Abstract
Species of genus Aelia are important pests of wheat crops in arid areas. In this study, the mitogenomes of A. sibirica and A. fieberi were sequenced using high-throughput sequencing technology. The mitochondrial genome characteristics of both Aelia species were compared and analyzed, and the phylogenetic relationships of Pentatomidae were constructed based on protein-coding genes. In addition, the taxonomic status of the genus Aelia was confirmed. The results showed that the total length of the mitogenome sequences of A. sibirica and A. fieberi were 15,372 bp and 15,450 bp, respectively, including 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a control region. By comparing the mitochondrial genome structure, base composition, codon usage, RNA secondary structure, and other characteristics, it was found that the mitochondrial genome characteristics of the two species were similar. Phylogenetic analysis showed that Phyllocephalinae and Asopinae both formed monophyletic groups, but the relationship between Podopinae and Pentatominae was not resolved. Within the subfamily Pentatominae, (Nezarini + Antestiini), (Aeliini + Carpocorini), and (Strachiini + Pentatoma) formed stable clades. Aelia sibirica and A. fieberi were found to be a stable sibling pair, and the clade was closely related to Dolycoris baccarum.
1. Introduction
The mitochondrial genome is characterized by its compact size, genetic stability, rapid evolutionary rate, and maternal inheritance, and has emerged as one of the most commonly used molecular markers in evolutionary studies [1,2,3,4,5,6]. In hemipteran insects, the mitochondrial genome typically consists of a circular double-stranded DNA ranging from 14 to 20 kb and encompassing 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a control region [7,8,9]. The known structure of hemipteran mitochondrial genomes exhibits a high degree of conservation, with a tight arrangement of genes and only sporadic occurrences of tRNA gene rearrangements observed among certain species [10]. The overall gene order remains largely consistent with that of ancestral insects [11,12]. The advent of high-throughput sequencing technologies has allowed the mitochondrial genome to be widely used in molecular evolutionary studies, phylogenetic analyses, and investigations pertaining to biogeography within hemipteran taxa [13,14,15].
The insect family Pentatomidae comprises a diverse range of species, and is the largest family in the superfamily Pentatomoidea. It is widely distributed across the world, with nearly 5000 species belonging to 896 genera and ten subfamilies [16]. The classification of higher taxonomic ranks within Pentatomidae has been the source of considerable disagreement among researchers, despite the increasing number of studies and abundant data. Based on morphological and anatomical characteristics, Pentatomidae has been successively divided into 11, 9, 8, and 10 subfamilies [17,18,19,20]. Originally, seven subfamilies were identified for Pentatomidae species in China; however, with the development of molecular biology technology and the comprehensive analysis with multiple characters, the classification of Pentatomidae has been changed a lot [21,22,23]. Therefore, Chinese Pentatomidae currently includes four subfamilies: Phyllocephalinae, Asopinae, Pentatominae, and Podopinae. Previous phylogenetic studies of Pentatomidae subfamilies and tribes were constructed based on morphological characters. With the wide application of multiple molecular data, morphological characters were combined with molecular data for species classification [20,24]. Studies on the phylogenetic relationships within the subfamilies of Pentatomidae and the tribes within Pentatominae are insufficient, with most studies predominantly based on morphological characteristics or limited molecular data. Only a few taxa have been studied, and there is currently no widely recognized viewpoint [25,26,27,28]. Despite the vast number of Pentatomidae species, there is a lack of mitochondrial genomic research specifically targeting this important taxonomic group. To date, mitochondrial genome data are available for only 72 species (https://www.ncbi.nlm.nih.gov/, accessed on 2 January 2024); therefore, it is important to conduct mitochondrial genomic studies on Pentatomidae insects and gain comprehensive insights into the diversity and evolutionary characteristics of their mitochondrial genomes, with a view to enabling more accurate taxonomic assignments.
The genus Aelia belongs to the family Pentatomidae, and is widely distributed in the Palearctic region and North America [29,30]. It is an important agricultural pest in the arid areas of northern China and Central Asia, and primarily infests wheat crops, sucking on the ears of wheat and causing serious harm to wheat sprouts. It is essential to conduct comprehensive studies on this pest, which would be conducive to the long-term development of the agricultural economy. This study aimed to sequence the mitochondrial genomes of A. sibirica Reuter, 1884 and A. fieberi Scott, 1874 using Illumina MiSeq next-generation sequencing, and analyze the genomic composition and structural characteristics. By incorporating the mitochondrial sequence data from representative species of the Pentatomidae obtained from the GenBank database, Bayesian inference (BI) and maximum likelihood (ML) phylogenetic trees were constructed based on the PCG sequences.
2. Materials and Methods
2.1. Sampling, DNA Extraction, and Sequencing
The adult specimens of A. sibirica used in this experiment were collected on 14 August 2019, from the Sanjiangyuan National Nature Reserve in Qinghai Province. The adult specimens of A. fieberi were collected on 10 August 2019 from Shanfanao in Henan Province. Specimens were preserved in 100% ethanol and stored at −20 °C until used for DNA extraction. The leg muscle tissues of A. sibirica and A. fieberi were extracted, and genomic DNA extraction was performed following the instructions of the TIANamp Genomic DNA Kit (Tiangen biotechnology, Beijing, China). Voucher specimens were deposited at the Animal Systematics Laboratory, Department of Biology, Xinzhou Normal University, Xinzhou, China. The quality of the extracted DNA was assessed using a NanoDrop 2000 nucleic acid analyzer (Thermo Fisher Scientific, Waltham, MA, USA). The mitogenome was sequenced using the whole-genome shotgun method on an Illumina Miseq platform (Personalbio, Shanghai, China).
2.2. Genome Annotations and Sequence Analyses
The sequencing data was assembled using A5-miseq v. 20150522 [31] and SPAdes v. 3.9 [32] software. The mitochondrial genome sequences were annotated by Geneious 10.1.3 [33]. The annotation of protein-coding genes was conducted by Open Reading Frame Finder on the NCBI website (http://www.ncbi.nlm.nih.gov/gorf/gorf.html, accessed on 13 December 2021), as well as identifying start and stop codons. The tRNA genes were automatically annotated by MITOS (http://mitos.bioinf.uni-leipzig.de/index.py/, accessed on 13 December 2021) [34] with the invertebrate mitochondrial code. The annotation of rRNA genes was based on published reference sequence (Eurydema dominulus (Scopoli, 1763)) and combined with their putative secondary structures. The locations of the control region were identified by the boundary of neighboring genes. The newly sequenced and annotated mitogenomes were submitted to GenBank with accession numbers NC_067883 (A. sibirica) and ON059969 (A. fieberi). The mitochondrial genome maps were generated by GCView Server [35]. The base composition and relative synonymous codon usage (RSCU) were performed using MEGA 7.0 [36]. Additionally, AT skew was calculated using the formula (A − T)/(A + T), while GC skew was calculated using (G − C)/(G + C) [37]. The number of tandem repeats of control region was investigated with Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.html, accessed on 20 March 2022) [38].
2.3. Phylogenetic Analysis
In order to investigate the phylogenetic relationships among different taxonomic levels within Pentatomidae, and the phylogenetic relationship between the genus Aelia and its closely related genera, this study selected 27 representative species from the family Pentatomidae to construct phylogenetic trees (Table 1). The selected 27 taxa covered 4 subfamilies of Pentatomidae and 13 tribes of Pentatominae which existed in the NCBI database. In addition, Sastragala esakii (Pentatomoidea: Acanthosomatidae) was selected as the outgroup. The maximum likelihood method (ML) and Bayesian inference (BI) were used to construct phylogenetic trees based on 13 protein-coding genes. Multiple alignments of protein-coding gene sequences were performed using MEGA 7.0, and the aligned gene sequence matrices were concatenated using the SequenceMatrix 1.8 [39]. The BI tree was generated using MrBayes v. 3.2 [40], with the GTR + I + G model selected and run for 10,000,000 generations. The program was stopped when the convergence diagnostic value was below 0.01. The ML tree was constructed using RAxML v. 7.0.3 [41], and branch support was assessed using 1000 bootstrap replicates.

Table 1.
List of sequences used to reconstruct the phylogenetic tree.
3. Results
3.1. Genomic Features
The mitochondrial genomes of two Aelia species were obtained using high-throughput sequencing. The complete mitochondrial genome sequence of A. sibirica was 15,372 bp (Figure 1A), whereas that of A. fieberi was 15,450 bp (Figure 1B). Both genomes consisted of 13 PCGs, 22 tRNA, and two rRNA genes and a control region (Table 2). This gene arrangement was consistent with the typical order observed in Heteroptera insects without any specific rearrangements. The N-strand encoded fourteen genes, including four protein-coding, nad1, nad4, nad4L, and nad5; eight tRNA, trnQ, trnC, trnY, trnF, trnH, trnP, trnL1, and trnV; and the 12S rRNA and 16S rRNA. The remaining genes were encoded on the J-strand.
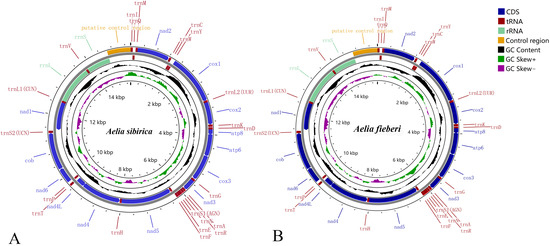
Figure 1.
Mitochondrial genome structures of A. sibirica (A) and A. fieberi (B).

Table 2.
Organization of the mitochondrial genomes of A. sibirica and A. fieberi.
The mitochondrial genome of A. sibirica contained ten overlapping regions, ranging from 1 to 8 bp, and seventeen intergenic spacer regions, ranging from 1 to 27 bp. In contrast, the mitochondrial genome of A. fieberi had eight overlapping regions, ranging from 1 to 8 bp, and nineteen intergenic spacer regions, ranging from 1 to 24 bp.
3.2. Nucleotide Composition and Codon Usage
In the complete mitogenomes, nucleotide composition analysis revealed that both A. sibirica and A. fieberi had significantly higher AT than GC content. The nucleotide composition in the mitochondrial genomes of both species were similar, and the overall base composition was identical (Table 3). The AT content in the control region was slightly lower than that in the complete mitochondrial genome, PCGs, rRNAs, and tRNAs with values of 70.5% and 69.2%, respectively. Among the PCGs, nad2 had the highest AT content in A. sibirica (79.8%), and nad6 had the highest AT content in A. fieberi (80.2%). The gene with the lowest AT content was cox1 (67.6% and 67.0%, respectively). The AT-skew values of the complete mitochondrial genome, 22 tRNAs, and the control region were positive in both Aelia species, whereas the AT-skew values for the two rRNAs and thirteen PCGs were negative. Among the PCGs, AT-skew values were negative for nad1, nad4, nad4L, nad5, and cytb in both Aelia species; nad6 also had a negative AT-skew value in A. fieberi. The GC-skew values for the complete mitochondrial genome, thirteen PCGs, and the control region were negative, whereas the GC-skew values for the two rRNAs and twenty-two tRNAs were positive. Among the PCGs, the AT-skew values were negative, and the GC-skew values were positive for nad1, nad4, nad4L, nad5, and cytb in both Aelia species.

Table 3.
Nucleotide composition and skewness of mitochondrial genome of A. sibirica and A. fieberi.
3.3. PCGs
The lengths of the 13 PCGs in A. sibirica and A. fieberi were 11,033 and 11,082 bp, respectively, accounting for 71.77% and 71.73% of the complete mitochondrial genome sequence, respectively, with the exception of cox1, nad1, and atp8, which started with the TTG, TTG/GTG, and TTG codons, respectively; the genes in both species started with the ATN codon. In A. sibirica, the numbers of genes starting with the ATC, ATG, ATT, and ATA codons were one, five, two, and two, respectively; whereas in A. fieberi, the numbers of genes starting with the ATC, ATG, ATT, and ATA codons were one, four, two, and three, respectively. With the exception of nad3 and nad5, the PCGs in both species had identical stop codons. The preferred stop codon was TAA; however, some genes ended with TAG or an incomplete T.
The relative synonymous codon usage (RSCU) of PCGs for A. sibirica and A. fieberi were calculated (Figure 2). Excluding stop codons, A. sibirica contained 3456 codons, whereas A. fieberi contained 3478 codons. The most frequently used codons in A. sibirica were AUU (I), UAU (Y), and UUU (F), with 262, 251, and 243 occurrences, respectively. In contrast, the most frequently used codons in A. fieberi were UUU (F), AUU (I), and UUA (L), with 284, 248, and 224 occurrences, respectively. Codons with A/U nucleotides were predominant among those with higher usage frequencies.
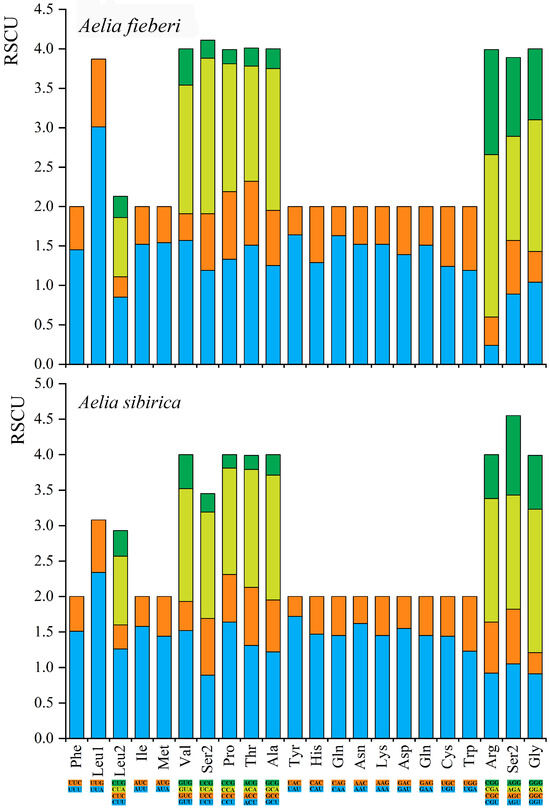
Figure 2.
The relative synonymous codon usage (RSCU) in the mitogenomes of A. sibirica and A. fieberi.
3.4. Ribosomal and Transfer RNA Genes
The mitogenomes of A. sibirica and A. fieberi contained 22 tRNA genes with total lengths of 1458 and 1453 bp, respectively. The length ranges of the tRNA genes in A. sibirica were from 62 bp (trnC) to 71 bp (trnK), whereas in A. fieberi, they ranged from 61 bp (trnR) to 70 bp (trnK). A comparison of the tRNA secondary structures found minimal differences between the two Aelia species. Apart from trnS1 and trnV, which lacked a dihydrouridine arm, the remaining 20 tRNA genes exhibited a typical cloverleaf structure (Figure 3). The number of nonclassical pairings of A. sibirica and A. fieberi was 18 and 26, respectively, with multiple occurrences of G-U non-canonical pairing.
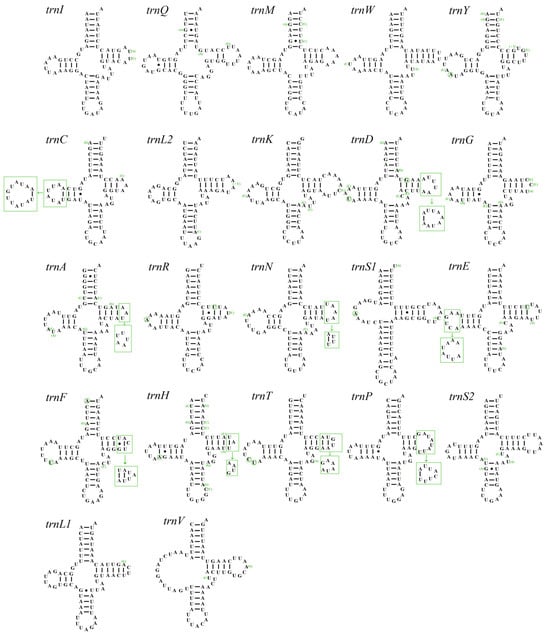
Figure 3.
Secondary structures of tRNAs of A. sibirica and A. fieberi (A. sibirica as the template).
The lengths of 12S rRNA and 16S rRNA genes in A. sibirica were 796 and 1282 bp, respectively, whereas those in A. fieberi were 795 and 1272 bp, respectively. The 16S rRNA gene was located between trnL1 and trnV, whereas the 12S rRNA gene was located between trnV and the control region. The secondary structures of the rRNA in A. sibirica were compared with those in A. fieberi and identified five domains in the 16S rRNA, whereas the 12S rRNA contained three domains (Figure 4 and Figure 5). The 12S rRNA secondary structure in both Aelia species showed minimal changes in the structural domains and nucleotide positions, particularly in the loop region. In contrast, domains IV and VI in the secondary structure of 16S rRNA were more conserved than domains I, II, and V, with variable sites concentrated in various loop regions.
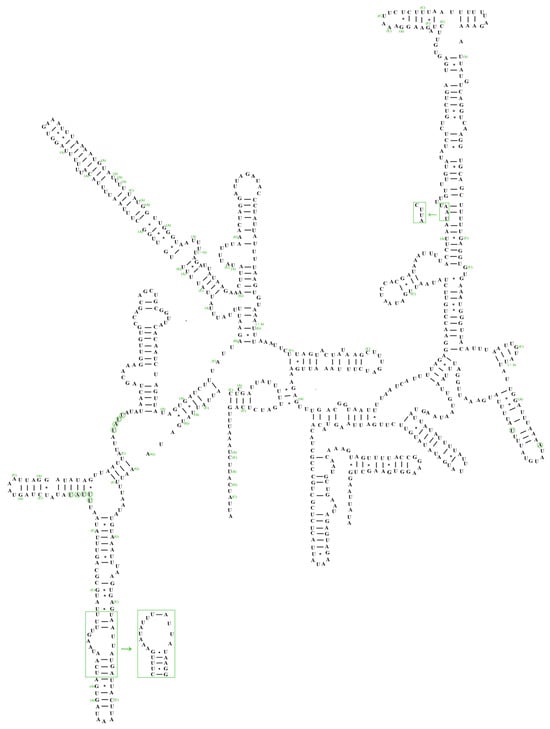
Figure 4.
Secondary structure of 12S rRNA gene of A. sibirica and A. fieberi (A. sibirica as the template).
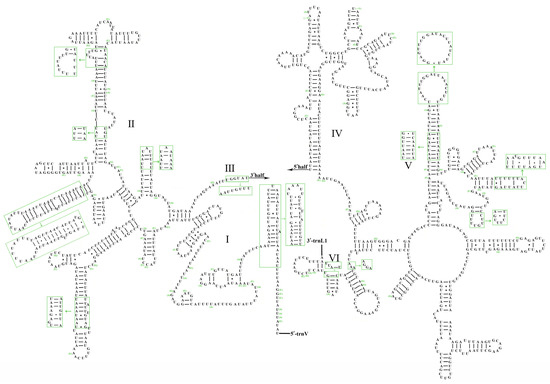
Figure 5.
Secondary structure of 16S rRNA gene of A. sibirica and A. fieberi (A. sibirica as the template).
3.5. Control Region
Complete sequences of the control region in A. sibirica and A. fieberi were obtained, resulting in lengths of 717 bp and 768 bp, respectively. The control region was located between the 12S rRNA gene and trnI. The control region of A. sibirica had an AT content of 70.5%, whereas the control region of A. fieberi had an AT content of 69.2%, both of which were lower than the overall AT content of the complete mitogenomes. The repeat unit of A. fieberi was 54 bp, with five copies located between 462 and 731 bp; the repeat unit of A. sibirica was 55 bp, with four copies located between 487 and 706 bp.
3.6. Phylogenetic Analyses
Phylogenetic trees of the Pentatomidae were constructed based on 13 PCGs using both the ML and BI methods (Figure 6). The topological structures of the phylogenetic trees were consistent, and most internal nodes had high posterior values. The results showed that the subfamily Phyllocephalinae was the earliest diverging lineage and formed the basal clade of the Pentatomidae. Phyllocephalinae were a sister group to Placosternum, currently classified in Pentatominae. Both the subfamilies Phyllocephalinae and Asopinae were found to be monophyletic and showed clear relationships within the subfamilies, supported by high branch support values. In the subfamily Podopinae, Scotinophara lurida, Graphosoma rubrolineata, and Deroploa parva did not cluster into a monophyletic group, but formed sister-group relationships with Pentatominae species. Within the subfamily Pentatominae, some tribes formed stable sibling clades, such as (Aeliini + Carpocorini), (Nezarini + Antestiini), and (Strachiini + Pentatoma).
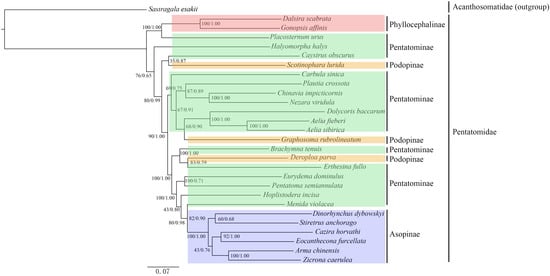
Figure 6.
ML and BI phylogenetic trees of Pentatomidae based on the protein-coding genes. Numbers above each node indicate Bayesian posterior probabilities values and ML bootstrap values.
4. Discussion
The sequence lengths of insect mitochondrial genomes are relatively stable and primarily influenced by the length of the control region [42,43]. In this study, the complete mitochondrial genome sequences of two species belonging to the genus Aelia were obtained, with lengths of 15,372 and 15,450 bp. The lengths of the control regions were 717 and 768 bp, showing only minor differences. The 37 genes were tightly arranged, and their genomic structure was consistent with that of most Pentatomidae insects, without gene rearrangements or losses [14,44,45,46]. The longest overlapping region was located between trnC and trnY, and the longest intergenic region was located between nad1 and trnL1, which is a common feature of the mitochondrial genomes of Pentatomidae [47]. The A + T content of both species of Aelia was significantly higher than the G + C content, indicating a clear AT bias. For most genes, the usage of T exceeded that of A bases, whereas the usage of G exceeded that of C bases. The compositional bias of bases has important implications for studying the mechanisms of mitochondrial genome replication, transcription, and phylogenetic relationships [48].
In addition to TTG and GTG, other start codons are preferred by Pentatomidae [49,50], and ATN is the most commonly used start codon in Aelia species. In the present study, cox1, nad1, and atp8 used TTG, TTG/GTG, and TTG start codons, respectively. In total, 11 of the 13 PCGs had identical stop codons, and the most frequently used stop codon was TAA, followed by TAG and incomplete T. PCGs ending with a T codon are completed with a TAA stop codon during transcription through polyadenylation [51]. The AT bias in the mitochondrial genome was also reflected in the frequency of synonymous codon usage by PCGs. Codons with higher frequencies often ended with an A or U, and their relative synonymous codon usage values were greater than one.
The lengths and nucleotide compositions of RNA genes showed high similarity between A. sibirica and A. fieberi. The secondary structures of rRNA genes play a crucial role in the functionality of PCGs. Most nucleotide position variations existed in the ring region, whereas the stem regions tended to be more conserved. In addition, the 12S rRNA gene was more conserved than the 16S rRNA gene. G-U pairing was observed multiple times in the secondary structure of tRNA, and such mismatches have been shown to play an important role in maintaining secondary structure stability [52].
In the phylogenetic analysis, there were relatively few species from Asopinae and Phyllocephalinae, and they exhibited distinct differences from other subfamilies within the family Pentatomidae, making their classification less controversial and more stable. The phylogenetic relationships obtained by different methods supported the monophyly of Asopinae and Phyllocephalinae, which was consistent with the results based on mitochondrial RNA genes and aligned with current mainstream views [20,53].
Species in the subfamily Pentatominae are abundant, and their morphological variations are great. Owing to the lack of stable and reliable identification characters for phylogenetic reconstructions, different researchers have had different opinions on the division of tribes and the monophyly of Pentatominae, and the relationships between the various groups within Pentatominae remain uncertain [24,54]. Based on morphological studies, Yang divided the Chinese Pentatominae species into seven tribes [55]. Whereas Rider recorded 42 tribes about the worldwide Pentatominae species [18]. In the 7-tribe classification system, the genus Dolycoris was classified under the tribe Dolycorini, whereas in the 42-tribe classification system, it was placed in the tribe Carpocorini, together with Carpocoris, Palomena, and Rubiconia. In this study, Carpocorini and Aeliini were found to be sister groups, this is congruent with Roca-Cusachs et al. [20]. However, Lian et al. support the finding that Eysarcorini and Carpocorini are sister groups based on mitochondrial genomes [56]. The external morphological features of Placosternum urus are similar to those of the tribe Pentatomini; however, shorter or ear-shaped scent gland openings are observed. The results of this study indicated that Placosternum urus forms a sister group relationship with the subfamily Phyllocephalinae, whereas Pentatoma semiannulata forms a sister group relationship with Eurydema dominulus (belonging to Strachiini). Graphosoma rubrolineata and Scotinophara lurida, both of which belong to the subfamily Podopinae, have a complex relationship with Pentatominae. Some Podopinae species were previously assigned to Scutelleridae or Pentatominae, and were even separately promoted to family level; however, there is no widely accepted view of Podopopinae based on a comprehensive analytical approach. Yang previously classified Graphosoma and Eysarcoris in the tribe Graphosomini of the subfamily Pentatominae [55]. In addition, Zhao et al. and Li et al. also support a close relationship between the genus Graphosoma and species of Pentatominae, which is corroborated by the results of this study [57,58]. In this study, the representative species of Podopinae mixed in the Pentatominae species, the result combined with previous studies indicate that the classified position of Graphosoma rubrolineata could not be determined [26,56]. Owing to the limitations of the sample size in this study, further investigation of the monophyly of the subfamily Pentatominae and the validity of taxonomic units within the subfamily Podopinae require additional mitochondrial sequence information.
5. Conclusions
Currently, there are few phylogenetic studies within Pentatomidae, and the use of molecular data is limited. This study determined the mitochondrial genome sequences of A. sibirica and A. fieberi. The sequence features, base composition, codon usage, and RNA secondary structures were found to be highly similar between the two species, with no significant differences observed within the genus. The comprehensive analysis of the Aelia mitochondrial genomes performed in this study contributes new information which can be used in molecular evolutionary studies and phylogenetic analyses. More mitogenomes or other molecular data are needed to conduct further exploration of the taxonomic status and phylogenetic history of Pentatomidae species.
Author Contributions
Conceptualization, W.Z. and Q.Z.; methodology, W.Z. and D.L.; software, D.L.; validation, D.L., Y.W. and W.Z.; formal analysis, S.F. and H.Z.; investigation, S.F. and Q.Z.; resources, W.Z.; data curation, D.L.; writing—original draft preparation, D.L. and W.Z.; writing—review and editing, H.Z. and S.F.; visualization, D.L. and Y.W.; supervision, W.Z. and Q.Z.; project administration, W.Z.; funding acquisition, W.Z. and H.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundamental Research Program of Shanxi Province, grant number 20210302124140 and 202103021224331, and funded by Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi, grant number 2023L299, and funded by the Fund for Shanxi “1331 Project” Improving the quality and efficiency Plan Project subtopic, grant number 2023WTS06.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data presented in this study are deposited in GenBank with accession numbers NC_067883 (A. sibirica) and ON059969 (A. fieberi).
Acknowledgments
The authors acknowledge any support given that is not covered by the author contributions or by funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wolstenholme, D.R. Animal Mitochondrial DNA: Structure and Evolution. In International Review of Cytology; Wolstenholme, D.R., Jeon, K.W., Eds.; Academic Press: Cambridge, MA, USA, 1992; Volume 141, pp. 173–216. [Google Scholar]
- Boore, J.L. Animal Mitochondrial Genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Simon, C.; Buckley, T.R.; Frati, F.; Stewart, J.B.; Beckenbach, A.T. Incorporating Molecular Evolution into Phylogenetic Analysis, and a New Compilation of Conserved Polymerase Chain Reaction Primers for Animal Mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 545–579. [Google Scholar] [CrossRef]
- Liu, Y.; Li, H.; Song, F.; Zhao, Y.; Wilson, J.-J.; Cai, W. Higher-Level Phylogeny and Evolutionary History of Pentatomomorpha (Hemiptera: Heteroptera) Inferred from Mitochondrial Genome Sequences. Syst. Entomol. 2019, 44, 810–819. [Google Scholar] [CrossRef]
- Jeong, J.S.; Park, J.S.; Sohn, J.-C.; Kim, M.J.; Kim, I. Complete Mitochondrial Genome of the Hawthorn Moth Scythropia Crataegella Linnaeus, 1767 (Lepidoptera: Scythropiidae). J. Asia-Pac. Entomol. 2023, 26, 102103. [Google Scholar] [CrossRef]
- Song, N.; Wu, Q.; Lin, X.; Zhang, Y.; Zhao, T. The Complete Mitochondrial Genome of Nephrotoma Scalaris Parvinotata (Brunetti 1918) (Diptera: Tipulidae) and Insights into the Phylogeny of Tipulomorpha. J. Asia-Pac. Entomol. 2023, 26, 102133. [Google Scholar] [CrossRef]
- Song, N.; Zhang, H.; Zhao, T. Insights into the Phylogeny of Hemiptera from Increased Mitogenomic Taxon Sampling. Mol. Phylogenet. Evol. 2019, 137, 236–249. [Google Scholar] [CrossRef]
- Mu, Y.-L.; Zhang, C.-H.; Zhang, Y.-J.; Yang, L.; Chen, X.-S. Characterizing the Complete Mitochondrial Genome of Arma Custos and Picromerus Lewisi (Hemiptera: Pentatomidae: Asopinae) and Conducting Phylogenetic Analysis. J. Insect Sci. 2022, 22, 6. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, D.; Jia, Q.; Wu, X.; Zhang, H. Characterization of the Complete Mitochondrial Genome of Myrmus Lateralis (Heteroptera, Rhopalidae) and Its Implication for Phylogenetic Analyses. ZooKeys 2021, 1070, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Dowton, M.; Castro, L.R.; Austin, A.D. Mitochondrial Gene Rearrangements as Phylogenetic Characters in the Invertebrates: The Examination of Genome “Morphology”. Invert. Syst. 2002, 16, 345–356. [Google Scholar] [CrossRef]
- Françoso, E.; Gomes, F.; Arias, M.C. A Protocol for Isolating Insect Mitochondrial Genomes: A Case Study of NUMT in Melipona Flavolineata (Hymenoptera: Apidae). Mitochondrial DNA Part A 2016, 27, 2401–2404. [Google Scholar] [CrossRef] [PubMed]
- Djoumad, A.; Nisole, A.; Zahiri, R.; Freschi, L.; Picq, S.; Gundersen-Rindal, D.E.; Sparks, M.E.; Dewar, K.; Stewart, D.; Maaroufi, H.; et al. Comparative Analysis of Mitochondrial Genomes of Geographic Variants of the Gypsy Moth, Lymantria Dispar, Reveals a Previously Undescribed Genotypic Entity. Sci. Rep. 2017, 7, 14245. [Google Scholar] [CrossRef]
- Talavera, G.; Vila, R. What Is the Phylogenetic Signal Limit from Mitogenomes? The Reconciliation between Mitochondrial and Nuclear Data in the Insecta Class Phylogeny. BMC Evol. Biol. 2011, 11, 315. [Google Scholar] [CrossRef]
- Fedorov, V.B.; Trucchi, E.; Goropashnaya, A.V.; Stenseth, N.C. Conflicting Nuclear and Mitogenome Phylogenies Reveal Ancient Mitochondrial Replacement between Two North American Species of Collared Lemmings (Dicrostonyx Groenlandicus, D. Hudsonius). Mol. Phylogenet. Evol. 2022, 168, 107399. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, D.; Gao, Z.; Liu, J.; Yi, W.; Zhang, H. Complete mitochondrial genome sequence and phylogenetic implications of Chorosoma macilentum (Heteroptera: Rhopalidae). Mitochondrial DNA Part B 2019, 4, 3882–3883. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.Y.; Gui, H. Insect Classification; Nanjing Normal University Press: Nanjing, China, 1999. [Google Scholar]
- McPherson, J.E.; Esquivel, J.F.; Musolin, D.L.; Jones, W.A.; Hoelmer, K.A. Invasive Stink Bugs and Related Species (Pentatomoidea): Biology, Higher Systematics, Semiochemistry, and Management; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Rider, D.A.; Schwertner, C.F.; Vilimová, J.; Redei, D.; Kment, P.; Thomas, D.B. Higher Systematics of the Pentatomoidea. In Invasive Stink Bugs and Related Species (Pentatomoidea); CRC Press: Boca Raton, CA, USA, 2018. [Google Scholar]
- Genevcius, B.C.; Greve, C.; Koehler, S.; Simmons, R.B.; Rider, D.A.; Grazia, J.; Schwertner, C.F. Phylogeny of the Stink Bug Tribe Chlorocorini (Heteroptera, Pentatomidae) Based on DNA and Morphological Data: The Evolution of Key Phenotypic Traits. Syst. Entomol. 2021, 46, 327–338. [Google Scholar] [CrossRef]
- Roca-Cusachs, M.; Schwertner, C.F.; Kim, J.; Eger, J.; Grazia, J.; Jung, S. Opening Pandora’s Box: Molecular Phylogeny of the Stink Bugs (Hemiptera: Heteroptera: Pentatomidae) Reveals Great Incongruences in the Current Classification. Syst. Entomol. 2022, 47, 36–51. [Google Scholar] [CrossRef]
- Xu, Z.Q. Phylogeny and Cladistic Analysis of the Pentatomoidea (Hemiptera: Heteroptera). Acta Agric. Univ. Pekin. 1993, 19, 49–53. [Google Scholar]
- Weirauch, C.; Schuh, R.T. Systematics and Evolution of Heteroptera: 25 Years of Progress. Annu. Rev. Entomol. 2011, 56, 487–510. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.; Li, M.; Dong, P.; Cui, Y.; Xie, Q.; Bu, W. Comparative and Phylogenomic Studies on the Mitochondrial Genomes of Pentatomomorpha (Insecta: Hemiptera: Heteroptera). BMC Genom. 2008, 9, 610. [Google Scholar] [CrossRef] [PubMed]
- Grazia, J.; Schuh, R.T.; Wheeler, W.C. Phylogenetic Relationships of Family Groups in Pentatomoidea Based on Morphology and DNA Sequences (Insecta: Heteroptera). Cladistics 2008, 24, 932–976. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Z.; Rédei, D.; Eger, J., Jr.; Wang, Y.H.; Wu, H.Y.; Carapezza, A.; Kment, P.; Cai, B.; Sun, X.-Y.; Guo, P.-L.; et al. Phylogeny and the Colourful History of Jewel Bugs (Insecta: Hemiptera: Scutelleridae). Cladistics 2018, 34, 502–516. [Google Scholar] [CrossRef]
- Yuan, M.-L.; Zhang, Q.-L.; Guo, Z.-L.; Wang, J.; Shen, Y.-Y. Comparative Mitogenomic Analysis of the Superfamily Pentatomoidea (Insecta: Hemiptera: Heteroptera) and Phylogenetic Implications. BMC Genom. 2015, 16, 460. [Google Scholar] [CrossRef]
- Zhao, W.; Zhao, Q.; Li, M.; Wei, J.; Zhang, X.; Zhang, H. DNA Barcoding of Chinese Species of the Genus Eurydema Laporte, 1833 (Hemiptera: Pentatomidae). Zootaxa 2017, 4286, 151–175. [Google Scholar] [CrossRef]
- Ding, X.; Chen, C.; Wei, J.; Gao, X.; Zhang, H.; Zhao, Q. Comparative Mitogenomics and Phylogenetic Analyses of the Genus Menida (Hemiptera, Heteroptera, Pentatomidae). ZooKeys 2023, 1138, 29–48. [Google Scholar] [CrossRef]
- Derzhansky, V.; Péricart, J.; Pluot-Sigwalt, D. Notes on Synonymy and Distribution of Some Species of the Genus Aelia (Heteroptera: Pentatomidae). Zoosyst. Ross. 2007, 15, 298. [Google Scholar] [CrossRef]
- Gibicsár, S.; Keszthelyi, S. Topographical Based Significance of Sap-Sucking Heteropteran in European Wheat Cultivations: A Systematic Review. Diversity 2023, 15, 109. [Google Scholar] [CrossRef]
- Coil, D.; Jospin, G.; Darling, A.E. A5-Miseq: An Updated Pipeline to Assemble Microbial Genomes from Illumina MiSeq Data. Bioinformatics 2014, 31, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de Novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A Comparative Genomics Tool for Circular Genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of Nucleotide Composition at Fourfold Degenerate Sites of Animal Mitochondrial Genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation Software for the Fast Assembly of Multi-Gene Datasets with Character Set and Codon Information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Stamatakis, A. Using RAxML to Infer Phylogenies. Curr. Protoc. Bioinform. 2015, 51, 6–14. [Google Scholar] [CrossRef]
- Yan, L.; Xu, W.; Zhang, D.; Li, J. Comparative Analysis of the Mitochondrial Genomes of Flesh Flies and Their Evolutionary Implication. Int. J. Biol. Macromol. 2021, 174, 385–391. [Google Scholar] [CrossRef]
- Yuan, L.; Ge, X.; Xie, G.; Liu, H.; Yang, Y. First Complete Mitochondrial Genome of Melyridae (Coleoptera, Cleroidea): Genome Description and Phylogenetic Implications. Insects 2021, 12, 87. [Google Scholar] [CrossRef]
- Lee, W.; Kang, J.; Jung, C.; Hoelmer, K.; Lee, S.H.; Lee, S. Complete Mitochondrial Genome of Brown Marmorated Stink Bug Halyomorpha Halys (Hemiptera: Pentatomidae), and Phylogenetic Relationships of Hemipteran Suborders. Mol. Cells 2009, 28, 155–165. [Google Scholar] [CrossRef]
- Mi, Q.; Zhang, J.; Gould, E.; Chen, J.; Sun, Z.; Zhang, F. Biology, Ecology, and Management of Erthesina Fullo (Hemiptera: Pentatomidae): A Review. Insects 2020, 11, 346. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ji, Y.; Li, H.; Song, F.; Zhang, L.; Wang, M. Characterization of the Complete Mitochondrial Genome of Pentatoma Semiannulata (Hemiptera: Pentatomidae). Mitochondrial DNA Part B 2021, 6, 750–752. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Cassis, G.; Zhao, L.; He, Y.; Zhang, H.; Wei, J. The Complete Mitochondrial Genome of Zicrona caerulea (Linnaeus) (Hemiptera: Pentatomidae: Asopinae) and Its Phylogenetic Implications. Zootaxa 2020, 4747, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wei, J.; Zhao, W.; Chen, C.; Gao, X.; Zhao, Q. The Complete Mitochondrial Genome of Pentatoma Rufipes (Hemiptera, Pentatomidae) and Its Phylogenetic Implications. ZooKeys 2021, 1042, 51–72. [Google Scholar] [CrossRef]
- Hassanin, A.; Leger, N.; Deutsch, J. Evidence for Multiple Reversals of Asymmetric Mutational Constraints during the Evolution of the Mitochondrial Genome of Metazoa, and Consequences for Phylogenetic Inferences. Syst. Biol. 2005, 54, 277–298. [Google Scholar] [CrossRef]
- Jiang, P. Studies on the Comparative Mitochondrial Genomics and Phylogeny of Heteroptera (Insecta: Hemiptera). Ph.D. Thesis, China Agricultural University, Beijing, China, 2017. [Google Scholar]
- Sun, C.-H.; Zhang, Y.-N.; Zeng, X.-S.; Liu, D.-W.; Huang, Q.; Zhang, X.-L.; Zhang, Q. Mitogenome of Knodus Borki (Cypriniformes: Characidae): Genomic Characterization and Phylogenetic Analysis. Mol. Biol. Rep. 2022, 49, 1741–1748. [Google Scholar] [CrossRef]
- He, H.; Yan, B.; Yu, X.; Yang, M. Characterization and Phylogenetic Implications of Newly Sequenced Mitogenomes of Five Mileewa and Processina Species from China (Hemiptera: Cicadellidae: Mileewinae). Sci. Rep. 2022, 12, 20852. [Google Scholar] [CrossRef]
- Xu, S.; Wu, Y.; Liu, Y.; Zhao, P.; Chen, Z.; Song, F.; Li, H.; Cai, W. Comparative Mitogenomics and Phylogenetic Analyses of Pentatomoidea (Hemiptera: Heteroptera). Genes 2021, 12, 1306. [Google Scholar] [CrossRef] [PubMed]
- Lian, D.; Wei, J.; Ding, X.; Liu, Y.; Zhao, Q. Comparison and application of tRNA and rRNA genes in the mitochondrial genome of Pentatomidae (Hemiptera: Pentatomoidea). J. Fujian Agric. For. Univ. (Nat. Sci. Ed.) 2022, 51, 782–791. [Google Scholar]
- Lian, D.; Wei, J.; Chen, C.; Niu, M.; Zhang, H.; Zhao, Q. Comparative analysis and phylogeny of mitochondrial genomes of Pentatomidae (Hemiptera: Pentatomoidea). Front. Genet. 2022, 13, 1045193. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.I. Hemiptera, Pentatomidae. In Economic Insect Fauna of China; Academia Sinica, Science Press: Beijing, China, 1962; Volume 2, pp. 67–128. [Google Scholar]
- Zhao, W.; Zhao, Q.; Li, M.; Wei, J.; Zhang, X.; Zhang, H. Comparative mitogenomic analysis of the Eurydema genus in the context of representative Pentatomidae (Hemiptera: Heteroptera) taxa. J. Insect Sci. 2019, 19, 20. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, M.; Yan, J.; Bai, M.; Zhang, H. Five mitochondrial genomes of the genus Eysarcoris Hahn, 1834 with phylogenetic implications for the Pentatominae (Hemiptera: Pentatomidae). Insects 2021, 12, 597. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).