Abstract
Klebsiella oxytoca is an emerging pathogen that can cause life-threatening infectious diseases in humans. Recently, we firstly reported for the first time the presence of K. oxytoca in edible aquatic animals. In this study, we further investigated its bacterial environmental fitness and genome evolution signatures. The results revealed that K. oxytoca isolates (n = 8), originating from eight species of aquatic animals, were capable of growing under a broad spectrum of environmental conditions (pH 4.5–8.5, 0.5–6.5% NaCl), with different biofilm formation and swimming mobility profiles. The genome sequences of the K. oxytoca isolates were determined (5.84–6.02 Mb, 55.07–56.06% GC content). Strikingly, numerous putative mobile genetic elements (MGEs), particularly genomic islands (GIs, n = 105) and prophages (n = 24), were found in the K. oxytoca genomes, which provided the bacterium with specific adaptation traits, such as resistance, virulence, and material metabolism. Interestingly, the identified prophage-related clusters were derived from Burkholderia spp., Enterobacter spp., Klebsiella spp., Pseudomonas spp., and Haemophilus spp., suggesting phage transmission across Klebsiella and the other four genera. Many strain-specific (n = 10–447) genes were present in the K. oxytoca genomes, whereas the CRISPR-Cas protein-encoding gene was absent, indicating likely active horizontal gene transfer (HGT) and considerable genome variation in K. oxytoca evolution. Overall, the results of this study are the first to demonstrate the environmental compatibility and genome flexibility of K. oxytoca of aquatic animal origins.
1. Introduction
Klebsiella oxytoca was first isolated from a specimen of sour milk in 1886. The organism was classified as a member of the genus Klebsiella in 1963 [1]. K. oxytoca has been implicated in human infectious diseases, including diarrhea [2], ventriculitis [3], keratitis [4], antibiotic-associated hemorrhagic colitis [5], necrotizing enterocolitis [6], bacteremia [7], meningitis [8], spontaneous spondylodiscitis [9], and pyogenic liver abscess [10]. The bacterium also infects other organs, leading to pneumonia as well as urinary tract and skin infections [2].
Antibiotics such as β-lactam drugs can effectively control K. oxytoca infection [11]. Nevertheless, due to the inappropriate use of antimicrobials, the increasing resistance of pathogenic bacteria poses an alarming health threat and complicates options for clinical therapy [12]. Antibiotic-resistant K. oxytoca isolates have also been reported. For instance, Yang et al. [13] analyzed 5724 K. oxytoca clinical isolates from North America (n = 3501), Europe (n = 1783), the Asia–West Pacific region (n = 257), and Latin America (n = 183) reported in 2013–2019. They found that the rates of resistance to carbapenems, ceftriaxone, ciprofloxacin (CIP), colistin, and tigecycline were 1.8%, 12.5%, 7.1%, 0.8%, and 0.1%, respectively. Resistance to carbapenems was increasingly alarming [13]. To the best of our knowledge, reports in the current literature on K. oxytoca isolates of environmental or aquatic animal origins are rare. Recently, Abdurehman Damissie et al. [14] isolated and identified Klebsiella species in the gut of honey bees collected from worker honey bees (Apis mellifera) on Haramaya University bee farm in March–October of 2021 in Ethiopia. K. oxytoca was identified from 23.3% of the isolates (n = 60). The K. oxytoca isolates (n = 14) were resistant to ampicillin (54.5%), erythromycin (54.5%), and gentamycin and amoxicillin (18.2%) [14]. In our recent research, K. oxytoca was for the first time found in 14 species of aquatic animals, which were sampled in July–September of 2018–2019 in Shanghai, and Fuzhou, China. Approximately 8.0% of the K. oxytoca isolates (n = 125) displayed multidrug-resistant (MDR) phenotypes [15].
K. oxytoca is a Gram-negative bacterium and inhabits water and soil environments [16], where diverse microbial communities exist as well as pools of naturally occurring antibiotic resistance genes (ARGs). This facilitates rapid antibiotic resistance transmission via horizontal gene transfer (HGT) [17]. Previous studies have also indicated co-selection between antibiotics and heavy metals [18], due to the increasing heavy metal pollution in these environments [19]. For instance, we found that high percentages of the K. oxytoca isolates (n = 125) in aquatic animals tolerated the heavy metals Cu2+ (84.8%), Pb2+ (80.8%), Cr3+ (66.4%), Zn2+ (66.4%), and Hg2+ (49.6%) [15].
Currently, complete genome sequences of over 34 K. oxytoca isolates are available in the National Center for Biotechnology Information (NCBI) genome database (https://www.ncbi.nlm.nih.gov/, accessed on 1 October 2023). Of these, most strains were isolated from human specimens (n = 27), and only a few from the environment (n = 4). In this study, based on our recent research findings [15], we further investigated the environmental fitness and genome evolution signatures of K. oxytoca isolates originating from eight species of aquatic animals. The major objectives of this study were (1) to characterize the survival traits of the K. oxytoca isolates (n = 8) under different environmental conditions; (2) to determine the genome sequences of the K. oxytoca isolates and identify mobile genetic elements (MGEs) and virulence- and resistance-related genes in the K. oxytoca genomes; and (3) to analyze the phylogenetic relationships of the K. oxytoca isolates. The results of this study will fill prior gaps in the K. oxytoca genomes of aquatic animal origins and improve our understanding of the evolution and pathogenesis of the emerging pathogen worldwide.
2. Materials and Methods
2.1. K. oxytoca Isolates and Cultural Conditions
K. oxytoca strains 7-7-27, 8-2-3-6, 8-2-11, 8-3-38, 8-6-19, 8-8-40 8-1-12-7, and 8-11-1 were isolated from six species of crustaceans (Mytilus eduli, Sinonovacula constricta, Scapharca subcrenata, Arca granosa, Neptunea cumingi Crosse, and Anodonta woodiana); one species of shellfish (Procambarus clarkia); and one species of fish (Carassius auratus), respectively [15]. The K. oxytoca isolates (Supplementary Materials: Table S1) were identified in our recent report [15], and were stored in a −80 °C freezer in the laboratory of Shanghai Ocean University, Shanghai, China. The K. oxytoca isolates were routinely incubated in tryptic soybean broth (TSB) medium (pH 7.2, 0.5% NaCl) (Beijing Land Bridge Technology, Beijing, China) aerobically at 37 °C with shaking at 175 rpm [15].
2.2. Antibiotic Susceptibility and Heavy Metal Tolerance Assays
Antibiotic susceptibility of the K. oxytoca isolates was determined according to the disco diffusion method approved by Clinical and Laboratory Standards Institute (CLSI, M100-S28, 2018, USA) [15,20]. Mueller–Hinton (MH) agar medium and antibiotic discs were purchased from OXOID, Basingstoke, UK [15,20]. Heavy metal tolerance of the K. oxytoca isolates was examined according to broth dilution testing (microdilution, CLSI) [15,20,21]. CdCl2, CrCl3, CuCl2, HgCl2, MnCl2, NiCl2, PbCl2, and ZnCl2 (3200–3.125 μg/mL, Analytical Reagent) were purchased from Sinopharm Chemical Reagent Co., Ltd., Shanghai, China. Escherichia coli ATCC25922 and K12 strains (Institute of Industrial Microbiology, Shanghai, China) were used as quality control strains [15,20,21].
2.3. Growth Curve Assay
Growth curves of the K. oxytoca isolates were individually determined at different NaCl concentrations (0.5%, 2.5%, 4.5%, 6.5%, 8.5%) in the TSB medium (pH 7.2), or at different pH values (3.5, 4.5, 5.5, 6.5, 7.5, 8.5) in the TSB (0.5% NaCl) at 37 °C for 24 h, using a Multimode Microplate Reader (BioTek Instruments, Winooski, VT, USA) [18,22].
2.4. Swimming Mobility Analysis
The K. oxytoca isolates were individually incubated in the TSB medium (pH 8.5, 0.5% NaCl) at 37 °C until the logarithmic growth stage (LGS). The bacterial culture was inoculated onto semi-solid TSB agar plates (0.5% NaCl, pH 8.5, 0.25% agar). The agar plates were cultured at 37 °C for 72 h. The colony diameters were measured and photographed to analyze the swimming mobility of the K. oxytoca isolates [23].
2.5. Biofilm Formation Analysis
Biofilm formation was examined using the crystal violet staining method as described in our recent report [23]. Briefly, the bacterial cell culture at the LGS was inoculated into 24-well bacterial culture plates (1 mL/well), and cultured at 37 °C for 72 h, fixed and stained using the crystal violet, and eluted with 95% ethanol every 12 h. The absorbance values at OD600 were measured using the Multimode Microplate Reader (BioTek Instruments, Winooski, VT, USA), and calculated [23].
2.6. Genome Sequencing, Assembly, and Annotation
Genomic DNA of the K. oxytoca isolates was individually extracted using a TIANamp Bacteria DNA Kit (Tiangen Biochemical Technology Co., Ltd., Beijing, China) according to the manufacture’s instruction. The quality and quantity of the DNA samples for genome sequencing were controlled as described in our recent reports [18,22,23].
Genomes of the K. oxytoca isolates were sequenced by Shanghai Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China, using the Illumina Hiseq × 10 (Illumina, San Diego, CA, USA) platform. Three separately produced DNA samples were used for each of the K. oxytoca isolates. Sequence assembly, gene prediction, and Clusters of Orthologous Groups (COG) of proteins were employed using the same software as described in our recent reports [18,22,23] with default parameters. The Virulence Factor database (http://www.mgc.ac.cn/VFs, accessed on 1 October 2023) and ARGs database (http://arpcard.Mcmaster.ca, accessed on 1 October 2023) were used to detect virulence- and resistance-related genes, respectively, via the cloud platform of Shanghai Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China [18,22].
2.7. Comparative Genome Analysis
MGEs, including genomic islands (GIs), prophages, integrons (Ins), and insertion sequences (ISs), as well as CRISPR-Cas sequences, were predicted in the K. oxytoca genomes using the same software as described in our recent reports [18,22] with default parameters. Core genes shared by the eight K. oxytoca isolates, and strain-specific genes in single genomes, were predicted using the same software with default parameters [18,22].
A phylogenetic tree was constructed on the basis of 42 K. oxytoca isolates, of which the complete genome sequences of 34 K. oxytoca isolates are available in the NCBI GenBank database so far. To generate the phylogenomic tree, 1482 single-copy orthologues present in all the genomes were inferred using the OrthoFinder software (version 2.5.5) (https://doi.org/10.1186/s13059-019-1832-y, 19 October 2023). Each sequence of single-copy orthologues was aligned separately using the mafft software (version 7.520) (https://doi.org/10.1093/bioinformatics/bty121, 19 October 2023). Alignments were concatenated via Perl script (https://github.com/nylander/catfasta2phyml, 19 October 2023), and a maximum likelihood tree (1000 bootstraps) was constructed using the IQ-Tree software (version 2.2.3) (https://doi.org/10.1093/molbev/msaa015, 19 October 2023). Similarly, a phylogenetic tree was constructed on the basis of the identified prophage sequences using the MEGA software (https://www.megasoftware.net, 11 December 2023).
Multilocus sequence typing (MLST) analysis of the K. oxytoca isolates was performed on the seven conserved core genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) in K. oxytoca against the MLST database (https://cge.food.dtu.dk/services/MLST/, accessed on 1 October 2023) [20]
2.8. Statistical Analysis
The SPSS software (version 17.0, SPSS Inc., Chicago, IL, USA) was used to analyze the data. All tests were conducted in triplicate.
3. Results and Discussion
3.1. Growth of the K. oxytoca Isolates in Different NaCl and pH Conditions
K. oxytoca 7-7-27, 8-1-12-7, 8-2-3-6, 8-2-11, 8-3-38, 8-6-19, 8-8-40, and 8-11-1 isolates were recovered from eight species of edible aquatic animals, including M. edulis, P. clarkii, S. constricta, S. subcrenata, A. granosa, N. cumingi Crosse, C. auratus, and C. auratus, respectively (Table S1). The 16S rRNA gene sequencing and analysis confirmed the K. oxytoca strains, whose sequences were deposited in the NCBI GenBank database (Table S1).
The bacterial hosts P. clarkii, C. auratus, and A. woodiana were produced in freshwater, while M. edulis, A. granosa, N. cumingi Crosse, S. constricta, and S. subcrenata were produced in seawater. Therefore, we examined the growth of the K. oxytoca isolates at different NaCl concentrations (0.5–8.5% NaCl) in the TSB (pH 7.2) at 37 °C, and the results are illustrated in Figure 1 and Supplementary Figure S1. All the K. oxytoca isolates were fully inhibited at 8.5% NaCl; however, they were all capable of growing at 6.5% NaCl, but with a retardation phase (RP) of 4–7 h. Under the 4.5–0.5% NaCl conditions, the K. oxytoca isolates grew vigorously, but reached the maximum biomass at 0.5% NaCl (OD600 = 1.4).
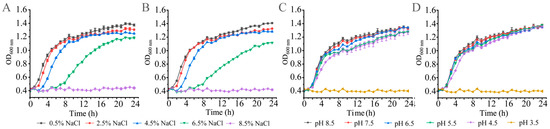
Figure 1.
Growth curves of the representative K. oxytoca isolates of aquatic animal origins under different concentrations of NaCl and pH conditions. (A,C) K. oxytoca 7-7-27; (B,D) K. oxytoca 8-11-1.
The human acidic stomach environment is a defense barrier that does not allow pathogenic bacteria to pass through. Nevertheless, current studies on the acid or alkaline adaptation of K. oxytoca are rare. Therefore, in this study, the growth of K. oxytoca isolates was also examined in different pH conditions (pH 3.5–8.5) in the TSB (0.5% NaCl) at 37 °C. As shown in Figure 1 and Supplementary Figure S2, although the growth of all the isolates was completely inhibited at pH 3.5, remarkably, the isolates were all able to grow well in acidic pH 4.5–6.5 conditions. Moreover, under alkaline conditions (pH 7.5–8.5), all the K. oxytoca isolates grew vigorously, as well, and reached maximum biomass at pH 8.5, with the OD600 values ranging from 1.31 to 1.45.
The results of this study provide the first experimental evidence for a broad spectrum of pH (4.5–8.5) conditions under which K. oxytoca isolates of aquatic animal origins were capable of survival. Moreover, the K. oxytoca isolates were all capable of growing vigorously at 0.5–6.5% NaCl. Most recently, we reported the growth traits of Klebsiella pneumoniae isolates recovered from seven species of commonly consumed aquatic animals, including M. veneriformis, Cipangopaludina cahayensis, T. granosa, Eriocheir sinensis, P. clarkii, Epinephelus fuscoguttatus, and Misgurnus anguillicaudatus [18]. These K. pneumoniae isolates were found to be able to grow at pH 4.5–7.5 and 0.5–1.0% NaCl in the TSB at 37 °C [18]. Compared with these, the results of this study highlight wide ranges of saline concentrations (0.5–6.5%) and pH (4.5–8.5) in which K. oxytoca isolates originating from aquatic animals were capable of survival, indicating their notable compatibility and fitness in their niches.
3.2. Biofilm Formation of K. oxytoca Isolates of Aquatic Animal Origins
A biofilm is a population of bacterial cells growing on a surface and is enclosed in an exopolysaccharide matrix. Biofilm formation allows bacteria to survive in hostile environments and colonize in new niches [24]. Therefore, in this study, for the first time, we examined the biofilm formation dynamics of K. oxytoca isolates of aquatic animal origins (Figure 2).
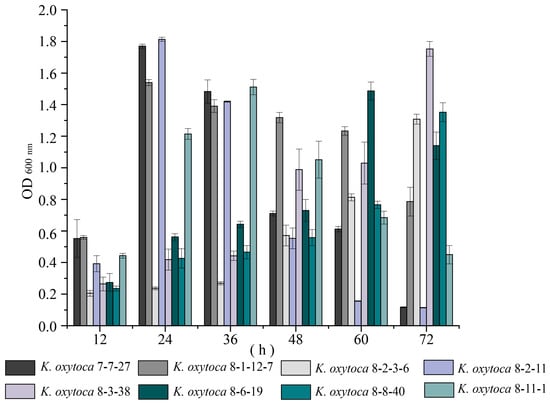
Figure 2.
Biofilm formation of K. oxytoca isolates of aquatic animal origins. The K. oxytoca 7-7-27, 8-1-12-7, 8-2-3-6, 8-2-11, 8-3-38, 8-6-19, 8-8-40, and 8-11-1 isolates were incubated in the TSB (pH 8.5, 0.5% NaCl) at 37 °C for 72 h.
As shown in Figure 2, the isolates displayed distinct biofilm formation dynamics. For example, the biofilm of K. oxytoca 7-7-27, 8-1-12-7, and 8-2-11 isolates developed rapidly, and reached their maximum biomass at 24 h; then, they decreased sharply at 48 h. Conversely, the biofilm formation of K. oxytoca 8-2-3-6, 8-3-38, and 8-8-40 isolates was relatively slower, reaching their maximum biomass at 72 h. Among all the isolates, K. oxytoca 8-2-11 from S. subcrenata showed the strongest ability to form a biofilm with the highest production at 24 h (OD600 = 1.81). It will be interesting to investigate its potential virulence in future research.
3.3. Swimming Mobility of K. oxytoca Isolates of Aquatic Animal Origins
Motility is involved in the interaction between microorganisms and their host, specifically in colonization or infectious pathogenic processes [25]. Therefore, we examined the swimming motility of the K. oxytoca isolates in the TSB (pH 8.5, 0.5% NaCl, 0.25% agar) at 37 °C for 72 h (Supplementary Materials: Figure S3).
As shown in Figure S3, swimming motility differed among the K. oxytoca isolates. For example, K. oxytoca 8-3-38 from A. granosa displayed maximum swimming diameters of 8 mm, 15.5 mm, and 21 mm at 24 h, 48 h, and 72 h, respectively. In contrast, K. oxytoca 8-8-40 swam the most slowly, with swimming diameters of 5.5 mm, 9 mm, and 10 mm at 24 h, 48 h, and 72 h, respectively, which were 0.69-fold, 0.58-fold, and 0.48-fold smaller than those of K. oxytoca 8-3-38.
3.4. Genome Features of K. oxytoca Isolates of Aquatic Animal Origins
Based on the obtained results, we further determined draft genome sequences of the eight K. oxytoca isolates using the Illumina Hiseq × 10 sequencing platform, which generated 20,195–204,452 clean single reads. The sequence assembly generated 34–90 scaffolds. The obtained genome sizes were in the range of 5,837,340–6,018,771 bp with GC contents of 55.07–56.06% (Table 1, Figure 3). The 5367–5595 protein-coding genes were predicted, of which 4713–4856 genes were classified into 24 functional catalogs in the COG database.

Table 1.
Genome features of the K. oxytoca isolates of aquatic animal origins.
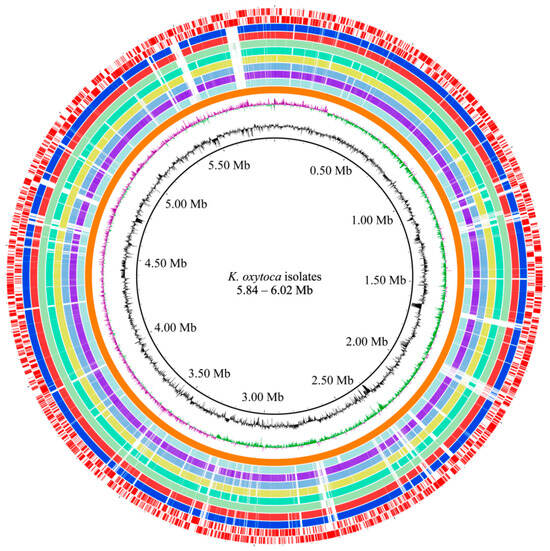
Figure 3.
Genome circle map of the eight K. oxytoca isolates of aquatic animal origins. Circles from the inside to the outside: the first circle (in black)—GC contents (outward part means higher than average, inward part means lower than average); the second circle—GC-skew (purple value is greater than zero, green value is less than zero); the third circle (in orange)—the reference genome of K. oxytoca NCTC13727; the fourth to eleventh circles—K. oxytoca 7-7-27, 8-1-12-7, 8-2-3-6, 8-2-11, 8-3-38, 8-6-19, 8-8-40 and 8-11-1 genomes, respectively; and the twelfth to thirteenth circles (in red)—CDSs on the positive and negative chains (inward and outward parts), respectively. The maps were obtained by aligning the sequences of multiple contigs obtained for the individual strains to the genome sequence of the reference strain NCTC13727. The GC content, GC-skew, and marked CDSs were applied to the NCTC13727 strain.
A typical Poisson distribution is shown in the sequencing data, indicating less repetitive DNA in the K. oxytoca genomes (Supplementary Materials: Figure S4). The obtained genomes contained 5369–5601 predicted genes, consistent with the 34 K. oxytoca strains encoding 4840–6521 predicted genes, whose complete genome sequences (5.39–6.25 Mb) were available in the GenBank database (accessed on 1 October 2023). Of these, only a few were isolated from the environment (n = 4). The results of this study enrich the K. oxytoca genome database, and fill the prior gap in the genomes of K. oxytoca of aquatic animal origins.
The draft genomes of the K. oxytoca 7-7-27, 8-1-12-7, 8-2-3-6, 8-2-11, 8-3-38, 8-6-19, 8-8-40, and 8-11-1 isolates were deposited in the GenBank database under accession numbers SAMN37879549 to SAMN37879556.
Strikingly, the K. oxytoca genomes carried a large number of putative MGEs, particularly GIs (n = 105), and prophages (n = 24), which may constitute an important driving force in the shaping and reshuffling of the K. oxytoca genome. To the best of our knowledge, reports in the current literature on the GIs and prophages in K. oxytoca are rare. Moreover, Ins (n = 2) and ISs (n = 16) were also found in the K. oxytoca isolates.
Additionally, genes of plasmid origin were identified in some contigs of the K. oxytoca genomes, suggesting possible HGT via plasmids. Moreover, small plasmid DNA was extracted from all the strains. It will be interesting to determine their sequences and evolution origin in future research.
3.5. GIs
GIs are large genomic regions (typically 10–200 Kb) that play a crucial role in bacterial genome evolution. They carry variable genes, such as antibiotic resistance and virulence genes, leading to the generation of hospital ‘superbugs’, as well as catabolic genes, leading to the formation of new metabolic pathways [26]. In this study, strikingly, a total of 105 GIs were identified in the eight K. oxytoca genomes, each of which contained 10–19 GIs in the range of 3328–99,236 bp and carrying 7–81 genes (Supplementary Materials: Figures S5 and S6, Tables S2 and S3). The genome of K. oxytoca 8-3-38 originating from A. granosa contained the maximum number of GIs (n = 19), while that of K. oxytoca 8-2-11 from S. subcrenata contained the minimum number (n = 10).
Interestingly, various function-related genes were identified in the GIs, e.g., virulence, resistance, metabolism, and phage and stress regulation. For example, GI 5 (15,235 bp) of the K. oxytoca 8-6-19 genome encoded multidrug efflux-related proteins, e.g., multidrug efflux resistance–nodulation–division (RND) transporter periplasmic adaptor subunit AcrA (K. oxytoca 8-6-19_1871), multidrug efflux RND transporter permease subunit AcrB (K. oxytoca 8-6-19_1870), and multidrug efflux transporter transcriptional repressor AcrR (K. oxytoca 8-6-19_1872). GI 11 (7106 bp) carried nine genes encoding stress-related proteins, e.g., envelope stress response membrane protein PspB (K. oxytoca 8-6-19_4104), envelope stress response membrane protein PspC (K. oxytoca 8-6-19_4105), phage shock protein operon transcriptional activator (K. oxytoca 8-6-19_4102), phage shock protein PspA (K. oxytoca 8-6-19_4103), and phage shock protein PspD (K. oxytoca 8-6-19_4106).
Notably, there were 27 identified GIs carrying virulence-related genes in the eight K. oxytoca genomes. For example, K. oxytoca 8-8-40 contained the maximum number of GIs (GI 1, GI 3, GI 4, GI 7, and GI 8) with virulence-related genes. GI 1 encoded a type II toxin–antitoxin system PemK/MazF family toxin (K. oxytoca 8-8-40_0365), and an antitoxin (K. oxytoca 8-8-40_0366); GI 3 encoded a type II toxin–antitoxin system RatA family toxin (K. oxytoca 8-8-40_1464); GI 4 encoded a type IV toxin–antitoxin system YeeU family antitoxin (K. oxytoca 8-8-40_1892), and a TA system toxin CbtA family protein (K. oxytoca 8-8-40_1893); GI 7 encoded a toxin YdaT family protein (K. oxytoca 8-8-40_2990); GI 8 encoded an inovirus Gp2 family protein (K. oxytoca 8-8-40_3456), a virulence factor SrfC family protein (K. oxytoca 8-8-40_3464), a virulence factor SrfB (K. oxytoca 8-8-40_3465), a type IV toxin-antitoxin system YeeU family antitoxin (K. oxytoca 8-8-40_3481), and a TA system toxin CbtA family protein (K. oxytoca 8-8-40_3482). Additionally, there were 14 identified GIs carrying T6SS genes, including GI 10 in K. oxytoca 7-7-27 genome; GI 6 and GI 7 in K. oxytoca 8-1-12-7; GI 8 in K. oxytoca 8-2-3-6; GI 3 and GI 4 in K. oxytoca 8-8-40; GI 4 and GI 5 in K. oxytoca 8-3-38; and GI 3 in K. oxytoca 8-11-1.
Additionally, some identified GIs carried phage regulation-related genes in the K. oxytoca genomes. For example, GI4 in K. oxytoca 8-8-40 encoded an AlpA family phage regulatory protein (K. oxytoca 8-8-40_1881), and GI 8 in K. oxytoca 8-6-19 encoded a phage regulatory CII family protein (K. oxytoca 8-6-19_2771).
3.6. Putative MGEs
3.6.1. Prophages
Prophages are viral genomes integrated into host bacterial genomes. They can confer various phenotypic traits to their hosts, such as enhanced pathogenicity [27]. In this study, a total of 24 prophages were identified in the eight K. oxytoca genomes, each of which contained 1–6 prophages (21,338–108,967 bp) carrying 13–71 genes (Supplementary Materials: Figure S7). The predicted prophages contained genetic modules involved in integration/excision, head and tail assembly, cell lysis, DNA modification, and immunity (Supplementary Materials: Tables S4 and S5).
The genome of K. oxytoca 7-7-27 from M. edulis contained the maximum number of prophage gene clusters (n = 6), which displayed sequence similarity to Burkholderia_phage_BcepC6B (42,415 bp, NCBI accession number: NC_005887), Enterobacter_phage_HK97 (39,732 bp, NCBI accession number: NC_002167), Enterobacter_phage_PsP3 (30,636 bp, NCBI accession number: NC_005340), Klebsiella_phage_phiKO2 (51,601 bp, NCBI accession number: NC_005857), Phage_phiO18P (43,101 bp, NCBI accession number: NC_009542), and Haemophilus_phage_Aaphi23 (43,033 bp, NCBI accession number: NC_004827), respectively.
The K. oxytoca 8-2-3-6 and K. oxytoca 8-2-11 genomes contained the minimum number of prophage gene clusters (n = 1), which showed sequence similarity to Klebsiella_phage_phiKO2 (51,601 bp, NCBI accession number: NC_005857) and Enterobacter_phage_P2 (33,593 bp, NCBI accession number: NC_001895), respectively.
Remarkably, the identified Klebsiella_phage_phiKO2 (46,610 bp) in K. oxytoca 8-3-38 was also found in the K. oxytoca 8-11-1, 7-7-27, 8-2-3-6, 8-8-40, and 8-1-12-7 genomes, with varying length (46,610 bp to 18,994 bp). Similarly, the identified Phage_phiO18P (43,101 bp) in K. oxytoca 7-7-27 was also found in K. oxytoca 8-6-19, but with a truncated version (27,839 bp). Additionally, the identified Enterobacter_phage_ES18 (46,900 bp, NCBI accession number: NC_006949) in K. oxytoca 8-8-40 was also present in K. oxytoca 8-1-12-7, while the identified Enterobacter_phage_P2 (33,593 bp, NCBI accession number: NC_001895) in K. oxytoca 8-1-12-7 was found in K. oxytoca 8-2-11, as well. These results suggested extensive genome rearrangement during K. oxytoca evolution.
Taken together, the identified prophages in the eight K. oxytoca genomes were derived from five different genera, including Burkholderia spp., Enterobacter spp., Haemophilus spp., Klebsiella spp., and Pseudomonas spp., indicating HGT of the phages across different genera and Klebsiella. A phylogenetic tree was constructed to show the evolutionary relationship of the identified prophages (Supplementary Materials: Figure S8). Moreover, similar prophages of different sizes, e.g., the identified Klebsiella_phage_phiKO2 in the K. oxytoca 8-3-38, 8-11-1, 7-7-27, 8-2-3-6, 8-8-40, and 8-1-12-7 genomes and the identified Phage_phiO18P in the K. oxytoca 7-7-27 and 8-6-19 genomes, were present in different K. oxytoca isolates, suggesting extensive genome rearrangement during K. oxytoca evolution. Furthermore, several prophages originating from different genera co-existed in one K. oxytoca isolate. For instance, K. oxytoca 7-7-27 contained the predicted Burkholderia_phage_BcepC6B, Enterobacter_phage_HK97, Enterobacter_phage_PsP3, Klebsiella_phage_phiKO2, Haemophilus_phage_Aaphi23, and phage_phiO18P. These results highlighted the considerable compatibility and flexibility of K. oxytoca genomes.
3.6.2. Ins
Ins are also genetic hotspots for bacterial genome evolution [28]. They have three essential core features: intI, integron-associated recombination site attI, and an integron-associated promoter Pc. The class 1 Ins are major players in the dissemination of antibiotic resistance genes across pathogens and commensals [29]. In this study, one class 1 In was identified in the K. oxytoca 8-6-19 and K. oxytoca 7-7-27 genomes, respectively, but absent from the other six genomes (Supplementary Materials: Figure S9 and Table S6).
For instance, the K. oxytoca 7-7-27 genome contained a complete In (1815 bp), encoding a trimethoprim-resistant dihydrofolate reductase DfrA14 (K. oxytoca 7-7-27_5666) and a class 1 integrase IntI 1 (K. oxytoca 7-7-27_5667).
Similarly, the K. oxytoca 8-6-19 genome also contained a complete In. However, the total length of the gene cassette array (3365 bp) was larger than that found in the K. oxytoca 7-7-27 genome, which encoded a quaternary ammonium compound efflux SMR transporter QacE delta 1 (K. oxytoca 8-6-19_5484), an AadA family aminoglycoside 3’-O-nucleotidyltransferase (K. oxytoca 8-6-19_5485), a trimethoprim-resistant dihydrofolate reductase DfrA12 (K. oxytoca 8-6-19_5486), and a class 1 integrase IntI 1 (K. oxytoca 8-6-19_5487).
Acquiring class 1 Ins can enable the development of MDR phenotypes in Gram-negative enterobacteria [30,31]. The results of this study provide additional evidence for this finding, as the K. oxytoca 7-7-27 and 8-6-19 isolates were resistant to three and four antibiotics, respectively.
3.6.3. ISs
ISs are the smallest and most numerous autonomous transposable elements in shaping host genomes [32]. In this study, ISs (n = 1 to 7) were found in the K. oxytoca 7-7-27, 8-1-12-7, 8-2-11, 8-3-38, 8-6-19, and 8-8-40 genomes, in the range of 594–1448 bp (Supplementary Materials: Table S7).
For example, the K. oxytoca 7-7-27 genome contained the maximum number of ISs (n = 7). Notably, the IS (915 bp), belonging to the IS91 family, carried a tyrosine-type DNA invertase (K. oxytoca 7-7-27_0499). This IS was also found in the K. oxytoca 8-2-11, 8-6-19, and 8-1-12-7 genomes, respectively.
Additionally, the K. oxytoca 8-3-38 genome contained only one IS (1114 bp), belonging to the IS5 family, encoding an SDR family oxidoreductase (K. oxytoca 8-3-38_3540), suggesting that this IS was a transporter ISs (tIS). The K. oxytoca 8-6-19 genome contained four ISs (IS1–IS4), among which the identified IS3 (763 bp) belonged to the IS1-family elements that contained two CDSs for the fusion transposase: the InsA (K. oxytoca 8-6-19_5478) for the proximal part of a fusion functional enzyme, and an IS1 family transposase (K. oxytoca 8-6-19_5479).
3.7. CRISPR-Cas Arrays
CRISPR-Cas is an adaptive immune system that exists in most bacteria and archaea, preventing them from being infected by foreign genetic elements [33,34]. In this study, a total of 79 CRISPR cassette arrays were identified in the eight K. oxytoca genomes; however, none of them contained the Cas protein-encoding gene, suggesting inactive CRISPR-Cas systems in these isolates (Supplementary Materials: Figure S10). Cas is an endonuclease that can cleave foreign DNA, and then, integrate it into the CRISPR array as new spacers [33]. K. oxytoca 8-6-19 had the maximum number of CRISPR cassette arrays (n = 14), while K. oxytoca 8-2-36 had the minimum (n = 6). Additionally, different repeated sequences were found in the CRISPR cassette arrays of the K. oxytoca genomes. These results provide indirect evidence for an inactive adaptive immunity system but possible active HGT in K. oxytoca isolates.
3.8. Strain-Specific Genes of the K. oxytoca Isolates of Aquatic Animal Origins
Approximately 4295 core genes were identified in the eight K. oxytoca genomes, which accounted for 74.3% of the pan genes (n = 5778). Meanwhile, many strain-specific genes (n = 10–403) were identified in the K. oxytoca genomes (Figure 4).
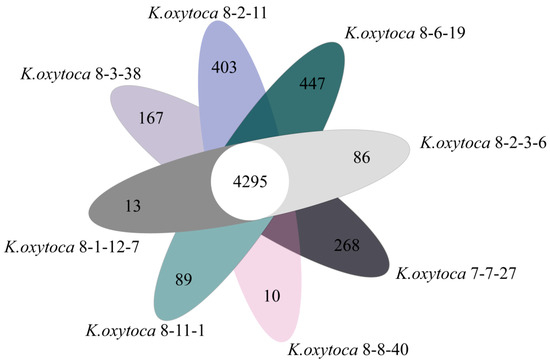
Figure 4.
Venn diagram showing the core genes and strain-specific genes in the eight K. oxytoca genomes.
The genome of K. oxytoca 8-2-11 from S. subcrenata harbored the maximum number of strain-specific genes (n = 403), whereas K. oxytoca 8-8-40 from A. woodiana had the minimum (n = 10). Notably, approximately 30.0–86.5% of the strain-specific genes encoded hypothetical proteins with unknown proteins in the current databases. These results provide additional genome-level evidence for the genome diversity of K. oxytoca isolates of aquatic animal origins.
3.9. Putative Virulence-Associated Genes in the K. oxytoca Genomes
K. oxytoca is associated with human diseases [35]. Nevertheless, few studies are currently available on the virulence of K. oxytoca of aquatic animal origins. In this study, based on the obtained K. oxytoca genomes, comparative genomic analysis revealed putative virulence-related genes (n = 97–104) in the eight K. oxytoca genomes (Supplementary Materials: Table S8). K. oxytoca 8-3-38 from A. granosa contained the maximum number of such genes (n = 104).
For example, the virulence-related fimABCDEFGHI, entABCDEFS, and mrkABCDFHJ gene clusters were present in all the K. oxytoca genomes. The former is involved in the adhesion and colonization of K. oxytoca [36], while the latter two mediate biofilm formation upon biotic and abiotic surfaces [36]. The K. oxytoca 8-2-11 and 8-6-19 genomes also contained the allABCDRS gene cluster, which provides a nitrogen source to increase the virulence of bacteria at infection sites [37]. Specifically, the sciN/tssJ gene was only found in the K. oxytoca 8-3-38 and 8-11-1 genomes, while the ureA and dotU/tssL genes were present in the K. oxytoca 8-2-11 and 8-3-38 genomes, respectively.
Additionally, the RelE, SymE, and IlpA genes, which are involved in adhesion, colonization, the secretion system, and gene regulation, were also identified in the eight isolates of aquatic animal origins. For example, the RelE protein in Escherichia colicytotoxin exhibited ribosome-binding activity in vitro, suggesting that it is an inhibitor of translation [38], while the overproduction of SymE in E. coli led to cell growth inhibition, decreased protein synthesis, and increased RNA degradation [39]. IlpA was an adhesion and immune stimulator [40], while mrkA gene expression was related to biofilm formation in carbapenemase-producing K. pneumoniae [41]. The periodontal disease-associated bacterium Porphyromonas gingivalis primarily uses FimA fimbriae for adhesion to and colonization in the gingival tissues. FimC, FimD, and FimE were associated with the fimbriae as minor components [42]. The potential virulence-related genes may be candidate targets for the development of new diagnostics, vaccines, and treatments to control K. oxytoca infection. It will be interesting to investigate their potential virulence using cell and animal models in future research.
3.10. Antibiotic and Heavy Metal Resistance-Associated Genes in the K. oxytoca Genomes and Their Bacterial Resistance Phenotypes
The increasing prevalence of infections caused by MDR pathogens poses a serious threat to global public health and places a heavy burden on health-care systems [43]. In this study, comparative genomic analysis also revealed putative antibiotic resistance-related genes (n = 27–62) in the K. oxytoca genomes (Supplementary Materials: Table S9), e.g., arcABDEF, aph3-1, arnAC, bacA, baeRS, blaOXY, cpxA, crp, emrABR, eptA, kdpE, marA, mdfA, mdtABCGHIJMN, msbA, nmpC, ompCN, oqxAB, phoE, ramA, sdiA, soxR, tolC, ugd, and yojI, which are involved in resistance to cephalosporin, fluoroquinolone, tetracycline (TET), aminoglycoside, macrolide, phenicol, sulfonamide, rifamycin, and fosfomycin. K. oxytoca 7-7-27 contained the maximum number of such genes (n = 62), whereas K. oxytoca 8-2-3-6 and 8-3-38 isolates had the fewest (n = 27).
For instance, the multidrug-effluxing or transporter-related genes were found in all the K. oxytoca genomes, e.g., membrane fusion protein of RND family multidrug efflux pump (acrA), multidrug efflux RND transporter permease subunit AcrB (acrB), multidrug efflux RND transporter permease AcrD (acrD), efflux RND transporter periplasmic adaptor subunit (acrE), efflux RND transporter permease subunit (acrF), multidrug ABC transporter permease/ATP-binding protein (yojl), lipid A ABC transporter ATP-binding protein/permease MsbA (msbA), MFS transporter (mdfA), phosphoethanolamine transferase EptA (mptA), cAMP-activated global transcriptional regulator CRP (crp), efflux RND transporter periplasmic adaptor subunit (oqxA), multidrug efflux RND transporter permease subunit OqxB (oqxB), multidrug efflux MFS transporter permease subunit EmrB (emrB), multidrug efflux MFS transporter periplasmic adaptor subunit EmrA (emrA), and transcriptional repressor MprA (emrR).
The multiple efflux transporter transcriptional reporter AcrR gene was found in the K. oxytoca 7-7-27, 8-2-11, 8-1-12-7, and 8-11-1 genomes, while the aadA, ebr, and dfrA12 genes were present in K. oxytoca 8-6-19, which encoded AadA family aminoglycoside 3″-O-nucleotidyltransferase, quaternary ammonium compound efflux SMR transporter QacE delta 1, and trimethoprim-resistant dihydrofolate reductase DfrA12, respectively.
Additionally, the aadA, ebr, and dfrA12 genes encoded AadA family aminoglycoside 3’-O-nucleotidyltransferase, quaternary ammonium compound efflux SMR transporter QacE delta 1, and trimethoprim-resistant dihydrofolate reductase DfrA12, respectively, which confer resistance to aminoglycoside [44], cephalosporin [45], and trimethoprim [46], respectively. Notably, K. oxytoca 7-7-27 from M. edulis contained the maximum number of such genes (n = 62), suggesting antibiotic exposure risk of its host M. edulis.
Heavy metal tolerance-related genes were also identified in some GIs in the K. oxytoca genomes as well. For example, the kdpE gene, encoding a transcriptional activator that is part of the two-component system KdpD/KdpE, was found in all the K. oxytoca isolates. KdpE has been identified as an adaptive regulator involved in the potassium transport, virulence, and intracellular survival of pathogenic bacteria [47].
To confirm the in silico predicted resistance genes, the resistance phenotypes of the K. oxytoca isolates were examined experimentally. The results revealed that the K. oxytoca isolates harbored different antibiotic resistance profiles (Table S1). For instance, three isolates displayed MDR phenotypes: K. oxytoca 8-6-19 isolated from N. cumingi Crosse was resistant to chloramphenicol (CHL), sulphamethoxazole-trimethoprim (SXT), kanamycin (KAN), and TET; K. oxytoca 7-7-27 from M. edulis to CHL, SXT, and TET; and K. oxytoca 8-11-1 from C. auratus to CIP, norfloxacin (NOR), and SXT. Meanwhile, the K. oxytoca isolates also displayed different heavy metal tolerance profiles. For instance, K. oxytoca 8-8-40 from A. woodiana and K. oxytoca 8-2-11 from S. subcrenata were tolerant to the maximum number of heavy metal ions evaluated in this study: Cd2+/Cr3+/Cu2+/Hg2+/Pb2+/Zn2+ and Cr3+/Cu2+/Hg2+/Mn2+/Pb2+/Zn2+, respectively.
3.11. Phylogenetic Relatedness of the K. oxytoca Isolates of Aquatic Animal Origins
Based on the eight K. oxytoca genomes obtained in this study, we constructed a phylogenetic tree, combined with 34 K. oxytoca strains, whose complete genome sequences are currently available in the GenBank database (Supplementary Materials: Table S10). The majority of these K. oxytoca strains (n = 27) were isolated from human specimens, while only a few were isolated from the environment (n = 4) and the other sources (n = 3), from the period of 1980–2022. Seven different clusters, designated as Clusters A–G, were classified (Figure 5).
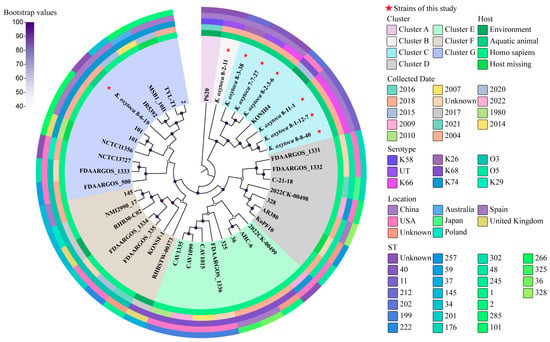
Figure 5.
A phylogenetic tree showing the relationships of the 42 K. oxytoca genomes. Complete genome sequences of the 34 K. oxytoca isolates were retrieved from the GenBank database, with accession numbers shown in Table S10. The sequenced K. oxytoca genomes in this study are marked with red stars. The maximum likelihood method was used to build the tree, with 1000 bootstrap replications and a cut-off threshold of ≥50% bootstrap values.
K. oxytoca 7-7-27, 8-1-12-7, 8-2-3-6, 8-3-38, 8-8-40, and 8-11-1 isolates, originating from M. edulis, P. clarkii, S. constricta, A. granosa, A. woodiana and C. auratus, respectively, fell into Cluster C, together with K. oxytoca KONIH4 (GenBank accession no. NZ_CP026269.1), which was isolated from waste water in 2015 in the USA.
K. oxytoca 8-2-11 from S. subcrenata was classified into a single Cluster (B), while K. oxytoca 8-6-19 from N. cumingi Crosse was grouped into Cluster G, together with the other ten K. oxytoca strains, but showed the closest phylogenetic distance to the K. oxytoca strain (GenBank accession no. NZ_CP064108.1), which was isolated from a human specimen in 2015 in China. Notably, K. oxytoca 8-6-19 was phylogenetically distant from the other seven K. oxytoca isolates of aquatic animal origins.
3.12. MLST of the K. oxytoca Isolates of Aquatic Animal Origins
The MLST analysis against the MLST database revealed that K. oxytoca 7-7-27, 8-1-12-7, 8-3-38, 8-8-40, and 8-11-1 isolates belonged to ST-40, ST-11, ST-212, ST-11, and ST-11, respectively, while K. oxytoca 8-2-3-6, 8-2-11, and 8-6-19 isolates were new STs, which have not been classified so far.
4. Conclusions
K. oxytoca is an emerging pathogen that can cause life-threatening infectious diseases in humans. Recently, we reported for the first time the presence of K. oxytoca in edible aquatic animals sampled in Shanghai and Fuzhou, China, in July–September of 2018–2019. In this study, we further investigated the environmental fitness and genome evolution signatures of such K. oxytoca isolates (n = 8), which originated from six species of crustaceans, one species of shellfish, and one species of fish. The results revealed that the K. oxytoca isolates were capable of growing under a broad spectrum of environmental conditions (pH 4.5–8.5, 0.5–6.5% NaCl) in TSB at 37 °C, indicating their remarkable compatibility and fitness in their niches. Among the isolates, K. oxytoca 8-2-11 from S. subcrenata showed the strongest capability to form a biofilm, while K. oxytoca 8-3-38 from A. granosa displayed the fastest swimming mobility.
The genome sequences of the eight K. oxytoca isolates were determined (5.84–6.02 Mb, 55.07–56.06% GC content), which contained 5367–5595 protein-encoding genes. Strikingly, numerous putative MGEs, particularly GIs (n = 105), and prophages (n = 24), were for the first time found in the K. oxytoca genomes, which provided the bacterium with specific adaptation traits, such as resistance, virulence, and material metabolism. Interestingly, the identified prophage homologues were derived from Burkholderia spp., Enterobacter spp., Klebsiella spp., Pseudomonas spp., and Haemophilus spp., suggesting phage transmission across Klebsiella and the other four genera. Moreover, some prophage homologues, such as the Klebsiella_phage_phiKO2, Enterobacter_phage_ES18, Enterobacter_phage_P2, and Phage_phiO18P, were found in different K. oxytoca genomes. Notably, several prophage homologues originating from different genera co-existed in one K. oxytoca isolate. CRISPR cassette arrays (n = 75) were also identified in the eight K. oxytoca genomes. However, no Cas protein-encoding gene was found, which provided indirect evidence for an inactive adaptive immunity system but possible active HGT in the K. oxytoca isolates. These results indicate considerable compatibility and flexibility of the bacterial genomes.
Comparative genomic analyses also revealed many ARGs (n = 27–62) and virulence (n = 97–104)-related genes in the K. oxytoca genomes. K. oxytoca 7-7-27 from M. edulis contained the highest number of ARGs (n = 97), while K. oxytoca 8-3-38 carried the most virulence-related genes (n = 104). These genes may be candidate targets for the development of new diagnostics, vaccines, and treatments to control K. oxytoca infection. In addition, numerous strain-specific (n = 10–447) genes were also present in the K. oxytoca genomes, approximately 30.0–86.5% of which encoded unknown proteins. K. oxytoca 8-6-19 from N. cumingi Crosse contained the highest number of strain-specific genes (n = 447), whereas K. oxytoca 8-8-40 from A. woodiana had the fewest (n = 10).
Overall, the results of this study enrich the K. oxytoca genome database and fill prior gaps in K. oxytoca genomes of aquatic animal origins, and also demonstrate for the first time the environmental compatibility and genome flexibility of K. oxytoca isolates.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d16010030/s1. Table S1: The resistance phenotypes of the K. oxytoca isolates used in this study; Table S2: The identified GIs in the K. oxytoca genomes; Table S3: The predicted genes in the identified GIs in the K. oxytoca genomes; Table S4: The identified prophages in the K. oxytoca genomes; Table S5: The predicted genes in the identified prophages in the K. oxytoca genomes; Table S6: The identified Ins in the K. oxytoca genomes; Table S7: The identified ISs in the K. oxytoca genomes; Table S8: The putative virulence-related genes identified in the K. oxytoca genomes; Table S9: The putative antibiotic and heavy metal resistance-related genes identified in the K. oxytoca genomes; Table S10: The thirty-four K. oxytoca strains with complete genomes used in the phylogenetic tree. Figure S1: Growth curves of the K. oxytoca isolates of aquatic animal origins under different concentrations of NaCl. A-H: K. oxytoca 7-7-27, 8-1-12-7, 8-2-3-6, 8-2-11, 8-3-38, 8-6-19, 8-8-40, and 8-11-1 isolates were incubated in the TSB (pH 7.2, 0.5–8.5% NaCl) at 37 °C for 24 h, respectively. Figure S2: Growth curves of the K. oxytoca isolates of aquatic animal origins under different pH conditions. A–H: K. oxytoca 7-7-27, 8-1-12-7, 8-2-3-6, 8-2-11, 8-3-38, 8-6-19, 8-8-40, and 8-11-1 isolates were incubated in the TSB (0.5% NaCl, pH 3.5–8.5) at 37 °C for 24 h, respectively. Figure S3: The swimming loops of the K. oxytoca isolates of aquatic animal origins. The K. oxytoca 7-7-27, 8-1-12-7, 8-2-3-6, 8-2-11, 8-3-38, 8-6-19, 8-8-40, and 8-11-1 isolates were incubated in the TSB (0.5% NaCl, pH 8.5, 0.25% agar) at 37 °C for 72 h. Figure S4: The k-mer analysis for K. oxytoca sequencing reads based on the number of unique 17-mers. A–H: K. oxytoca 7-7-27, 8-1-12-7, 8-2-3-6, 8-2-11, 8-3-38, 8-6-19, 8-8-40, and 8-11-1 genomes, respectively. Figure S5: Gene organizations of the GIs identified in the K. oxytoca genomes (A–C). Different colors refer to COG classification to mark gene functions (Figure S6). Figure S6: The COG function classification of the genes in the putative MGEs. Figure S7: Gene organizations of the prophages identified in the K. oxytoca genomes. Different colors refer to COG classification to mark gene functions (Figure S6). Figure S8: Phylogenetic relationship of the identified prophages in the K. oxytoca genomes. Figure S9: A structure diagram of the Ins identified in the K. oxytoca genomes. Figure S10: Structural features of the CRISPR cassette arrays identified in the K. oxytoca genomes. The repeat sequences are shown as rectangles in different colors, and the spacer regions are represented by rhombuses in different colors.
Author Contributions
S.S.: investigation, data curation, and writing—original draft preparation; T.G.: data analysis and writing—original draft preparation; Y.O.: writing—original draft preparation; Y.W. and L.X.: discussion and supervision; L.C.: funding acquisition, conceptualization, and writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Shanghai Municipal Science and Technology Commission, grant number 17050502200.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Draft genome sequences of the seven K. oxytoca isolates were deposited in the GenBank database under accession numbers SAMN37879549—SAMN37879556.
Acknowledgments
The authors are grateful to Lianzhi Yang at Shanghai Ocean University for his help in the manuscript preparation.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Power, J.T.; Calder, M.A. Pathogenic significance of Klebsiella oxytoca in acute respiratory tract infection. Thorax 1983, 38, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Herzog, K.A.; Schneditz, G.; Leitner, E.; Feierl, G.; Hoffmann, K.M.; Zollner-Schwetz, I.; Krause, R.; Gorkiewicz, G.; Zechner, E.L.; Högenauer, C. Genotypes of Klebsiella oxytoca isolates from patients with nosocomial pneumonia are distinct from those of isolates from patients with antibiotic-associated hemorrhagic colitis. J. Clin. Microbiol. 2014, 52, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Soto-Hernández, J.L.; Soto-Ramírez, A.; Pérez-Neri, I.; Angeles-Morales, V.; Cárdenas, G.; Barradas, V.A. Multidrug-resistant Klebsiella oxytoca ventriculitis, successfully treated with intraventricular tigecycline: A case report. Clin. Neurol. Neurosurg. 2020, 188, 105592. [Google Scholar] [CrossRef] [PubMed]
- Dago, T.R.; Zewudie, A.; Mamo, Y.; Feyissa, D.; Geleta, S. Multi-drug resistant post corneal repair Klebsiella oxytoca’s keratitis. Int. Med. Case. Rep. J. 2020, 13, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Högenauer, C.; Langner, C.; Beubler, E.; Lippe, I.T.; Schicho, R.; Gorkiewicz, G.; Krause, R.; Gerstgrasser, N.; Krejs, G.J.; Hinterleitner, T.A. Klebsiella oxytoca as a causative organism of antibiotic-associated hemorrhagic colitis. N. Engl. J. Med. 2006, 355, 2418–2426. [Google Scholar] [CrossRef] [PubMed]
- Paveglio, S.; Ledala, N.; Rezaul, K.; Lin, Q.; Zhou, Y.; Provatas, A.A.; Bennett, E.; Lindberg, T.; Caimano, M.; Matson, A.P. Cytotoxin-producing Klebsiella oxytoca in the preterm gut and its association with necrotizing enterocolitis. Emerg. Microbes. Infect. 2020, 9, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.C.; Hung, Y.P.; Lin, W.T.; Dai, W.; Huang, Y.L.; Ko, W.C. Risk factors and clinical impact of bacteremia due to carbapenem-nonsusceptible Enterobacteriaceae: A multicenter study in southern Taiwan. J. Microbiol. Immunol. Infect. 2021, 54, 1122–1129. [Google Scholar] [CrossRef] [PubMed]
- Carrie, C.; Walewski, V.; Levy, C.; Alexandre, C.; Baleine, J.; Charreton, C.; Coche-Monier, B.; Caeymaex, L.; Lageix, F.; Lorrot, M.; et al. Klebsiella pneumoniae and Klebsiella oxytoca meningitis in infants. Epidemiological and clinical features. Arch. Pediatr. 2019, 26, 12–15. [Google Scholar] [CrossRef]
- Sabio, J.M.; López-Gómez, M.; Jiménez-Alonso, J. Spontaneous spondylodiscitis caused by Klebsiella oxytoca. Ann. Rheum. Dis. 2002, 61, 758–759. [Google Scholar] [CrossRef]
- Surani, A.; Slama, E.M.; Thomas, S.; Ross, R.W.; Cunningham, S.C. Raoultella ornithinolytica and Klebsiella oxytoca pyogenic liver abscess presenting as chronic cough. IDCases 2020, 20, e00736. [Google Scholar] [CrossRef]
- Gharavi, M.J.; Zarei, J.; Roshani-Asl, P.; Yazdanyar, Z.; Sharif, M.; Rashidi, N. Comprehensive study of antimicrobial susceptibility pattern and extended spectrum beta-lactamase (ESBL) prevalence in bacteria isolated from urine samples. Sci. Rep. 2021, 11, 578. [Google Scholar] [CrossRef] [PubMed]
- Yahya Abdulla, N.; Abduljabbar Jaloob Aljanaby, I.; Hayder Hasan, T.; Abduljabbar Jaloob Aljanaby, A. Assessment of ß-lactams and carbapenems antimicrobials resistance in Klebsiella oxytoca isolated from patients with urinary tract infections in Najaf, Iraq. Arch. Razi. Inst. 2022, 77, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Long, H.; Hu, Y.; Feng, Y.; McNally, A.; Zong, Z. Klebsiella oxytoca complex: Update on taxonomy, antimicrobial resistance, and virulence. Clin. Microbiol. Rev. 2022, 35, e0000621. [Google Scholar] [CrossRef] [PubMed]
- Abdurehman Damissie, A.; Abdurahman Musa, K. Isolation, assessments of risk factors, and antimicrobial susceptibility test of Klebsiella from gut of bee in and around Haramaya University bee farm, East Hararghe, Oromia regional state, Ethiopia. Vet. Med. Int. 2022, 2022, 9460543. [Google Scholar] [CrossRef]
- Ni, L.; Xu, Y.; Chen, L. First experimental evidence for the presence of potentially virulent Klebsiella oxytoca in 14 species of commonly consumed aquatic animals, and phenotyping and genotyping of K. oxytoca isolates. Antibiotics 2021, 10, 1235. [Google Scholar] [CrossRef] [PubMed]
- Singh, L.; Cariappa, M.P.; Kaur, M. Klebsiella oxytoca: An emerging pathogen? Med. J. Armed. Forces. India. 2016, 72, S59–S61. [Google Scholar] [CrossRef] [PubMed]
- Kunhikannan, S.; Thomas, C.J.; Franks, A.E.; Mahadevaiah, S.; Kumar, S.; Petrovski, S. Environmental hotspots for antibiotic resistance genes. Microbiology Open 2021, 10, e1197. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Xie, L.; Chen, L. Survival and genome evolution signatures of Klebsiella pneumoniae isolates originated in seven species of aquatic animals. Diversity 2023, 15, 527. [Google Scholar] [CrossRef]
- Pal, A.; Bhattacharjee, S.; Saha, J.; Sarkar, M.; Mandal, P. Bacterial survival strategies and responses under heavy metal stress: A comprehensive overview. Crit. Rev. Microbiol. 2022, 48, 327–355. [Google Scholar] [CrossRef]
- Xu, Y.; Ni, L.; Guan, H.; Chen, D.; Qin, S.; Chen, L. First report of potentially pathogenic Klebsiella pneumoniae from serotype K2 in mollusk Tegillarca granosa and genetic diversity of Klebsiella pneumoniae in 14 species of edible aquatic animals. Foods 2022, 11, 4058. [Google Scholar] [CrossRef]
- Chen, D.; Li, X.; Ni, L.; Xu, D.; Xu, Y.; Ding, Y.; Xie, L.; Chen, L. First experimental evidence for the presence of potentially toxic Vibrio cholerae in snails, and virulence, cross-resistance and genetic diversity of the bacterium in 36 species of aquatic food animals. Antibiotics 2021, 10, 412. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Peng, X.; Xie, L.; Chen, L. Survival and genome diversity of Vibrio parahaemolyticus isolated from edible aquatic animals. Diversity 2022, 14, 350. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Y.; Yu, P.; Ren, S.; Zhu, Z.; Jin, Y.; Yan, J.; Peng, X.; Chen, L. Prophage-related gene VpaChn25_0724 contributes to cell membrane integrity and growth of Vibrio parahaemolyticus CHN25. Front. Cell. Infect. Microbiol. 2020, 10, 595709. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Josenhans, C.; Suerbaum, S. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 2002, 291, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Hsiao, W.W.; Brinkman, F.S. Detecting genomic islands using bioinformatics approaches. Nat. Rev. Microbiol. 2010, 8, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Rezaei Javan, R.; Ramos-Sevillano, E.; Akter, A.; Brown, J.; Brueggemann, A.B. Prophages and satellite prophages are widespread in Streptococcus and may play a role in pneumococcal pathogenesis. Nat. Commun. 2019, 10, 4852. [Google Scholar] [CrossRef]
- Engelstädter, J.; Harms, K.; Johnsen, P.J. The evolutionary dynamics of integrons in changing environments. ISME J. 2016, 10, 1296–1307. [Google Scholar] [CrossRef]
- Sabbagh, P.; Rajabnia, M.; Maali, A.; Ferdosi-Shahandashti, E. Integron and its role in antimicrobial resistance: A literature review on some bacterial pathogens. Iran. J. Basic. Med. Sci. 2021, 24, 136–142. [Google Scholar] [CrossRef]
- Ahmadian, L.; Haghshenas, M.R.; Mirzaei, B.; Bazgir, Z.N.; Goli, H.R. Distribution and molecular characterization of resistance gene cassettes containing class 1 integrons in multi-drug resistant (MDR) clinical isolates of Pseudomonas aeruginosa. Infect. Drug. Resist. 2020, 13, 2773–2781. [Google Scholar] [CrossRef]
- Ghaly, T.M.; Chow, L.; Asher, A.J.; Waldron, L.S.; Gillings, M.R. Evolution of class 1 integrons: Mobilization and dispersal via food-borne bacteria. PLoS ONE 2017, 12, e0179169. [Google Scholar] [CrossRef] [PubMed]
- Patricia, S.; Edith, G.; Mick, C. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2015, 38, 865–891. [Google Scholar] [CrossRef]
- Deveau, H.; Garneau, J.E.; Moineau, S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010, 64, 475–493. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Iwadare, T.; Kimura, T.; Sugiura, A.; Takei, R.; Kamakura, M.; Wakabayashi, S.I.; Okumura, T.; Hara, D.; Nakamura, A.; Umemura, T. Pyogenic liver abscess associated with Klebsiella oxytoca: Mimicking invasive liver abscess syndrome. Heliyon 2023, 9, e21537. [Google Scholar] [CrossRef] [PubMed]
- Araújo, B.F.; Ferreira, M.L.; Campos, P.A.; Royer, S.; Gonçalves, I.R.; da Fonseca Batistão, D.W.; Fernandes, M.R.; Cerdeira, L.T.; Brito, C.S.; Lincopan, N.; et al. Hypervirulence and biofilm production in KPC-2-producing Klebsiella pneumoniae CG258 isolated in Brazil. J. Med. Microbiol. 2018, 67, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Arena, F.; Henrici De Angelis, L.; Pieralli, F.; Di Pilato, V.; Giani, T.; Torricelli, F.; D’Andrea, M.M.; Rossolini, G.M. Draft genome sequence of the first hypermucoviscous Klebsiella quasipneumoniae subsp. quasipneumoniae isolate from a bloodstream infection. Genome Announc. 2015, 3, e00952-15. [Google Scholar] [CrossRef]
- Galvani, C.; Terry, J.; Ishiguro, E.E. Purification of the RelB and RelE proteins of Escherichia coli: RelE binds to RelB and to ribosomes. J. Bacteriol. 2001, 183, 2700–2703. [Google Scholar] [CrossRef]
- Kawano, M.; Aravind, L.; Storz, G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007, 64, 738–754. [Google Scholar] [CrossRef]
- Zheng, H.Y.; Yan, L.; Yang, C.; Wu, Y.R.; Qin, J.L.; Hao, T.Y.; Yang, D.J.; Guo, Y.C.; Pei, X.Y.; Zhao, T.Y.; et al. Population genomics study of Vibrio alginolyticus. Hereditas 2021, 43, 350–361. [Google Scholar] [CrossRef]
- Gual-de-Torrella, A.; Delgado-Valverde, M.; Pérez-Palacios, P.; Oteo-Iglesias, J.; Rojo-Molinero, E.; Macià, M.D.; Oliver, A.; Pascual, Á.; Fernández-Cuenca, F. Prevalence of the fimbrial operon mrkABCD, mrkA expression, biofilm formation and effect of biocides on biofilm formation in carbapenemase-producing Klebsiella pneumoniae isolates belonging or not belonging to high-risk clones. Int. J. Antimicrob. Agents 2022, 60, 106663. [Google Scholar] [CrossRef] [PubMed]
- Nagano, K. FimA fimbriae of the periodontal disease-associated bacterium Porphyromonas gingivalis. Yakugaku Zasshi 2013, 133, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Cerceo, E.; Deitelzweig, S.B.; Sherman, B.M.; Amin, A.N. Multidrug-resistant gram-negative bacterial infections in the hospital setting: Overview, implications for clinical practice, and emerging treatment options. Microb. Drug. Resist. 2016, 22, 412–431. [Google Scholar] [CrossRef] [PubMed]
- Frank, K.L.; Bundle, S.F.; Kresge, M.E.; Eggers, C.H.; Samuels, D.S. aadA confers streptomycin resistance in Borrelia burgdorferi. J. Bacteriol. 2003, 185, 6723–6727. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Lei, T.; Zhou, Y.; Dai, Y.; Yang, Z.; Luo, H. EBR-5, a novel variant of metallo-β-lactamase EBR from multidrug-resistant Empedobacter stercoris. Microbiol. Spectr. 2023, 12, e0003923. [Google Scholar] [CrossRef]
- Sadek, M.; Poirel, L.; Nordmann, P.; Nariya, H.; Shimamoto, T.; Shimamoto, T. Draft genome sequence of an mcr-1/IncI2-carrying multidrug-resistant Escherichia coli B1:ST101 isolated from meat and meat products in Egypt. J. Glob. Antimicrob. Resist. 2020, 20, 41–42. [Google Scholar] [CrossRef]
- Freeman, Z.N.; Dorus, S.; Waterfield, N.R. The KdpD/KdpE two-component system: Integrating K⁺ homeostasis and virulence. PLoS Pathog. 2013, 9, e1003201. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).