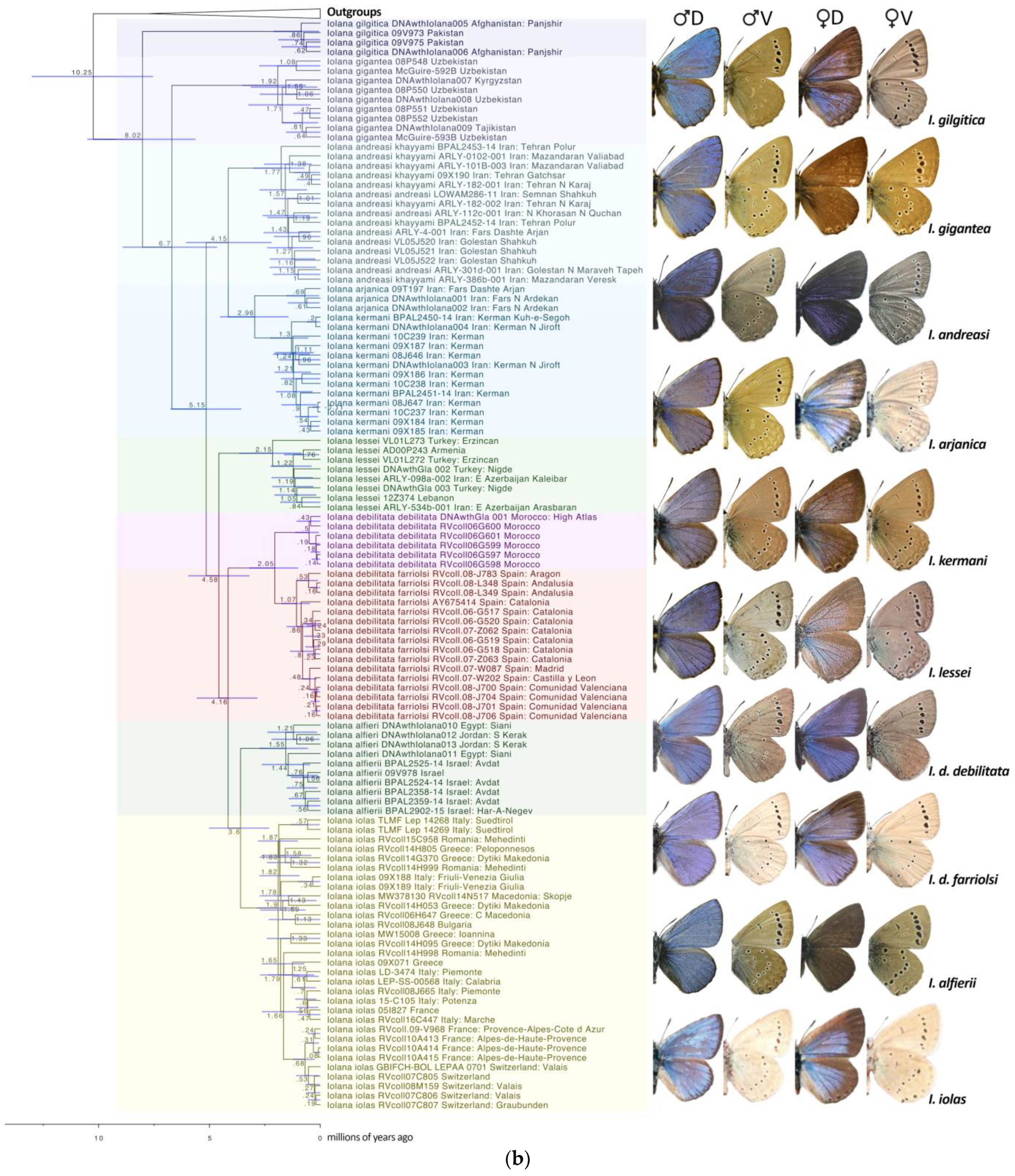
Figure 1.
(a) BEAST phylogeny of the combined dataset (9 genes) for Scolitantidina. Node values indicate the average node age estimated by BEAST; for node support values, see Supplementary Figure S2. (b) BEAST phylogeny of the combined dataset for Iolana. Node values indicate the average node age estimated by BEAST; for node support values, see Supplementary Figure S2. Upperside (D) and underside (V) of representative male (left) and female (right) specimens of Iolana species are depicted in phylogenetic order as they appear on the tree.
Throughout our analyses, Iolana always appeared monophyletic and sister to the Glaucopsyche clade; however, the taxon Glaucopsyche piasus Boisduval, 1852, recognized under subgenus Phaedrotes, always stayed far apart from the remaining Glaucopsyche and closer to the genera Euphilotes and Philotiella with good support (ML: 72, BPP: 0.97). In addition, one specimen of Iolana from the south of Iran (Fars: Dashte Arjan, coll. Naderi) appeared morphologically and genetically closer to I. andreasi. Since a single specimen is involved, the presence of I. andreasi in Fars remains to be confirmed.
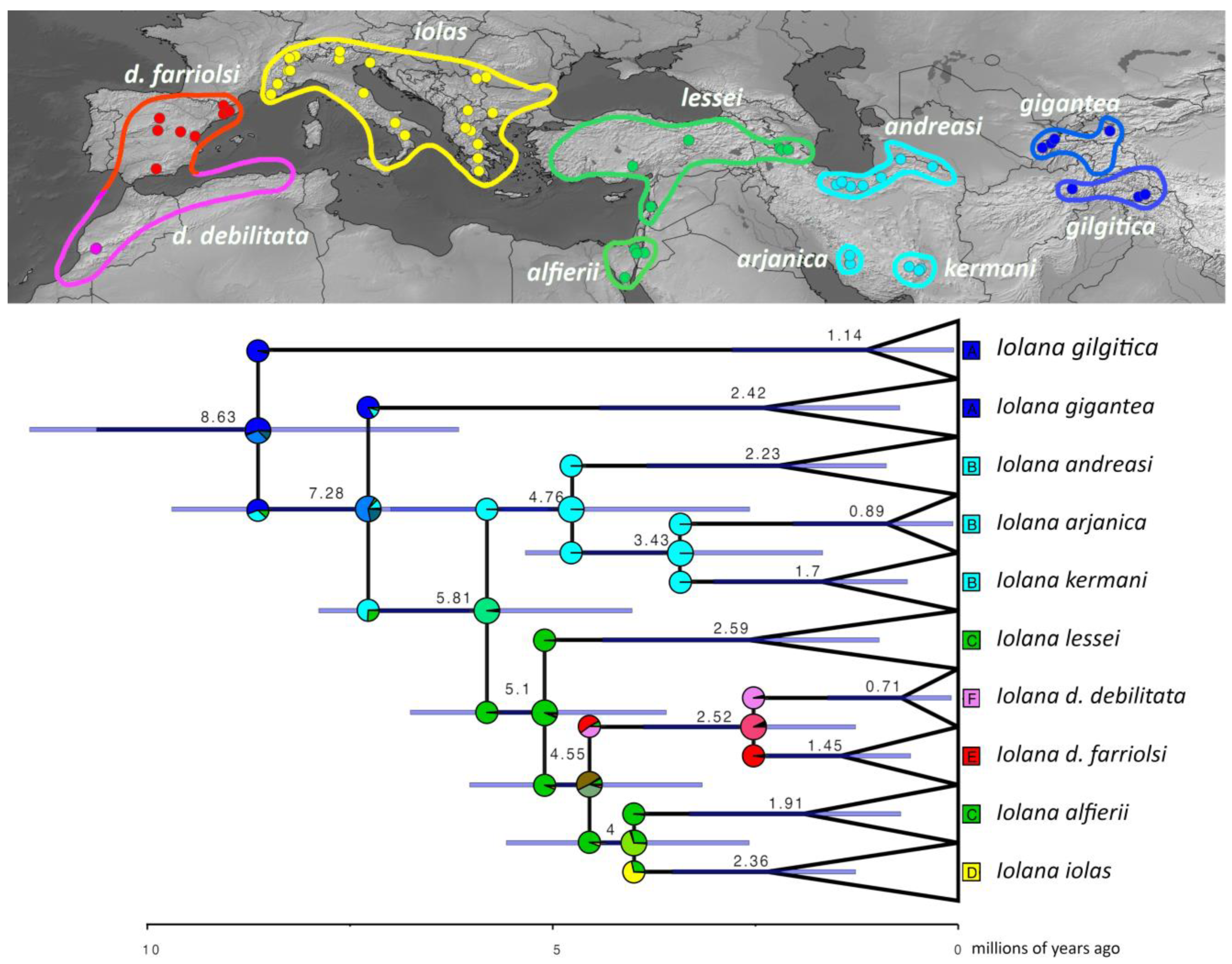
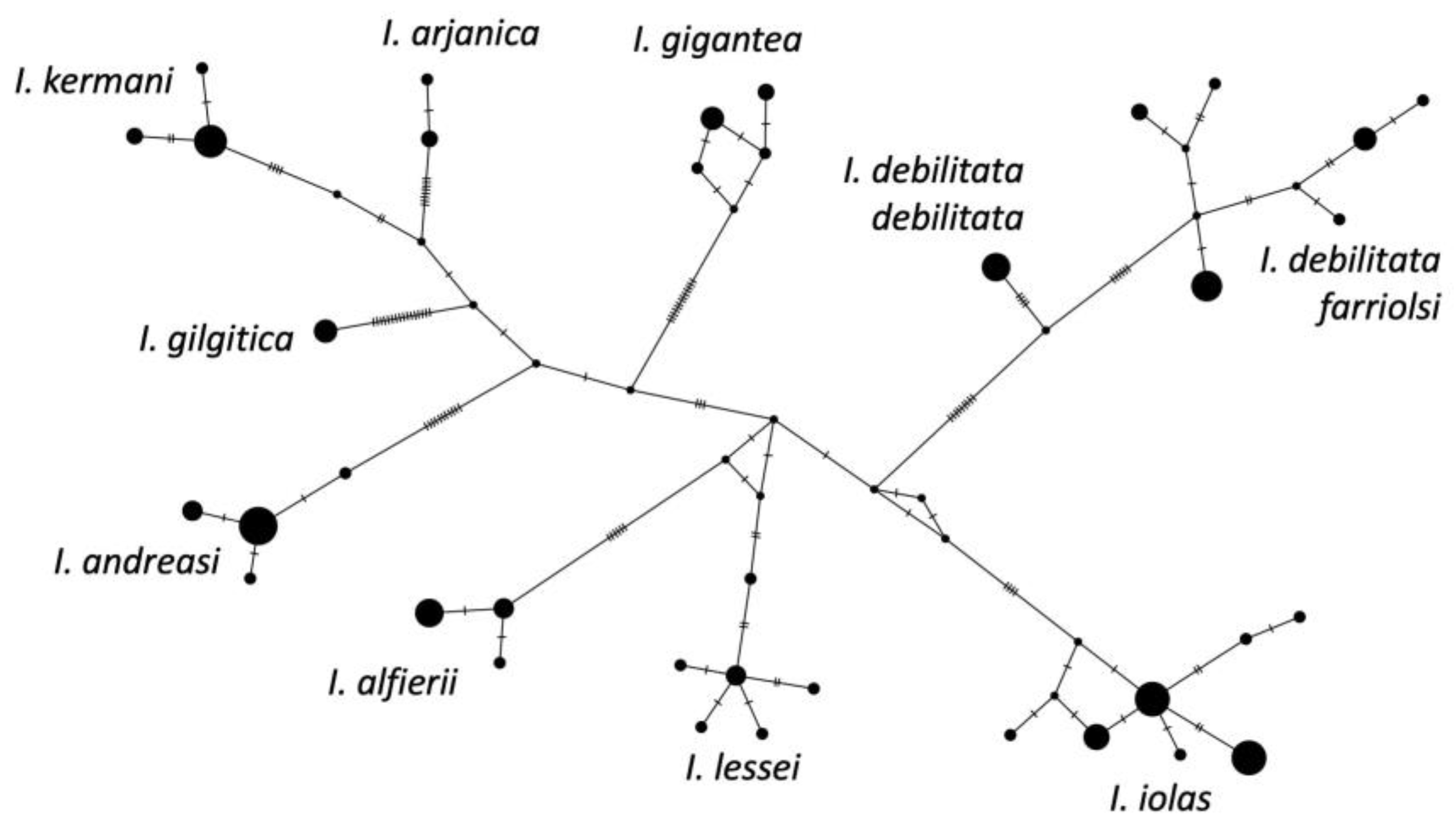
Our phylogeny placed Subsolanoides nagata Koiwaya, 1989 as a distinct lineage sister to Scolitantides orion, contrary to Kawahara et al. [16], who found it in a more basal position in the Scolitantidina phylogeny. As noted previously [17], we also found Sinia Forster, 1940 to be part of Glaucopsyche Scudder, 1872, and found Palaeophilotes Forster, 1938 to fall within Pseudophilotes Beuret, 1958. Our molecular clock analysis was also largely congruent with previous studies [16,24], with minor deviations (Figure 1). The split between ancestral Iolana and Glaucopsyche seems to have occurred about 10.25 mya, followed by gradual diversification of Iolana starting about 8 mya. The BioGeoBears analysis unambiguously assigned single ancestral areas to the majority of internal nodes in the Iolana phylogeny, with alternatives representing small proportions. The results clearly support an origin of the genus in Central Asia, and a gradual differentiation associated with east-to-west dispersal. Anatolia was colonized ca. 5.8 mya and in Europe ca. 4.6 mya (Figure 2). Finally, our COI Median-Joining Network of Iolana clearly separated the nine species, placing the taxa debilitata and farriolsi on the same branch, albeit substantially diverged (Figure 3).
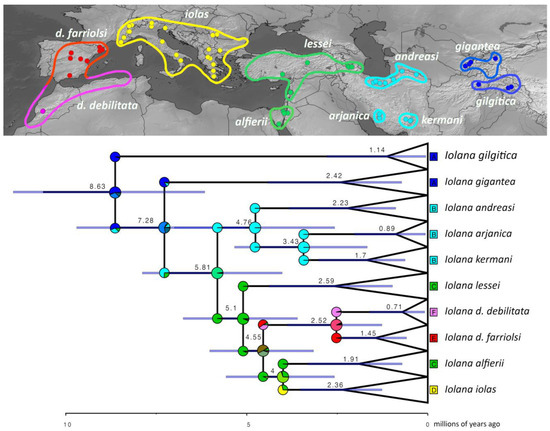
Figure 2.
Historical biogeography of Iolana. The most supported ancestral area reconstruction model (DIVALIKE+x) was estimated within the R package BioGeoBears. Pie charts on each node depict the relative probabilities of ancestral ranges corresponding to the colors on the map, which depict the distribution of the sequenced specimens (dots) and the approximate ranges for each species today. Median ages for each node and the error bars are inferred in BEAST.
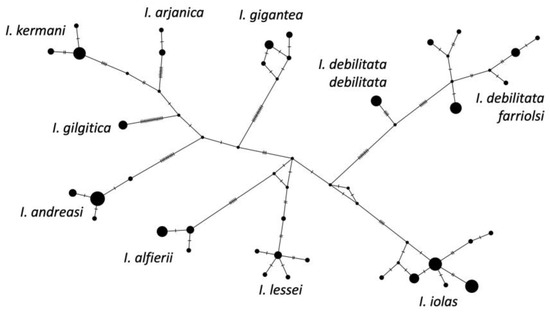
Figure 3.
Median-Joining Network of Iolana mitochondrial COI barcode sequences inferred using PopART (popart.otago.ac.nz, accessed on 10 December 2023). Circles are proportional to sample size for each haplotype. Smallest black dots represent unsampled but predicted haplotypes, and mutations are shown as dashes.
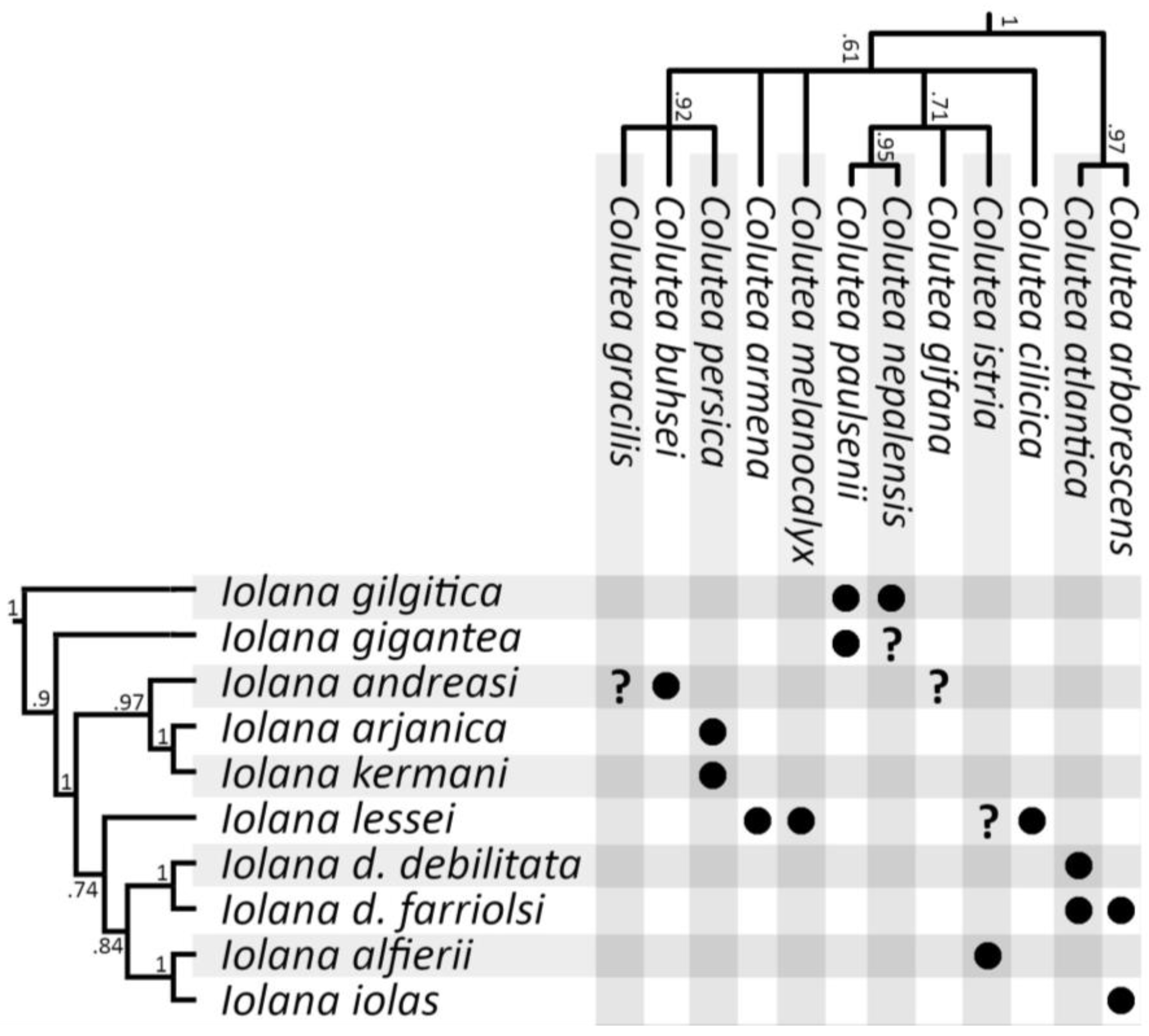
Host plant phylogeny. Despite the use of multiple ribosomal and chloroplast genes, the overall nucleotide variation within the host plant dataset was very low. Our reconstructed phylogeny did not differ significantly from the one inferred by Moghaddam et al. [33], with very little to no support for the deeper nodes in Colutea (see Supplementary Figure S1). We note that the split between Colutea and its Central Asian sister genera (Smirnowia, Eremosparton, and Sphaerophysa) also occurred around 10 mya [33], contemporaneous with the split between Iolana and Glaucopsyche. A simplified phylogeny of the Colutea clade was plotted against the Iolana phylogeny following Muto-Fujita et al. [39] (Figure 4).
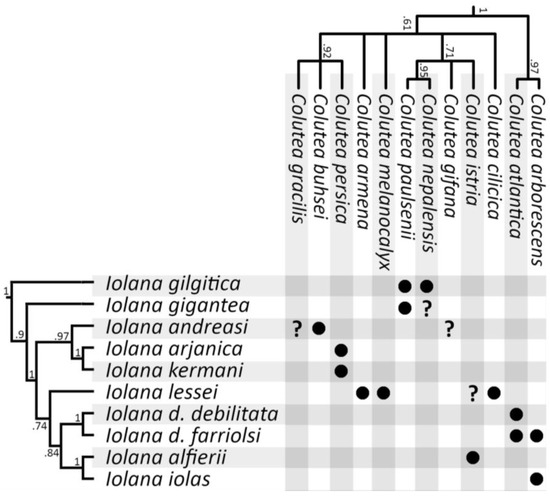
Figure 4.
Relationship between Iolana species and their Colutea host plants. Bayesian Posterior Probabilities are indicated at supported nodes. Additional Colutea species not recorded as hosts of Iolana are omitted (for a complete host plant phylogeny see Supplementary Figure S1). No sequences were available for some Colutea species recorded as hosts (e.g., C. atabajevii, C. jamnolenkoi) and their phylogenetic position remains unclear.
Genitalia. As noted previously [10,11,14], both the male and female genitalia in Iolana showed consistent differences between the nine recognized species, with some individual variation within each. We did not observe any meaningful differences in the male genitalia between I. debilitata ssp. debilitata from North Africa and the Iberian populations (here all treated as ssp. farriolsi); instead, we observed a considerable amount of individual variation in the genitalia of both the males and females without any clear pattern (Supplementary Figure S4).
4. Discussion
Fragmented distributions are a hallmark of species with highly specialized (e.g., monophagous or myrmecophilous) life histories. In the case of Iolana, both the butterflies and their larval host plants are characterized by localized, and often small, populations that evolved as a result of allopatric diversification. With about 30 species worldwide, Colutea are deciduous shrubby to sub-shrubby plants with inflated pods, distributed from Southern Europe and Northern Africa to Central Asia [33,40]. Recent studies have shown that Colutea diverged from Central Asiatic relatives around 10 mya in the Late Miocene, followed by a more recent burst of radiation about 3.2 mya [33]. This is interesting for two reasons: (1) the split between Iolana and its sister genus Glaucopsyche was estimated to have occurred around the same time (ca 10.25 mya), and (2) the ancestral area reconstruction unambiguously places the origin of the genus Iolana in Central Asia.
It is well established that host plant diversity generally sets the stage upon which Lepidoptera later diversify, and co-cladogenesis due to co-diversification is extremely rare [41]. The Iolana–Colutea partnership thus seems to represent a rare case to the contrary: the extreme dependence of the butterfly on its host plant, together with the fragmented distribution that does not facilitate dispersal between host plant populations, appears to have forced the simultaneous evolution of these butterflies together with their hosts. Our data show that Iolana originated in Central Asia from a common ancestor with Glaucopsyche and followed the dispersal and diversification of Colutea westwards, continuing to diversify in parallel, with additional speciation events interspersed due to other isolating factors (e.g., the split between I. kermani and I. arjanica in South Iran around 2.96 mya, both feeding on C. persica). The ancestral area reconstruction suggests that Iolana dispersed from the east Mediterranean to Iberia ca. 4.6 mya, and from there, reached the Maghreb later on. It is tempting to speculate that the Messinian salinity crisis could have facilitated the expansion across the Mediterranean. It is also worth noting that this dispersal event that reached Iberia seems to be unrelated to that of I. iolas, which is estimated as more recent (ca. 4 mya). Finally, it is hard to completely discard a route along the southern Mediterranean range that no longer exists due to recent aridification. Today, there is a distribution gap of Colutea in Tunisia, Libya, and most of Egypt; however, Colutea abyssinica is present in Eastern Africa (Ethiopia, Kenya, Tanzania, etc.), where no species of Iolana are known. The close phylogenetic relationship between C. abyssinica, C. atlantica, and C. arborescens suggests that Colutea previously had a much wider distribution in Northern Africa.
An interesting ecological aspect is the need for a precise emergence of the adult Iolana butterflies at the flowering and early seed development time of the Colutea host plants. Indeed, the flight period of Iolana is very short, and the caterpillars have a narrow time frame to feed on the tender seeds before they dry out. Such exquisite butterfly–plant phenological tuning necessarily involves local adaptation through natural selection, which could easily lead to co-evolution.
Despite the availability of several molecular markers, the phylogeny of Colutea remains poorly resolved. Even though without a robust Colutea phylogeny, it is not possible to infer a sensible phylogeographic scenario for the co-evolution of Iolana butterflies with respect to their hosts, given the coincidence in age, the specialization (monophagy) of Iolana on Colutea, and the highly allopatric distributions, this system is a potential candidate for butterfly–host plant parallel phylogeography. We predict that a comprehensive and well-resolved phylogeny of Colutea will likely closely mirror that of Iolana.
Species of Colutea are generally uncommon and very vulnerable to grazing and human activities, such as road work (e.g., [42]). Naturally, the monophagous Iolana that heavily depend on Colutea for survival are also vulnerable. In addition, climate change could potentially produce a mismatch between the phenologies of the butterflies and the host plants. These ecological specializations and the fragmented distribution patterns highlight the need to conserve both the host plants and the butterflies across their range. A similar extreme specialization with consequences for conservation can, for example, be seen in the endangered butterflies in the genus Phengaris [43]. As for Iolana, so far, only one species (I. iolas) has been assessed according to the IUCN criteria, and it is listed on the IUCN website (LC for the Mediterranean and NT for Europe). Based on the available information, I. debilitata is likely in a critical situation in North Africa [7], and the remaining Asiatic taxa (e.g., I. alfierii, I. lessei, I. kermani, and I. arjanica) are also extremely rare and threatened, and they seem to be locally extinct in some areas (WtH pers. obs., [44]). Future research should thus focus on identifying priority areas for the conservation of both Iolana and Colutea, as well as prospecting poorly studied areas.
5. Taxonomic Considerations
- Our results confirm many of the previously proposed synonymies (i.e., Glaucopsyche (=Shijimiaeoides); Turanana (=Otnjukovia; =Micropsyche); and Pseudophilotes (=Inderskia)) and taxonomic arrangements (Apelles, Bajluana, and Sinia as subgenera of Glaucopsyche; Palaeophilotes as the senior generic name for the Pseudophilotes clade; and Philotiella as the subgenus of Euphilotes) [9,15,17,45].
- Ugelvig et al. [9] separated Glaucopsyche (Phaedrotes) piasus from other Glaucopsyche species due to the paraphyly of the clade that also included Iolana spp. and S. divina. We also found G. piasus to belong to a distinct lineage closer to Euphilotes and far from the Glaucopsyche group (type species: G. lygdamus Doubleday, 1841). Nuclear genomic data from Zhang et al. [17] place this taxon as sister to all the rest of Glaucopsyche (including Sinia), but their mitochondrial phylogeny is supporting a different position, similar to the one we obtained. The same ambiguous situation was obtained in the study by Lukhtanov and Gagarina [15]. Thus, some kind of mito-nuclear discordance seems to be involved in this case. Even though Phaedrotes has been previously used as a stand-alone genus (cf. [46]), given its unstable phylogenetic position, and until better and more comprehensive methods are employed to address this question appropriately, we refrain from recognizing Phaedrotes as a monotypic genus.
- In our analyses, the monotypic Chinese endemic Subsolanoides nagata appears to be a distinct genus and species most closely related to Scolitantides orion. In the phylogeny presented by Kawahara et al. [16], Subsolanoides appears as sister to a clade consisting of Praephilotes, Scolitantides, Philotes, Iolana, and Glaucopsyche. Since the latter study used a larger amount of genomic information, we cannot exclude that the pattern observed in our study can change once additional genes become available.
- Lukhtanov and Gagarina [15] suggested that the African and Iberian populations attributed to Iolana debilitata Schultz, 1905 (TL: Algeria) may perhaps represent different species. García-Barros et al. [47] listed three subspecies present in the Iberian Peninsula: ssp. debilitata in Andalusia; ssp. thomasi Hemming, 1931 in Central Spain; and ssp. farriolsi Sagarra, 1931 in the northeast of Iberia. These authors, as well as Dumont [14], describe subtle differences in the male genitalia and the external morphology across the range of these populations. However, our molecular clock analysis, which estimates that the split between the Spanish populations and those in Morocco occurred around 2 mya (Figure 1), does not support the presence of the North African lineage (taxon debilitata) in Spain. On the other hand, a genetic structure roughly correlating with geography is observed within the Iberian populations. Here, we maintain Spanish and African populations under a single species, Iolana debilitata, but with the nominotypical populations restricted to North Africa, and tentatively consider a single subspecies in Spain, I. debilitata farriolsi Sagarra, 1931, possibly with a latitudinal morphological differentiation. A finer population genetics and morphometric study may shed light on the variation within the Iberian Peninsula.
- The distribution of I. iolas in Southern Europe is not continuous, and there are gaps, for example, between Südtirol and Slovenia [48]. The species is absent in Austria. The currently recognized subspecies include ssp. iolas (TL. Hungary) in Southeastern Europe, the Eastern Alps, and Italy; ssp. wullschlegeli (Oberthür, 1914) [49] (TL. Martigny, Switzerland) in France (Western Alps), Italy (Val d’Aoste), and Switzerland (Valais); and ssp. protogenes Fruhstorfer, 1917 [50] (TL. Digne, France) in Southern France [14]. Martigny and Digne are both in the Western Alps, about 200 km apart. In addition to the localities and haplotypes investigated by Groza et al. [42], we obtained sequences from two populations in Northern Italy (Südtirol [COI] and Friuli-Venezia Giulia [ND1]), both of which showed new and unique haplotypes (Figure 1). Based on current evidence, the presence of two subspecies in the Western Alps makes little sense. Dumont [14] mentions that these two taxa are weakly characterized, with the variation within subspecies being larger than that between subspecies. Our I. iolas haplogroup includes individuals from both ssp. wullschlegeli and ssp. protogenes. Therefore, here, we propose a synonymy: Iolana iolas wullschlegeli (Oberthür, 1914) (=protogenes Fruhstorfer, 1917 syn. nov.).
- We also did not find any reliable diagnostic differences in the morphology, genitalia, or DNA sequences between the nominotypical Iolana andreasi Sheljuzhko, 1919 [51] and the subspecies khayyami Bernardi, 1964 [52]. Therefore, here, we propose a synonymy: I. andreasi Sheljuzhko, 1919 (=khayyami Bernardi, 1964, syn. nov.).
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/d16020089/s1: Table S1. Primers and PCR protocols used for the amplification of COI (DNA barcode region), ND1, ITS2, and CAD; Table S2. Material examined and sequences used in this study; Table S3. Sequences used in the reconstruction of the host plant phylogeny; Figure S1. MrBayes reconstruction of the host plant phylogeny using a dataset composed on sequences in Supplementary Information Table S3; Figure S2. BEAST phylogeny of Scolitantidina combined dataset showing node posterior probabilities; Figure S3. Maximum Likelihood reconstruction with IQtree showing support for 1000 replicates of the same dataset; Figure S4. Genitalia dissections.
Author Contributions
Conceptualization, V.N., S.M.A., R.V. and V.D.; methodology, V.N., S.M.A. and R.V.; software, V.N. and L.S.; validation, S.M.A., L.S., V.D. and R.V.; formal analysis, V.N. and L.S.; investigation, V.N. and S.M.A.; resources, V.N., S.M.A., V.D., A.N., W.t.H. and R.V.; data curation, V.N., S.M.A. and V.D.; writing—original draft preparation and V.N.; writing—review and editing, S.M.A., V.D., A.N., W.t.H. and R.V.; visualization, V.N.; supervision, V.N. and R.V.; project administration, V.N.; funding acquisition, V.N., V.D. and R.V. All authors have read and agreed to the published version of the manuscript.
Funding
V.D. was supported by the Academy of Finland (Academy Research Fellow, decision nos. 324988 and 352652). R.V. was supported by grants PID2019-107078GB-I00 and PID2022-139689NB-I00, funded by MCIN/AEI/10.13039/501100011033 and by ERDF A way of making Europe, and by grant 2021 SGR 00420 from Departament de Recerca i Universitats de la Generalitat de Catalunya.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Detailed collection data, images, and COI barcode sequences for specimens used in this study are publicly available in the BOLD dataset “DS-IOLANA” (dx. https://doi.org/10.5883/DS-IOLANA). Additional data and sequences can be found on GenBank (for accessions, see Supplementary Table S2).
Acknowledgments
We thank Dubi Benyamini, Zdravko Kolev, Jean Claude Weiss, Dominique Dumont, Joaquin Baixeras, Akito Kawahara, David Plotkin, and Nick V. Grishin for materials and/or support, and three anonymous reviewers whose valuable comments improved an earlier draft of this paper.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Tolman, T.; Lewington, R. Collins Field Guide: Butterflies of Britain and Europe; Harper Collins Publications: New York, NY, USA, 1997; 320p. [Google Scholar]
- Fiedler, K. European and North West African Lycaenidae (Lepidoptera) and their associations with ants. J. Res. Lepid. 1991, 28, 239–257. [Google Scholar] [CrossRef]
- Gonseth, Y. Liste rouge des lépidoptères diurnes menacés de Suisse. In Listes Rouges des Espèces Animales Menacées de SUISSE; Duelli, P., Ed.; Office Fédéral de l’environnement, des Forêts et du Paysage (OFEFP): Berne, Switzerland, 1994; pp. 48–51. [Google Scholar]
- Carron, G.; Wermeille, E.; Schiesse, H.; Patocchi, N. Programme National de Conservation des Espèces Prioritaires de Papillons diurnes (Rhopalocera et Hesperiidae). Canton du Valais. 2001. 52p. Available online: http://home.page.ch/pub/insecta.carron@vtx.ch/ (accessed on 10 November 2023).
- Rákosy, L.; Corduneanu, C.; Crișan, A.; Dincă, V.; Kovács, S.; Stănescu, M.; Székely, L. Romanian Red List of Lepidoptera; Presa Universitară Clujeană: Cluj-Napoca, Romania, 2021; 187p. [Google Scholar]
- Vila, R.; Stefanescu, C.; Sesma, J.M. Guia de les Papallones Diürnes de Catalunya; Lynx Edicions: Barcelona, Spain, 2018; 509p. [Google Scholar]
- Tarrier, M.; Delacre, J. Les Papillons de Jour du Maroc; Mèze, Biotope, Ed.; Muséum National d’Histoire Naturelle: Paris, France, 2008; 480p. [Google Scholar]
- Bethune-Baker, G.T. Synonymic notes on the Ruralidae. Entomol. Rec. J. Var. 1914, 26, 159–164. [Google Scholar]
- Ugelvig, L.V.; Vila, R.; Pierce, N.E.; Nash, D.R. A phylogenetic revision of the Glaucopsyche section (Lepidoptera: Lycaenidae), with special focus on the Phengaris–Maculinea clade. Mol. Phylogenetics Evol. 2011, 61, 237–243. [Google Scholar] [CrossRef]
- Hemming, A.F. Revision of the genus Iolana, Bethune-Baker (Lepidoptera, Lycaenidae). Trans. Entomol. Soc. Lond. 1931, 79, 323–333. [Google Scholar] [CrossRef]
- Bernardi, G. Note sur la variation géographique de l’armure génilale mâle des Iolana (Lep. Lycaenidae). Bull. De La Société Entomol. De Fr. 1972, 77, 160–167. [Google Scholar] [CrossRef]
- Lukhtanov, V.A.; Lukhtanov, A.G. Die Tagfalter Nordwestasiens (Lepidoptera, Diurna); U. Eitschberger: Marktleuthen, Germany, 1994; 440p, ISBN 3-923807-02-3. [Google Scholar]
- Hesselbarth, G.; Van Oorschot, H.; Wagener, S. Die Tagfalter der Türkei; Selbstverlag Sigbert Wagener: Bocholt, Germany, 1995; Volume 1, 754p. [Google Scholar]
- Dumont, D. Révision du genre Iolana Bethune-Baker, 1914 (Lepidoptera: Lycaenidae). Description d’une nouvelle espèce: kermani n. sp. Linneana Belg. 2004, 19, 332–357. [Google Scholar]
- Lukhtanov, V.A.; Gagarina, A.V. Molecular Phylogeny and Taxonomy of the Butterfly Subtribe Scolitantidina with Special Focus on the Genera Pseudophilotes, Glaucopsyche and Iolana (Lepidoptera, Lycaenidae). Insects 2022, 13, 1110. [Google Scholar] [CrossRef]
- Kawahara, A.Y.; Storer, C.; Carvalho, A.P.S.; Plotkin, D.M.; Condamine, F.L.; Braga, M.P.; Ellis, E.A.; Laurent, R.A.S.; Li, X.; Barve, V.; et al. A global phylogeny of butterflies reveals their evolutionary history, ancestral hosts and biogeographic origins. Nat. Ecol. Evol. 2022, 7, 903–913. [Google Scholar] [CrossRef]
- Zhang, J.; Cong, Q.; Shen, J.; Song, L.; Opler, P.A.; Grishin, N.V. Additional taxonomic refinements suggested by genomic analysis of butterflies. The Taxonomic Report of the International Lepidoptera Survey 2023, 11, 1–46. [Google Scholar] [CrossRef]
- Eliot, J.N. The higher classification of the Lycaenidae (Lepidoptera): A tentative arrangement. Bull. Nat. Hist. Mus. Entomol. Ser. 1973, 28, 371–505. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Dewaard, J.R.; Ivanova, N.V.; Ratnasingham, S.; Dooh, R.T.; Kirk, S.L.; Mackie, P.M.; Hebert, P.D.N. Critical factors for assembling a high volume of DNA barcodes. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1959–1967. [Google Scholar] [CrossRef]
- Tamura, K.; Stechrer, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Bandelt, H.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. Available online: http://popart.otago.ac.nz (accessed on 10 December 2023). [CrossRef]
- Wiemers, M.; Chazot, N.; Wheat, C.W.; Schweiger, O.; Wahlberg, N. A complete time-calibrated multi-gene phylogeny of the European butterflies. ZooKeys 2020, 938, 97–124. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, 2010. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 10 December 2023).
- Van Dam, M.H.; Matzke, N.J. Evaluating the influence of connectivity and distance on biogeographical patterns in the south-western deserts of North America. J. Biogeogr. 2016, 43, 1514–1532. [Google Scholar] [CrossRef]
- Sanderson, M.J.; Wojciechowski, M.F. Diversification rates in a temperate legume clade: Are there so many species of Astragalus (Fabaceae)? Am. J. Bot. 1996, 83, 1488–1502. [Google Scholar] [CrossRef]
- Wojciechowski, M.F.; Lavin, M.; Sanderson, M.J. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot. 2004, 91, 1846–1862. [Google Scholar] [CrossRef]
- Duan, L.; Wen, J.; Yang, X.; Liu, P.L.; Arslan, E.; Ertugrul, K.; Chang, Z.Y. Phylogeny of Hedysarum and tribe Hedysareae (Leguminosae: Papilionoideae) inferred from sequence data of ITS, matK, trnL-F and psbA-trnH. Taxon 2015, 64, 49–64. [Google Scholar] [CrossRef]
- Moghaddam, M.; Kazempour Osaloo, S.; Hosseiny, H.; Azimi, F. Phylogeny and divergence times of the Coluteoid clade with special reference to Colutea (Fabaceae) inferred from nrDNA ITS and two cpDNAs, matK and rpl32-trnL(UAG) sequences data. Plant Biosyst. 2017, 151, 1082–1093. [Google Scholar] [CrossRef]
- Azani, N.; Bruneau, A.; Wojciechowski, M.; Zarre, S. Miocene climate change as a driving force for multiple origins of annual species in Astragalus (Fabaceae, Papilionoideae). Mol. Phylogenetics Evol. 2019, 137, 210–221. [Google Scholar] [CrossRef]
- Nafisi, H.; Kazempour-Osaloo, S.; Mozaffarian, V.; Schneeweiss, G.M. Molecular phylogeny and divergence times of the genus Hedysarum (Fabaceae) with special reference to section Multicaulia in Southwest Asia. Plant Syst. Evol. 2019, 305, 1001–1017. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, C.; Sun, Y.; Wu, P.; Chen, X.; Dong, W.; Yang, X.; Zhou, S. Methods for Quick DNA Barcode Reference Library Construction. Ecol. Evol. 2021, 11, 11627–11638. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Higgins, L.G. The Classification European Butterflies; Collins: London, UK, 1975; 320p. [Google Scholar]
- Muto-Fujita, A.; Takemoto, K.; Kanaya, S.; Nakazato, T.; Tokimatsu, T.; Matsumoto, N.; Kono, M.; Chubachi, Y.; Ozaki, K.; Kotera, M. Data integration aids understanding of butterfly–host plant networks. Sci. Rep. 2017, 7, 43368. [Google Scholar] [CrossRef]
- Mirzaei, L.; Mehregan, I.; Nejadsatari, T.; Assadi, M. Phylogeny analysis of Colutea L. (Fabaceae) from Iran based on ITS sequence data. Biodiversitas 2015, 16, 168–172. [Google Scholar] [CrossRef]
- Quek, S.-P.; Davies, S.J.; Itino, T.; Pierce, N.E. Codiversification in an ant-plant mutualism: Stem texture and the evolution of host use in Crematogaster (Formicidae: Myrmicinae) inhabitants of Macaranga (Euphorbiaceae). Evolution 2004, 58, 554–570. [Google Scholar] [CrossRef]
- Groza, B.; Vodă, R.; Székely, L.; Vila, R.; Dincă, V. Genetics and extreme confinement of three overlooked butterfly species in Romania call for immediate conservation actions. J. Insect Conserv. 2021, 25, 137–146. [Google Scholar] [CrossRef]
- Thomas, J.A.; Schönrogge, K. Conservation of co-evolved interactions: Understanding the Maculinea–Myrmica complex. Insect Conserv. Divers. 2019, 12, 459–466. [Google Scholar] [CrossRef]
- Benyamini, D. Butterflies of the Levant and Neighbouring Areas (Southern Turkey, Syria, Lebanon, Israel, Jordan, Egypt, North-West Saudi Arabia & Cyprus); Volume IV: Lycaenidae; 4D Microrobotics: Beit-Aryeh, Israel, 2023; 192p. [Google Scholar]
- Zhang, J.; Cong, Q.; Shen, J.; Opler, P.A.; Grishin, N.V. Changes to North American butterfly names. Taxon. Rep. Int. Lepid. Surv. 2019, 8, 1–12. [Google Scholar]
- Coolidge, K.L. The life history of Phaedrotes piasus Boisd. (Lepidoptera: Lycaenidae). Entomol. News 1923, 34, 295–300. [Google Scholar]
- García-Barros, E.; Munguira, M.L.; Stefanescu, C.; Vives Moreno, A. Fauna Ibérica Volumen 37: Lepidoptera: Papilionoidea; Consejo Superior de Investigaciones Científicas: Madrid, Spain, 2013. [Google Scholar]
- Verovnik, R.; Lasan, M. On the presence of Iolana iolas (Ochsenheimer, 1816) in Slovenia. Nat. Slov. 2003, 5, 43–44. [Google Scholar] [CrossRef]
- Oberthür, C. Etudes de Lépidoptérologie Comparée; Smithsonian Institution: Washington, DC, USA, 1914; Volume 10, p. 392. [Google Scholar]
- Fruhstorfer, H. Neue palaearktische Lycaeniden. Dtsch. Entomol. Z. Iris 1917, 31, 24–43. [Google Scholar]
- Sheljuzhko, L. Neue palaearktische Lepidopteren-Formen. Neue Beiträge Zur Syst. Insektenkunde 1919, 1, 129–132. [Google Scholar]
- Bernardi, G. Lépidoptères Lycaenidae (sauf Agrodiaetus) récoltés en Iran par H. de Lesse en 1955 et 1958. Alexanor 1964, 3, 209–216, 273–278. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).