Risk Screening of Invasive Aquatic Species and a Survey of Fish Diversity Using Environmental DNA Metabarcoding Analysis in Shanghai
Abstract
1. Introduction
2. Materials and Methods
2.1. Risk Assessment Modeling of Non-Native Aquatic Species
2.2. Field Monitoring Using eDNA Metabarcoding Analysis
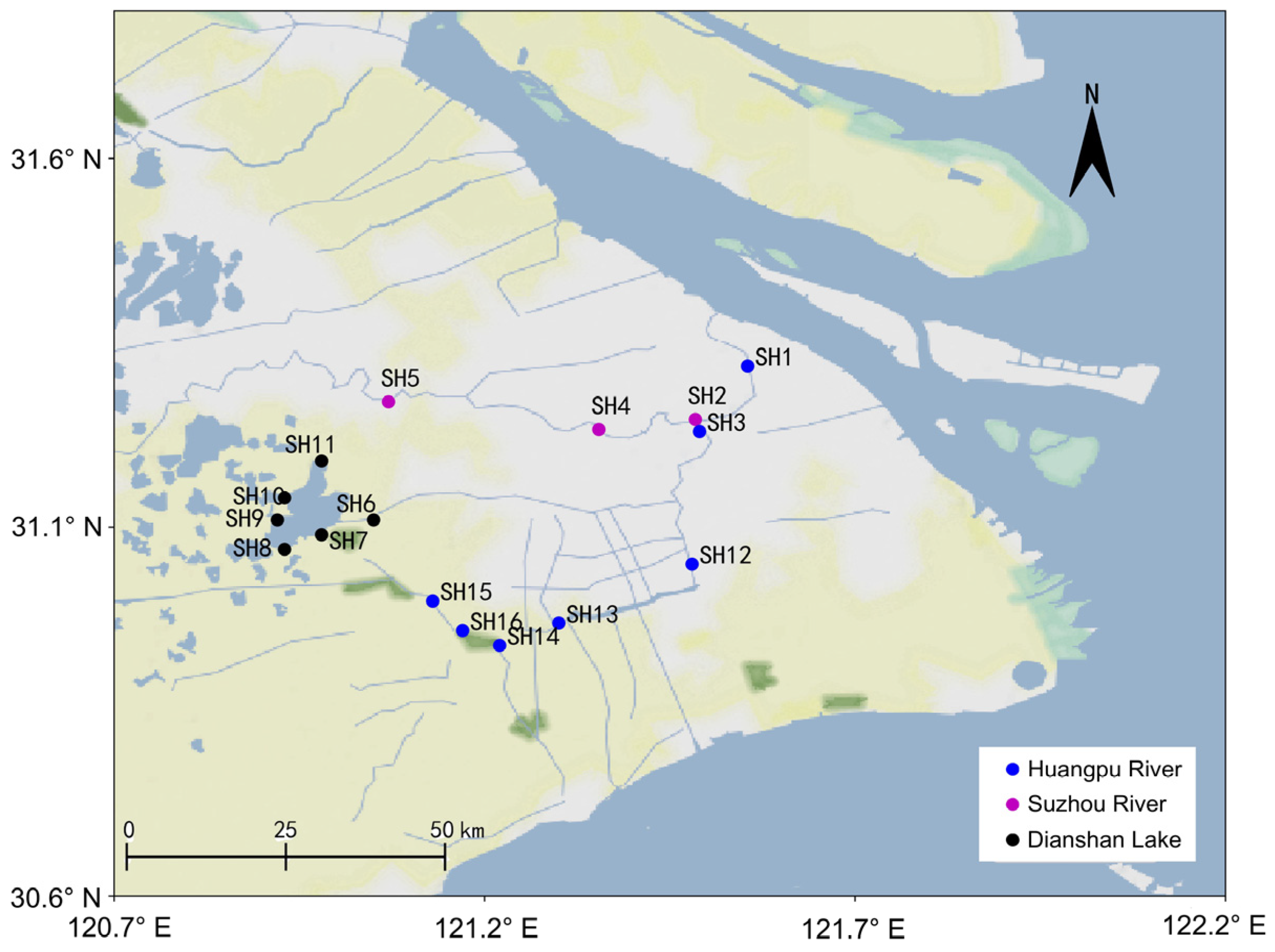
2.2.1. Water Sample Collection
2.2.2. Water Sample Processing and eDNA Extraction
2.2.3. Amplicon Library Preparation and Sequencing Run
2.2.4. Data Analysis
3. Results
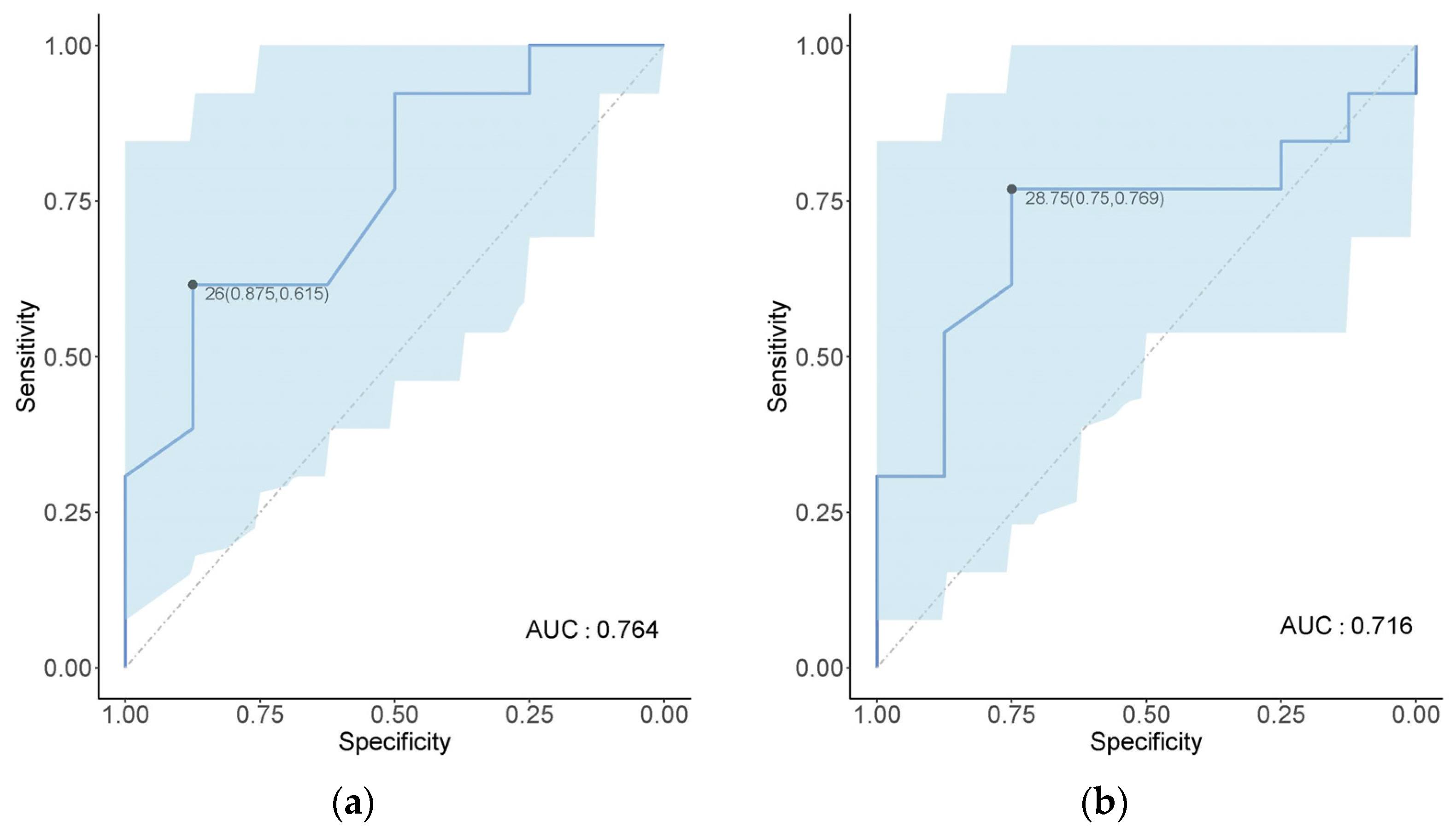
3.1. Risk Assessment of Non-Native Fish Species
3.2. eDNA Sampling Results
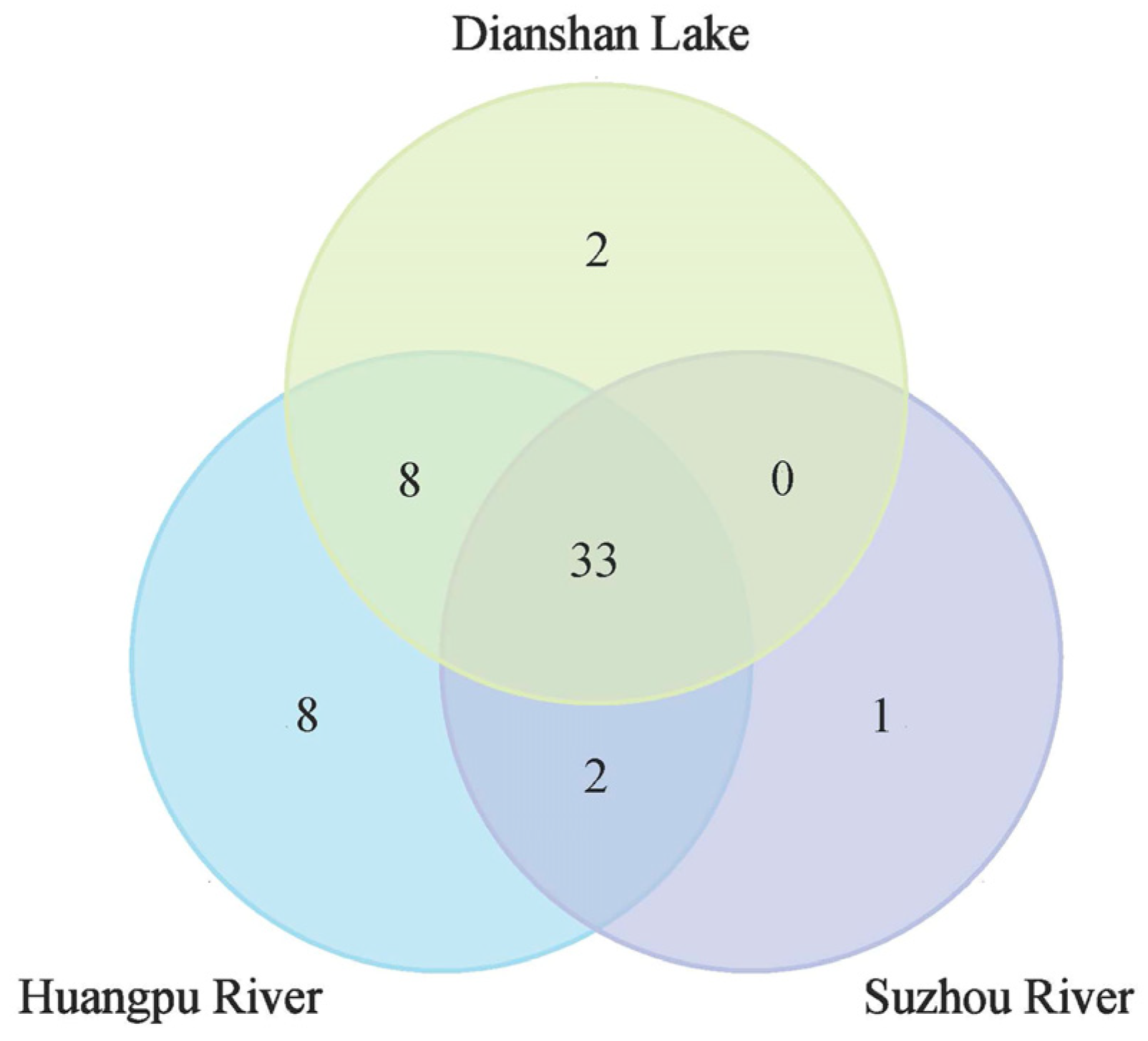
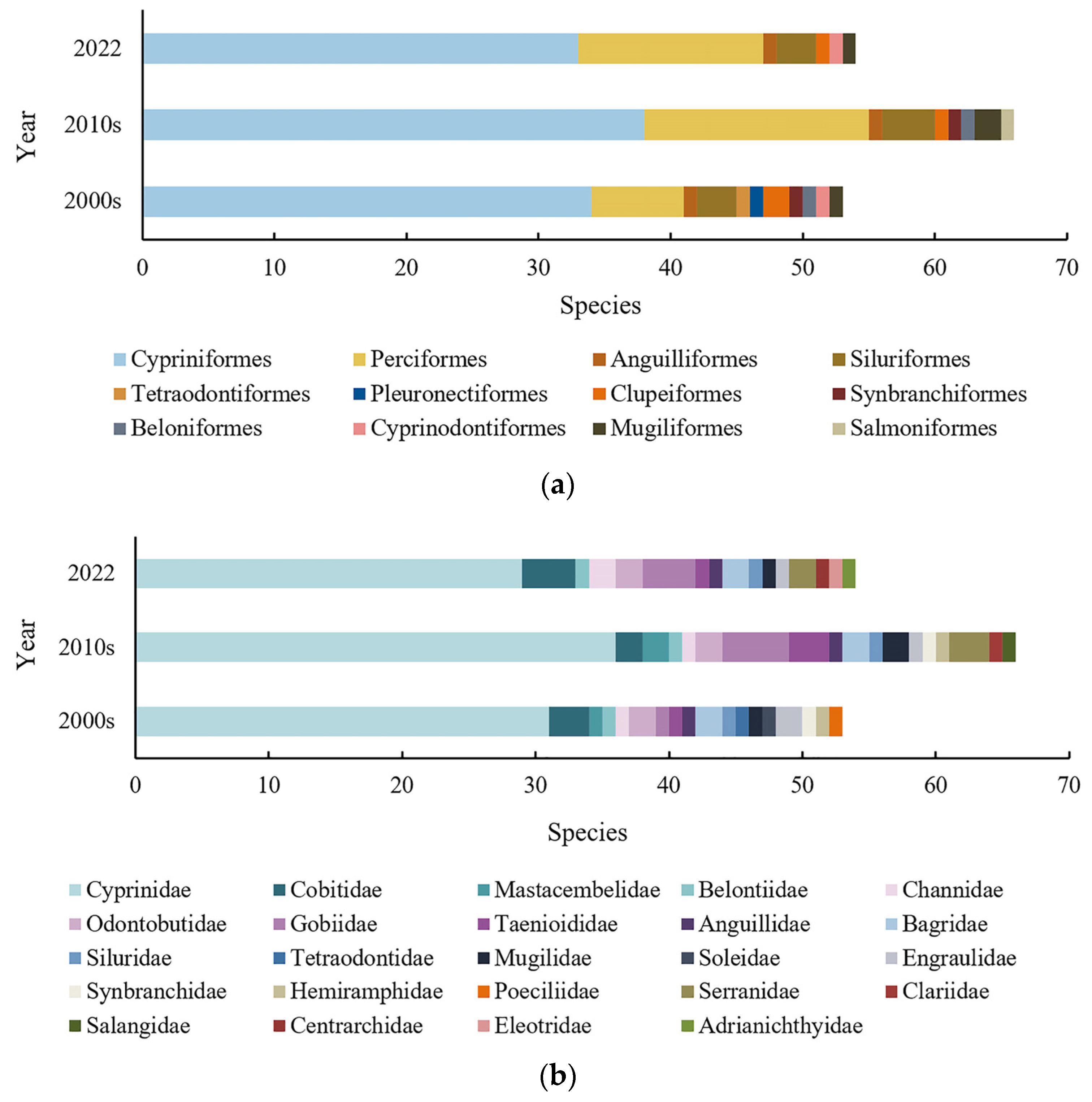
3.2.1. Fish Species Diversity in Shanghai
3.2.2. Native and Non-Native Fish Species
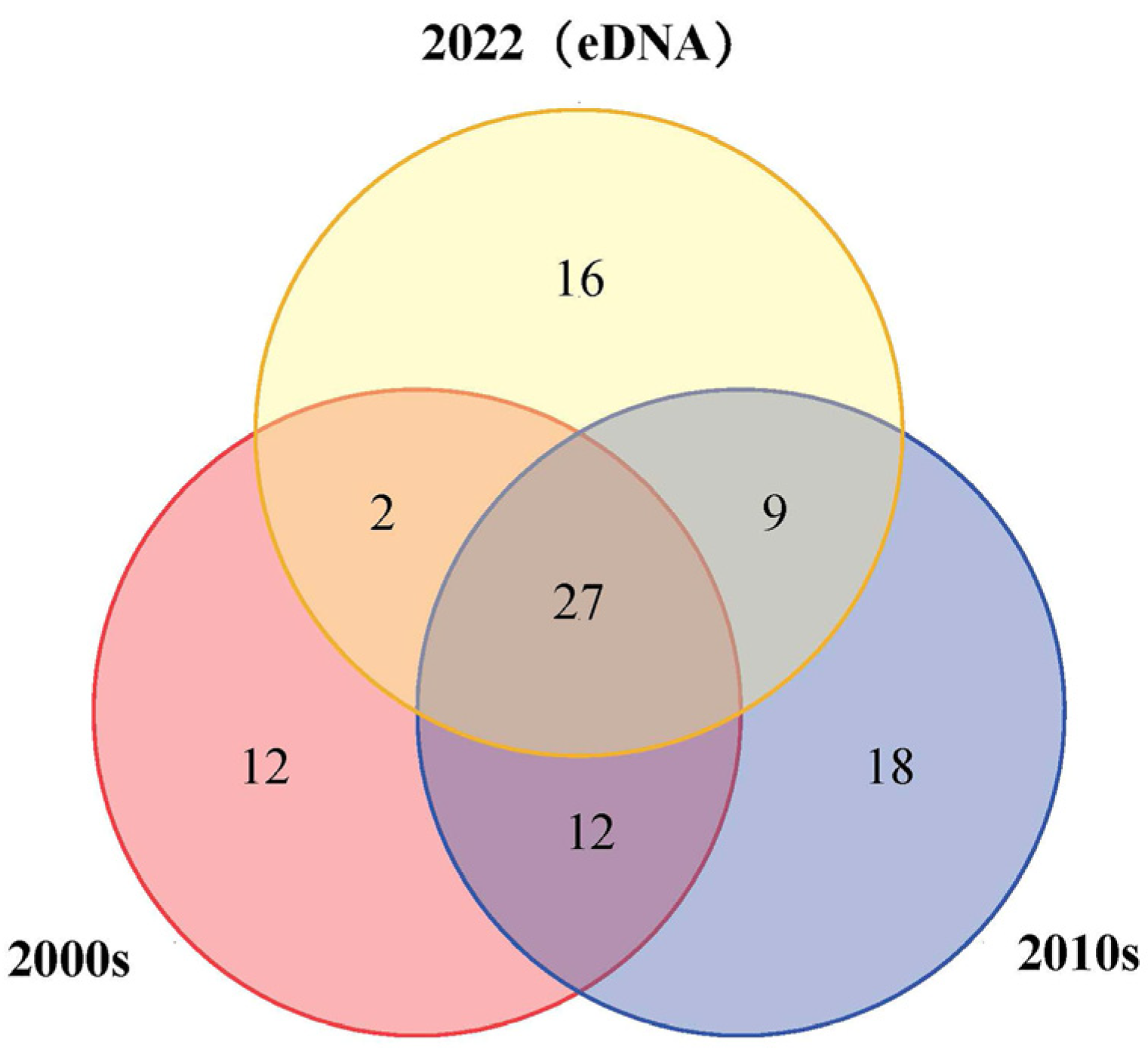
3.2.3. Comparison with Historical Data
4. Discussion
4.1. Invasion Risk Factors of Non-Native Fish Species in Shanghai
4.1.1. Introduction and Geographic Factors
4.1.2. Life History Traits
4.1.3. Climate Change
4.2. Discrepancies and Factors in Fish Diversity Survey Results between Traditional Sampling Methods and eDNA Metabarcoding Analysis
4.2.1. Fish Species Detected with eDNA Metabarcoding Analysis but Not with Traditional Methods
- (1)
- Environmental impact
- (2)
- Policy impact
- (3)
- Methodology impact
4.2.2. Fish Species Detected with Traditional Methods but Not with eDNA Metabarcoding Analysis
- (1)
- Manual identification in traditional sampling methods
- (2)
- Sampling intensity and location
4.3. Strategies for Prevention and Control of Invasive Fish Species
4.3.1. Enhancement of Regulatory Framework
4.3.2. Strengthening Responsibilities of All Parties
4.3.3. Improvement of Key Technologies
4.3.4. Raising Awareness and Education Quality
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Order | Family | Genus | Species | Dianshan Lake | Huangpu River | Suzhou River |
|---|---|---|---|---|---|---|
| Cypriniformes | Cyprinidae | Parabramis | Parabramis pekinensis | √ | √ | √ |
| Chanodichthys | Chanodichthys dabryi | √ | √ | |||
| Chanodichthys erythropterus | √ | √ | √ | |||
| Chanodichthys mongolicus | √ | √ | ||||
| Culter | Culter alburnus | √ | √ | |||
| Hemiculter | Hemiculter leucisculus | √ | √ | √ | ||
| Ctenopharyngodon | Ctenopharyngodon idella | √ | √ | √ | ||
| Megalobrama | Megalobrama terminalis | √ | ||||
| Elopichthys | Elopichthys bambusa | √ | √ | |||
| Rhynchocypris | Rhynchocypris oxycephalus | √ | √ | √ | ||
| Hemibarbus | Hemibarbus sp. | √ | √ | √ | ||
| Carassius | Carassius sp. | √ | √ | √ | ||
| Cyprinus | Cyprinus carpio | √ | √ | √ | ||
| Hypophthalmichthys | Hypophthalmichthys molitrix | √ | √ | |||
| Hypophthalmichthys nobilis | √ | √ | ||||
| Opsariichthys | Opsariichthys bidens | √ | √ | √ | ||
| Pseudorasbora | Pseudorasbora parva | √ | √ | √ | ||
| Rhodeus | Rhodeus ocellatus | √ | √ | √ | ||
| Mylopharyngodon | Mylopharyngodon piceus | √ | √ | |||
| Sarcocheilichthys | Sarcocheilichthys nigripinnis | √ | √ | √ | ||
| Sarcocheilichthys sinensis | √ | |||||
| Pseudobrama | Pseudobrama simoni | √ | √ | √ | ||
| Microphysogobio | Microphysogobio sp. | √ | √ | √ | ||
| Acheilognathus | Acheilognathus imberbis | √ | √ | √ | ||
| Acheilognathus macropterus | √ | √ | √ | |||
| Acheilognathus barbatulus | √ | √ | √ | |||
| Acheilognathus chankaensis | √ | √ | √ | |||
| Abbottina | Abbottina rivularis | √ | √ | √ | ||
| Zacco | Zacco acanthogenys | √ | √ | √ | ||
| Cobitidae | Paramisgurnus | Paramisgurnus dabryanus | √ | √ | √ | |
| Cobitis | Cobitis sp. | √ | √ | √ | ||
| Misgurnus | Misgurnus bipartitus * | √ | √ | √ | ||
| Misgurnus anguillicaudatus | √ | √ | √ | |||
| Perciformes | Belontiidae | Macropodus | Macropodus ocellatus | √ | ||
| Centrarchidae | Micropterus | Micropterus salmoides * | √ | √ | √ | |
| Serranidae | Lateolabrax | Lateolabrax maculatus | √ | |||
| Siniperca | Siniperca chuatsi | √ | ||||
| Channidae | Channa | Channa maculata * | √ | |||
| Channa argus | √ | √ | √ | |||
| Eleotridae | Eleotris | Eleotris oxycephala | √ | |||
| Odontobutidae | Odontobutis | Odontobutis potamophila | √ | √ | √ | |
| Micropercops | Micropercops swinhonis | √ | √ | √ | ||
| Gobiidae | Rhinogobius | Rhinogobius giurinus | √ | √ | √ | |
| Rhinogobius cliffordpopei | √ | √ | √ | |||
| Mugilogobius | Mugilogobius myxodermus | √ | ||||
| Tridentiger | Tridentiger bifasciatus | √ | √ | |||
| Taenioididae | Odontamblyopus | Odontamblyopus lacepedii | √ | √ | ||
| Anguilliformes | Anguillidae | Anguilla | Anguilla japonica | √ | ||
| Siluriformes | Bagridae | Tachysurus | Tachysurus fulvidraco | √ | √ | |
| Tachysurus nitidus | √ | |||||
| Siluridae | Silurus | Silurus asotus | √ | √ | √ | |
| Mugiliformes | Mugilidae | Mugil | Mugil cephalus | √ | ||
| Clupeiformes | Engraulidae | Coilia | Coilia sp. | √ | √ | √ |
| Cyprinodontiformes | Adrianichthyidae | Oryzias | Oryzias sinensis | √ | √ | √ |
Appendix B
| Order | Family | Genus | Species | 2000s | 2010s | 2022 |
|---|---|---|---|---|---|---|
| Cypriniformes | Cyprinidae | Parabramis | Parabramis pekinensis | √ | √ | √ |
| Chanodichthys | Chanodichthys dabryi | √ | √ | √ | ||
| Chanodichthys mongolicus | √ | √ | ||||
| Chanodichthys erythropterus | √ | √ | √ | |||
| Culter | Culter oxycephalus | √ | ||||
| Culter alburnus | √ | √ | ||||
| Erythroculter | Erythroculter ilishaeformis | √ | ||||
| Hemiculter | Hemiculter bleekeri | √ | √ | |||
| Hemiculter leucisculus | √ | √ | √ | |||
| Ctenopharyngodon | Ctenopharyngodon idella | √ | √ | √ | ||
| Squaliobarbus | Squaliobarbus curriculus | √ | ||||
| Megalobrama | Megalobrama mantschuricus | √ | √ | √ | ||
| Megalobrama amblycephala | √ | √ | ||||
| Elopichthys | Elopichthys bambusa | √ | ||||
| Xenocypris | Xenocypris microlepis | √ | ||||
| Xenocypris davidi | √ | |||||
| Xenocypris argentea | √ | |||||
| Rhynchocypris | Rhynchocypris oxycephalus | √ | ||||
| Hemibarbus | Hemibarbus maculatus | √ | √ | √ | ||
| Carassius | Carassius auratus | √ | √ | √ | ||
| Cyprinus | Cyprinus carpio | √ | √ | √ | ||
| Cyprinus carpio var. mirror * | √ | |||||
| Hypophthalmichthys | Hypophthalmichthys molitrix | √ | √ | √ | ||
| Hypophthalmichthys nobilis | √ | √ | √ | |||
| Opsariichthys | Opsariichthys bidens | √ | ||||
| Pseudorasbora | Pseudorasbora parva | √ | √ | √ | ||
| Rhodeus | Rhodeus lighti | √ | ||||
| Rhodeus ocellatus | √ | √ | √ | |||
| Pseudolaubuca | Pseudolaubuca engraulis | √ | √ | |||
| Pseudolaubuca sinensis | √ | √ | ||||
| Mylopharyngodon | Mylopharyngodon piceus | √ | √ | |||
| Sarcocheilichthys | Sarcocheilichthys nigripinnis | √ | √ | √ | ||
| Sarcocheilichthys sinensis | √ | √ | ||||
| Pseudobrama | Pseudobrama simoni | √ | √ | √ | ||
| Paracanthobrama | Paracanthobrama guichenoti | √ | ||||
| Toxabramis | Toxabramis swinhonis | √ | √ | |||
| Microphysogobio | Microphysogobio fukiensis | √ | ||||
| Microphysogobio microstomus | √ | |||||
| Acheilognathus | Acheilognathus taenianalis | √ | ||||
| Acheilognathus imberbis | √ | |||||
| Acheilognathus macropterus | √ | √ | √ | |||
| Acheilognathus barbatulus | √ | √ | ||||
| Acheilognathus chankaensis | √ | √ | √ | |||
| Acheilognathus tonkinensis | √ | |||||
| Distoechodon | Distoechodon hupeinensis | √ | ||||
| Distoechodon tumirostris | √ | |||||
| Squalidus | Squalidus argentatus | √ | ||||
| Abbottina | Abbottina rivularis | √ | √ | √ | ||
| Zacco | Zacco acanthogenys | √ | ||||
| Cobitidae | Paramisgurnus | Paramisgurnus dabryanus | √ | √ | √ | |
| Cobitis | Cobitis taenia | √ | √ | |||
| Misgurnus | Misgurnus bipartitus * | √ | ||||
| Misgurnus anguillicaudatus | √ | √ | √ | |||
| Perciformes | Mastacembelidae | Mastacembelus | Mastacembelus aculeatus | √ | √ | |
| Sinobdella | Sinobdella sinensis | √ | ||||
| Belontiidae | Macropodus | Macropodus ocellatus | √ | √ | √ | |
| Centrarchidae | Micropterus | Micropterus salmoides * | √ | |||
| Serranidae | Lateolabrax | Lateolabrax maculatus | √ | √ | ||
| Siniperca | Siniperca knerii | √ | ||||
| Siniperca chuatsi | √ | √ | ||||
| Channidae | Channa | Channa maculata * | √ | |||
| Channa argus | √ | √ | √ | |||
| Eleotridae | Eleotris | Eleotris oxycephala | √ | |||
| Odontobutidae | Odontobutis | Odontobutis potamophila | √ | √ | ||
| Odontobutis obscura | √ | |||||
| Micropercops | Micropercops swinhonis | √ | √ | √ | ||
| Gobiidae | Rhinogobius | Rhinogobius giurinus | √ | √ | √ | |
| Rhinogobius cliffordpopei | √ | √ | ||||
| Synechogobius | Synechogobius hasta | √ | ||||
| Mugilogobius | Mugilogobius myxodermus | √ | ||||
| Tridentiger | Tridentiger trigonocephalus | √ | ||||
| Tridentiger bifasciatus | √ | √ | ||||
| Taenioididae | Odontamblyopus | Odontamblyopus lacepedii | √ | √ | ||
| Odontamblyopus rubicundus | √ | √ | ||||
| Taenioides | Taenioides cirratus | √ | ||||
| Anguilliformes | Anguillidae | Anguilla | Anguilla japonica | √ | √ | √ |
| Siluriformes | Bagridae | Tachysurus | Tachysurus fulvidraco | √ | √ | √ |
| Tachysurus nitidus | √ | √ | √ | |||
| Siluridae | Silurus | Silurus asotus | √ | √ | √ | |
| Clariidae | Clarias | Clarias gariepinus * | √ | |||
| Tetraodontiformes | Tetraodontidae | Takifugu | Takifugu obscurus | √ | ||
| Mugiliformes | Mugilidae | Liza | Liza haematocheila | √ | ||
| Mugil | Mugil cephalus | √ | √ | √ | ||
| Pleuronectiformes | Soleidae | Cynoglossus | Cynoglossus gracilis | √ | ||
| Clupeiformes | Engraulidae | Coilia | Coilia nasus | √ | √ | √ |
| Coilia brachygnathus | √ | |||||
| Synbranchiformes | Synbranchidae | Monopterus | Monopterus albus | √ | √ | |
| Beloniformes | Hemiramphidae | Hyporhamphus | Hyporhamphus intermedius | √ | √ | |
| Salmoniformes | Salangidae | Salangichthys | Salangichthys tangkahkeii | √ | ||
| Cyprinodontiformes | Adrianichthyidae | Oryzias | Oryzias sinensis | √ | ||
| Poeciliidae | Gambusia | Gambusia affinis * | √ |
References
- Zhang, X.S.; Hill, W.G. Genetic variability under mutation selection balance. Trends Ecol. Evol. 2005, 20, 468–470. [Google Scholar] [CrossRef] [PubMed]
- Vilà, M.; Espinar, J.L.; Hejda, M.; Hulme, P.E.; Jarošík, V.; Maron, J.L.; Pergl, J.; Schaffner, U.; Sun, Y.; Pyšek, P. Ecological impacts of invasive alien plants: A meta-analysis of their effects on species, communities and ecosystems. Ecol. Lett. 2011, 14, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, J.K.; Wang, X.M. Global distribution, entry routes, mechanisms and consequences of invasive freshwater fish. Biodivers. Sci. 2016, 24, 672–685. [Google Scholar] [CrossRef]
- Gurevitch, J.; Padilla, D.K. Are invasive species a major cause of extinctions? Trends Ecol. Evol. 2004, 19, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Ehrenfeld, J.G. Ecosystem consequences of biological invasions. Annu. Rev. Ecol. Evol. Syst. 2010, 41, 59–80. [Google Scholar] [CrossRef]
- Doherty, T.S.; Glen, A.S.; Nimmo, D.G.; Ritchie, E.G.; Dickman, C.R. Invasive predators and global biodiversity loss. Proc. Natl. Acad. Sci. USA 2016, 113, 11261–11265. [Google Scholar] [CrossRef]
- Reid, W.V.; Mooney, H.A.; Cropper, A.; Capistrano, D.; Carpenter, S.R.; Chopra, K. Ecosystems and Human Well-Being, 1st ed.; Island Press: Washington, DC, USA, 2005; pp. 14–17. [Google Scholar]
- Brondízio, E.S.; Settele, J.; Diaz, S. The Global Assessment Report on Biodiversity and Ecosystem Services; IPBES: Bonn, Germany, 2019. [Google Scholar]
- Seebens, H.; Blackburn, T.M.; Dyer, E.E.; Genovesi, P.; Hulme, P.E.; Jeschke, J.M.; Pagad, S.; Pyšek, P.; Winter, M.; Arianoutsou, M.; et al. No saturation in the accumulation of alien species worldwide. Nat. Commun. 2017, 8, 14435. [Google Scholar] [CrossRef]
- Xing, W.Q.; Chen, R.S.; Lu, J.G.; Guo, X.N. lnternational progress of biological invasion research and the status quo in China: The bibliometric analysis based on CiteSpace. Acta Ecol. Sin. 2023, 43, 6912–6922. [Google Scholar]
- Ministry of Ecology and Environment of the People’s Republic of China Report on the State of the Ecology and Environment in China 2019. p. 39. Available online: https://www.mee.gov.cn/hjzl/sthjzk/zghjzkgb/202006/P020200602509464172096.pdf (accessed on 15 October 2023).
- Hu, Y.C.; Song, H.M.; Mou, X.D.; Luo, J.R. Invasion of exotic species and their control measures taken in China. J. Biosaf. 2012, 21, 256–261+242. [Google Scholar]
- Dominguez Almela, V.; Palmer, S.C.F.; Andreou, D.; Gillingham, P.K.; Travis, J.M.J.; Britton, J.R. Predicting the influence of river network configuration, biological traits and habitat quality interactions on riverine fish invasions. Divers. Distrib. 2022, 28, 257–270. [Google Scholar] [CrossRef]
- Xiao, Q.Z.; Chen, L.J.; Jin, J.J.; Qiu, Y.P.; Chen, G.Z. Ecomorphological traits explaining the competition exclusion between Oryzias and mosquitofish. Chin. J. Appl. Ecol. 2020, 31, 2087–2097. [Google Scholar]
- Zhong, L.; Yin, Y.Q.; Liao, H.; Kuang, G.X.; Yang, Y. Strictly prevent the Trachemys scripta elegans’s impact on Chongqing aquatic ecosystem. Guizhou J. Anim. Husb. Vet. Med. 2021, 45, 67–68. [Google Scholar]
- Li, B.; Xu, B.S.; Chen, J.K. Perspectives on general trends of plant invasions with special reference to alien weed flora of Shanghai. Biodivers. Sci. 2001, 9, 446–457. [Google Scholar]
- Zhang, Q.R.; Jiang, S.; Ju, R.T.; Pan, X.Y. Diversity of invasive species in Shanghai. Biodivers. Sci. 2014, 21, 732–737. [Google Scholar]
- Li, X.X.; Lv, H.; Wang, W.M.; Chen, Y.M.; Sun, P.; Wang, H.P.; Xu, X.; Yin, C.Y.; Xu, W. Review on the distribution status, hazards and control strategy of Pomacea canaliculate. Jiangxi Sci. 2023, 41, 236–243+265. [Google Scholar]
- Li, C.L. Trails on the growth speed and breeding potential of Pomacea canaliculata. Plant Prot. 1995, 21, 21. [Google Scholar]
- Carlsson, N.; Kestrup, A.; Martensson, M.; Nystrom, P. Lethal and non-lethal effects of multiple indigenous predators on the invasive golden apple snail (Pomacea canaliculata). Freshw. Biol. 2004, 49, 1269–1279. [Google Scholar] [CrossRef]
- Zhang, C.X.; Guo, J.; Zhang, J.E.; Chu, S.Y. Research progress on the interaction between alien invasive snail Pomacea canaliculata and native species. Ecol. Sci. 2017, 36, 226–235. [Google Scholar]
- Yu, H.; Bi, B.C.; Tang, W.Q.; Zhang, Y.; Guo, H.Y. Changes in fish diversity and assemblage during comprehensive restoration of the Suzhou River in Shanghai. Biodivers. Sci. 2021, 29, 32–42. [Google Scholar]
- Xia, J.H.; Lu, J.F.; Zhou, B.C. A preliminary study on fish communities in Suzhou Creek, Shanghai. J. Lake Sci. 2009, 21, 538–546. [Google Scholar]
- Copp, G.; Vilizzi, L.; Tidbury, H.; Stebbing, P.; Tarkan, A.S.; Miossec, L.; Goulletquer, P. Development of a generic decision-support tool for identifying potentially invasive aquatic taxa: AS-ISK. Manag. Biol. Invasions 2016, 7, 343–350. [Google Scholar] [CrossRef]
- Copp, G.H.; Godard, M.J.; Russell, I.C.; Peeler, E.J.; Gherardi, F.; Tricarico, E.; Miossec, L.; Goulletquer, P.; Almeida, D.; Britton, J.R.; et al. A preliminary evaluation of the European Non-native Species in Aquaculture Risk Assessment Scheme applied to species listed on Annex IV of the EU Alien Species Regulation. Fish. Manag. Ecol. 2016, 23, 12–20. [Google Scholar] [CrossRef]
- Li, S.; Chen, J.; Wang, X.; Copp, G.H. Invasiveness screening of non-native fishes for the middle reach of the Yarlung Zangbo River, Tibetan Plateau, China. River Res. Appl. 2017, 33, 1439–1444. [Google Scholar] [CrossRef]
- Li, X.; Tang, W.; Zhao, Y. Risk analysis of fish invasion in Haihe River Basin caused by the central route of the South-to-North Water Diversion Project. Biodivers. Sci. 2021, 29, 1336–1347. [Google Scholar] [CrossRef]
- Wei, H.; Chaichana, R.; Vilizzi, L.; Daengchana, P.; Liu, F.; Nimtim, M.; Zhu, Y.; Li, S.; Hu, Y.; Copp, G.H. Do non-native ornamental fishes pose a similar level of invasion risk in neighbouring regions of similar current and future climate? Implications for conservation and management. Aquat. Conserv.-Mar. Freshw. Ecosyst. 2021, 31, 2041–2057. [Google Scholar] [CrossRef]
- Tarkan, A.S.; Sari, H.M.; İlhan, A.; Kurtul, I.; Vilizzi, L. Risk screening of non-native and translocated freshwater fish species in a Mediterranean-type shallow lake: Lake Marmara (West Anatolia). Zool. Middle E. 2017, 63, 48–57. [Google Scholar] [CrossRef]
- Tarkan, A.S.; Vilizzi, L.; Top, N.; Ekmekçi, F.G.; Stebbing, P.D.; Copp, G.H. Identification of potentially invasive freshwater fishes, including translocated species, in Turkey using the Aquatic Species Invasiveness Screening Kit (AS-ISK). Int. Rev. Hydrobiol. 2017, 102, 47–56. [Google Scholar] [CrossRef]
- Bilge, G.; Filiz, H.; Yapici, S.; Tarkan, A.S.; Vilizzi, L. A risk screening study on the potential invasiveness of lessepsian fishes in the South-Eastern coasts of Anatolia. Acta Ichthyol. Piscat. 2019, 49, 23–31. [Google Scholar] [CrossRef]
- Dodd, J.A.; Vilizzi, L.; Bean, C.W.; Davison, P.I.; Copp, G.H. At what spatial scale should risk screenings of translocated freshwater fishes be undertaken—River basin district or climo-geographic designation? Biol. Conserv. 2019, 230, 122–130. [Google Scholar] [CrossRef]
- Suresh, V.R.; Ekka, A.; Biswas, D.K.; Sahu, S.K.; Yousuf, A.; Das, S. Vermiculated sailfin catfish, Pterygoplichthys disjunctivus (Actinopterygii: Siluriformes: Loricariidae): Invasion, biology, and initial impacts in East Kolkata Wetlands, India. Acta Ichthyol. Piscat. 2019, 49, 221–233. [Google Scholar] [CrossRef]
- Interesova, E.; Vilizzi, L.; Copp, G.H. Risk screening of the potential invasiveness of non-native freshwater fishes in the River Ob basin (West Siberian Plain, Russia). Reg. Environ. Chang. 2020, 20. [Google Scholar] [CrossRef]
- Haubrock, P.J.; Copp, G.H.; Johović, I.; Balzani, P.; Inghilesi, A.F.; Nocita, A.; Tricarico, E. North American channel catfish, Ictalurus punctatus: A neglected but potentially invasive freshwater fish species? Biol. Invasions 2021, 23, 1563–1576. [Google Scholar] [CrossRef]
- Paganelli, D.; Kamburska, L.; Zaupa, S.; Garzoli, L.; Boggero, A. Impacts analysis of alien macroinvertebrate species in the hydrographic system of a subalpine lake on the Italian–Swiss border. Water 2021, 13, 3146. [Google Scholar] [CrossRef]
- Paganelli, D.; Pandolfi, A.; Sconfietti, R.; Marchini, A.; Vilizzi, L. Potential invasiveness by non-indigenous macrozoobenthos in the secondary hydrographic system of a temperate-climate river catchment. Ecol. Indic. 2018, 88, 274–281. [Google Scholar] [CrossRef]
- Diao, C.Y.; Wang, W.; Xian, W.W.; Zhang, H. Role of the environmental DNA technology application in the biomass assessment of the fishery resource: Current status and future perpectives. Mar. Sci. 2022, 46, 135–144. [Google Scholar]
- Jiang, W.; Zhao, H.; Deng, J. Detection of aquatic species using environmental DNA. J. Hydroecol. 2016, 37, 1–7. [Google Scholar]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F.; et al. Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef]
- Liu, B.; Wang, H.; Qin, B. Environmental DNA metabarcoding-based monitoring of fish diversity and screening invasion risk of non-native fishes in the Beijing area. J. Biosaf. 2021, 30, 220–229. [Google Scholar]
- Jeunen, G.J.; Lipinskaya, T.; Gajduchenko, H.; Golovenchik, V.; Moroz, M.; Rizevsky, V.; Semenchenko, V.; Gemmell, N.J. Environmental DNA (eDNA) metabarcoding surveys extend the range of invasion for non-indigenous freshwater species in Eastern Europe. BioRxiv 2021. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef]
- Glamuzina, B.; Tutman, P.; Nikolić, V.; Vidović, Z.; Pavličević, J.; Vilizzi, L.; Copp, G.H.; Simonović, P. Comparison of taxon-specific and taxon-generic risk screening tools to identify potentially invasive non-native fishes in the river Neretva catchment (Bosnia and Herzegovina and Croatia). River Res. Applic. 2017, 33, 670–679. [Google Scholar] [CrossRef]
- Yamanaka, H.; Minamoto, T.; Matsuura, J.; Sakurai, S.; Tsuji, S.; Motozawa, H.; Hongo, M.; Sogo, Y.; Kakimi, N.; Teramura, I.; et al. A simple method for preserving environmental DNA in water samples at ambient temperature by addition of cationic surfactant. Limnology 2017, 18, 233–241. [Google Scholar] [CrossRef]
- Xia, Z.Q.; Gu, J.N.; Wen, Y.; Cao, X.K.; Gao, Y.C.; Li, S.G.; Haffner, G.D.; MacIsaac, H.J.; Zhan, A. eDNA-based detection reveals invasion risks of a biofouling bivalve in the world’s largest water diversion project. Ecol. Appl. 2023, e2826. [Google Scholar] [CrossRef] [PubMed]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.Q.; Sakata, M.K.; Wu, D.Y.; Yamanaka, H.; Minamoto, T. Application of environmental DNA metabarcoding in a lake with extensive algal blooms. Limnology 2021, 22, 363–370. [Google Scholar] [CrossRef]
- Hamady, M.; Walker, J.J.; Harris, J.K.; Gold, N.J.; Knight, R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 2008, 5, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Jeunen, G.J.; Lamare, M.D.; Knapp, M.; Spencer, H.G.; Taylor, H.R.; Stat, M.; Bunce, M.; Gemmell, N.J. Water stratification in the marine biome restricts vertical environmental DNA (eDNA) signal dispersal. Environ. DNA 2020, 2, 99–111. [Google Scholar] [CrossRef]
- Sun, J.Y.; Dai, X.J.; Zhu, J.F. Analysis of the fish species diversity in Dianshan Lake. J. Shanghai Fish. Univ. 2007, 16, 454–459. [Google Scholar]
- Wang, K. Temporal and Spatial Distrubition of Resources, and Trophic Levels of Main Species in the Dianshan Lake. Master’s Thesis, Shanghai Ocean University, Shanghai, China, 2017. [Google Scholar]
- Chen, X.H.; Li, X.P.; Cheng, X. Spatial-temporal distribution of fish assemblages in the upstreams of Huangpu River and Suzhou Creek. Biodivers. Sci. 2008, 16, 191. [Google Scholar]
- Zhou, T.S.; Zhang, Y.; Tang, W.Q.; Wang, L.Q. Ecological health assessment of Huangpu River based on fish index of biotic integrity. Resour. Environ. Yangtze Basin 2016, 25, 895–903. [Google Scholar]
- Chen, D.H.; Chen, Z.; Lin, J.B.; Qin, Z.Q.; Liang, P.; Li, X.G.; Qiu, M.L. Current situation and prospect of largemouth bass industry in Fujian Province. J. Aquacult. 2020, 41, 75–77. [Google Scholar]
- Bai, J.J.; Li, S.J. Current status and development trend on China largemouth bass industry. Chin. Fish. Econ. 2013, 31, 104–108. [Google Scholar]
- Gu, D.E.; Wang, J.W.; Xu, M.; Mu, X.D.; Wei, H.; Yu, F.D.; Fang, M.; Wang, X.J.; Song, H.M.; Yang, Y.X.; et al. Does aquaculture aggravate exotic fish invasions in the rivers of southern China? Aquaculture 2022, 547, 737492. [Google Scholar] [CrossRef]
- Hussein, G.H.G.; Chen, M.; Qi, P.P.; Cui, Q.K.; Yu, Y.; Hu, W.H.; Tian, Y.; Fan, Q.X.; Gao, Z.X.; Feng, M.W.; et al. Aquaculture industry development, annual price analysis and out-of-season spawning in largemouth bass Micropterus salmoides. Aquaculture 2020, 519, 734901. [Google Scholar] [CrossRef]
- Fang, K.; Zhang, Y.Y.; Shen, L.; Zhou, Q.; Shao, J.; Xu, Q.Q.; Gorfine, H.; Zhang, H. Increasing invasion risk from the northward expansion of largemouth bass (Micropterus salmoides) in china under multiple influences. Appl. Ecol. Environ. Res. 2023, 21, 835–852. [Google Scholar] [CrossRef]
- Peel, M.C.; Finlayson, B.L.; McMahon, T.A. Updated world Köppen-Geiger climate classification map. Hydrol. Earth Syst. Sci. 2007, 11, 259–263. [Google Scholar] [CrossRef]
- Wan, F.H.; Hou, Y.M.; Jiang, M.X. Invasion Biology, 1st ed.; Science Press: Beijing, China, 2015; pp. 19–41. [Google Scholar]
- Gross, M.R.; Sargent Craig, R. The evolution of male and female parental care in fishes. Am. Zool. 1985, 25, 807–822. [Google Scholar] [CrossRef]
- Liu, C.L.; Comte, L.; Olden, J.D. Heads you win, tails you lose: Life-history traits predict invasion and extinction risk of the world’s freshwater fishes. Aquat. Conserv. Mar. Freshw. Ecosyst. 2017, 27, 773–779. [Google Scholar] [CrossRef]
- Staub, B.P.; Hopkins, W.A.; Novak, J.; Congdon, J.D. Respiratory and reproductive characteristics of eastern mosquitofish (Gambusia holbrooki) inhabiting a coal ash settling basin. Arch. Environ. Contam. Toxicol. 2004, 46, 96–101. [Google Scholar] [CrossRef]
- Swingle, H.S.; Smith, E.V. Factors Affecting the Reproduction of Bluegill Bream and Largemouth Black Bass in Ponds; Alabama Polytechnic Institute: Auburn, AL, USA, 1943; Volume 87. [Google Scholar]
- Yan, Y.Z.; Chen, Y.F.; Tao, J. Ecological invasion of Gambusia affinis: A review. Chin. J. Ecol. 2009, 28, 950–958. [Google Scholar]
- Karp, C.A.; Tyus, H.M. Behavioral interactions between young Colorado squawfish and six fish species. Copeia 1990, 25–34. [Google Scholar] [CrossRef]
- Hoover, J.J.; Murphy, C.E.; Killgore, J. Ecological impacts of suckermouth catfishes (Loricariidae) in North America: A conceptual model. Aquat. Nuis. Species Res. Program 2014, 14, 1–13. [Google Scholar]
- Hellmann, J.J.; Byers, J.E.; Bierwagen, B.G.; Dukes, J.S. Five potential consequences of climate change for invasive species. Conserv. Biol. 2008, 22, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Britton, J.R.; Cucherousset, J.; Davies, G.D.; Godard, M.J.; Copp, G.H. Non-native fishes and climate change: Predicting species responses to warming temperatures in a temperate region. Freshw. Biol. 2010, 55, 1130–1141. [Google Scholar] [CrossRef]
- Li, B.; Zhou, T.J. Projected climate change over China under IPCC A1B scenario: Multi-model ensemble and uncertainties. Clim. Chang. Res. 2010, 6, 270–276. [Google Scholar]
- Zanatta, A.S.; Ramos, I.P.; Silva, R.J. Pisces, Siluriformes, Ictaluridae, Ictalurus punctatus (Rafinesque, 1818): First record in middle Paranapanema river reservoir, aquaculture and exotic species dispersion. Check List 2010, 6, 3069–3071. [Google Scholar] [CrossRef][Green Version]
- Chang, Z.C.; Wen, H.S.; Zhang, M.Z. Effects of dissolved oxygen levels on oxidative stress response and energy utilization of juvenile Chinese sea bass (Lateolabrax maculatus) and associate physiological mechanisms. Period. Ocean Univ. China 2018, 48, 20–28. [Google Scholar]
- Song, Y.D.; Zhang, M.D.; Zhou, H.T. Effects of different dissolved oxygen levels on respiratory metabolism and related gene expression in mandarinfish Siniperca chuatsi. Fish. Sci. 2022, 41, 438–444. [Google Scholar]
- Huang, S.L.; Dai, X.J.; Chen, Q. Current situation and existing problems of aquatic species enhancement and releasing in Shanghai water area. Chin. Fish. Econ. 2009, 4, 79–87. [Google Scholar]
- Li, H.X.; Huang, X.N.; Li, S.G.; Zhan, A.B. Environmental DNA (eDNA)-metabarcoding-based early monitoring and warning for invasive species in aquatic ecosystems. Biodivers. Sci. 2019, 27, 491–504. [Google Scholar]
- Xiong, W.; Li, H.T.; Zhan, A.B. Early detection of invasive species in marine ecosystems using high-throughput sequencing: Technical challenges and possible solutions. Mar. Biol. 2016, 163. [Google Scholar] [CrossRef]
- Zhan, A.B.; MacIsaac, H.J. Rare biosphere exploration using high-throughput sequencing: Research progress and perspectives. Conserv. Genet. 2015, 16, 513–522. [Google Scholar] [CrossRef]
- Liu, S.; Zhu, X.P.; Chen, K.C. Morphological variations of Channa maculata, Channa argus and their hybrid (C. maculata♀ × C. argus♂). J. Huazhong Agric. Univ. 2011, 30, 488–493. [Google Scholar]
- Lu, M.J.; Liu, H.B.; Jiang, T. Preliminary investigations on otolith microchemistry of Odontamblyopus rubicundus in the Daliao River Estuary, China. Mar. Fish. 2015, 37, 310–317. [Google Scholar]
- Liu, M.; Shi, H.T. The improvement of the prevention and control mechanism of the risk of alien species invasion in China. J. Fuyang Inst. Technol. 2023, 34, 97–101. [Google Scholar]
- Wang, R.; Huang, H.S.; Zhang, H.B. Analysis of gaps in regulations and management mechanisms for the prevention and control of invasive alien species in China. Plant Prot. 2022, 48, 2–9. [Google Scholar]
- Liu, R. Research on legal provisions on invasive alien species. L. Vision 2017, 109–110. [Google Scholar] [CrossRef]
- Dou, Y.; Wu, J.; Huang, C. Risk assessment system and method for invasion of alien fishes. J. Ecol. Rural Environ. 2011, 27, 12–16. [Google Scholar]
- Zhou, M.H.; Ding, Z.P.; Wang, M.S. Review on the current status of prevention and control of invasive alien species in China. Plant Quar. 2023, 37, 1–7. [Google Scholar]


| Species | Outcome | Confidence Factor | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Criteria | Common Name | Scientific Name | BRA Score | Invasion Risk | BRA + CCA Score | Invasion Risk | BRA | CCA | BRA + CCA |
| 1 | Mosquito fish | Gambusia affinis | 32 | high | 32 | high | 0.55 | 0.25 | 0.52 |
| 1 | Channeled applesnail | Pomacea canaliculata | 32 | high | 40 | high | 0.53 | 0.25 | 0.50 |
| 3 | Bluegill | Lepomis macrochirus | 31 | high | 33 | high | 0.56 | 0.25 | 0.53 |
| 3 | Redbelly tilapia | Coptodon zillii | 31 | high | 41 | high | 0.59 | 0.33 | 0.56 |
| 3 | Green sunfish | Lepomis cyanellus | 29.5 | high | 35.5 | high | 0.56 | 0.25 | 0.53 |
| 1 | Red swamp crayfish | Procambarus clarkii | 29.5 | high | 29.5 | high | 0.55 | 0.25 | 0.51 |
| 2 | Bullfrog | Rana catesbeiana | 28.5 | high | 34.5 | high | 0.54 | 0.25 | 0.50 |
| 4 | Suckermouth catfish | Hypostomus plecostomus | 28 | high | 36 | high | 0.47 | 0.38 | 0.46 |
| 4 | Largemouth black bass | Micropterus salmoides | 27 | high | 39 | high | 0.60 | 0.25 | 0.56 |
| 4 | Red-eared slider | Trachemys scripta elegans | 25 | low | 25 | low | 0.47 | 0.25 | 0.45 |
| 2 | North African catfish | Clarias gariepinus | 24 | low | 32 | high | 0.55 | 0.25 | 0.51 |
| 4 | American shad | Alosa sapidissima | 23.5 | low | 23.5 | low | 0.54 | 0.25 | 0.50 |
| 4 | Channel catfish | Ictalurus punctatus | 23.5 | low | 31.5 | high | 0.49 | 0.33 | 0.48 |
| 4 | Pirapitinga | Piaractus brachypomus | 23.5 | low | 33.5 | high | 0.58 | 0.25 | 0.55 |
| 4 | Amur sturgeon | Acipenser schrenckii | 21 | low | 17 | low | 0.51 | 0.29 | 0.49 |
| 4 | Blue tilapia | Oreochromis aureus | 20.5 | low | 22.5 | low | 0.58 | 0.25 | 0.55 |
| 3 | Red drum | Sciaenops ocellatus | 20 | low | 28 | low | 0.49 | 0.25 | 0.46 |
| 4 | European eel | Anguilla anguilla | 20 | low | 24 | low | 0.51 | 0.25 | 0.48 |
| 2 | Rainbow trout | Oncorhynchus mykiss | 18.5 | low | 10.5 | low | 0.56 | 0.38 | 0.54 |
| 4 | Tench | Tinca tinca | 15.5 | low | 21.5 | low | 0.53 | 0.33 | 0.50 |
| 4 | Roho labeo | Labeo rohita | 15 | low | 13 | low | 0.53 | 0.25 | 0.50 |
| Drainage Basin | Site | Species | ||
|---|---|---|---|---|
| Channa maculata | Misgurnus bipartitus | Micropterus salmoides | ||
| Huangpu River | SH1 | √ | √ | √ |
| SH3 | √ | √ | ||
| SH12 | √ | √ | ||
| SH13 | √ | √ | ||
| SH14 | √ | √ | ||
| SH15 | √ | |||
| SH16 | √ | √ | ||
| Suzhou River | SH2 | √ | ||
| SH4 | ||||
| SH5 | √ | √ | ||
| Dianshan Lake | SH6 | √ | ||
| SH7 | √ | √ | ||
| SH8 | √ | √ | ||
| SH9 | √ | |||
| SH10 | √ | √ | ||
| SH11 | √ | √ | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, R.; Wu, Q.; Li, F.; Zhan, A.; Zhou, J.; Li, S. Risk Screening of Invasive Aquatic Species and a Survey of Fish Diversity Using Environmental DNA Metabarcoding Analysis in Shanghai. Diversity 2024, 16, 29. https://doi.org/10.3390/d16010029
Yu R, Wu Q, Li F, Zhan A, Zhou J, Li S. Risk Screening of Invasive Aquatic Species and a Survey of Fish Diversity Using Environmental DNA Metabarcoding Analysis in Shanghai. Diversity. 2024; 16(1):29. https://doi.org/10.3390/d16010029
Chicago/Turabian StyleYu, Ruohan, Qianqian Wu, Fan Li, Aibin Zhan, Jinxin Zhou, and Shan Li. 2024. "Risk Screening of Invasive Aquatic Species and a Survey of Fish Diversity Using Environmental DNA Metabarcoding Analysis in Shanghai" Diversity 16, no. 1: 29. https://doi.org/10.3390/d16010029
APA StyleYu, R., Wu, Q., Li, F., Zhan, A., Zhou, J., & Li, S. (2024). Risk Screening of Invasive Aquatic Species and a Survey of Fish Diversity Using Environmental DNA Metabarcoding Analysis in Shanghai. Diversity, 16(1), 29. https://doi.org/10.3390/d16010029







