Elmis syriaca (Kuwert, 1890) and E. zoufali (Reitter, 1910) (Coleoptera: Elmidae) confirmed as distinct species based on molecular data, morphology and geographical distribution
Abstract
1. Introduction
2. Material and Methods
2.1. Morphological Examination of Specimens
2.2. DNA Extraction, Amplification and Sequencing
2.3. Sequence Data Analyses
3. Results
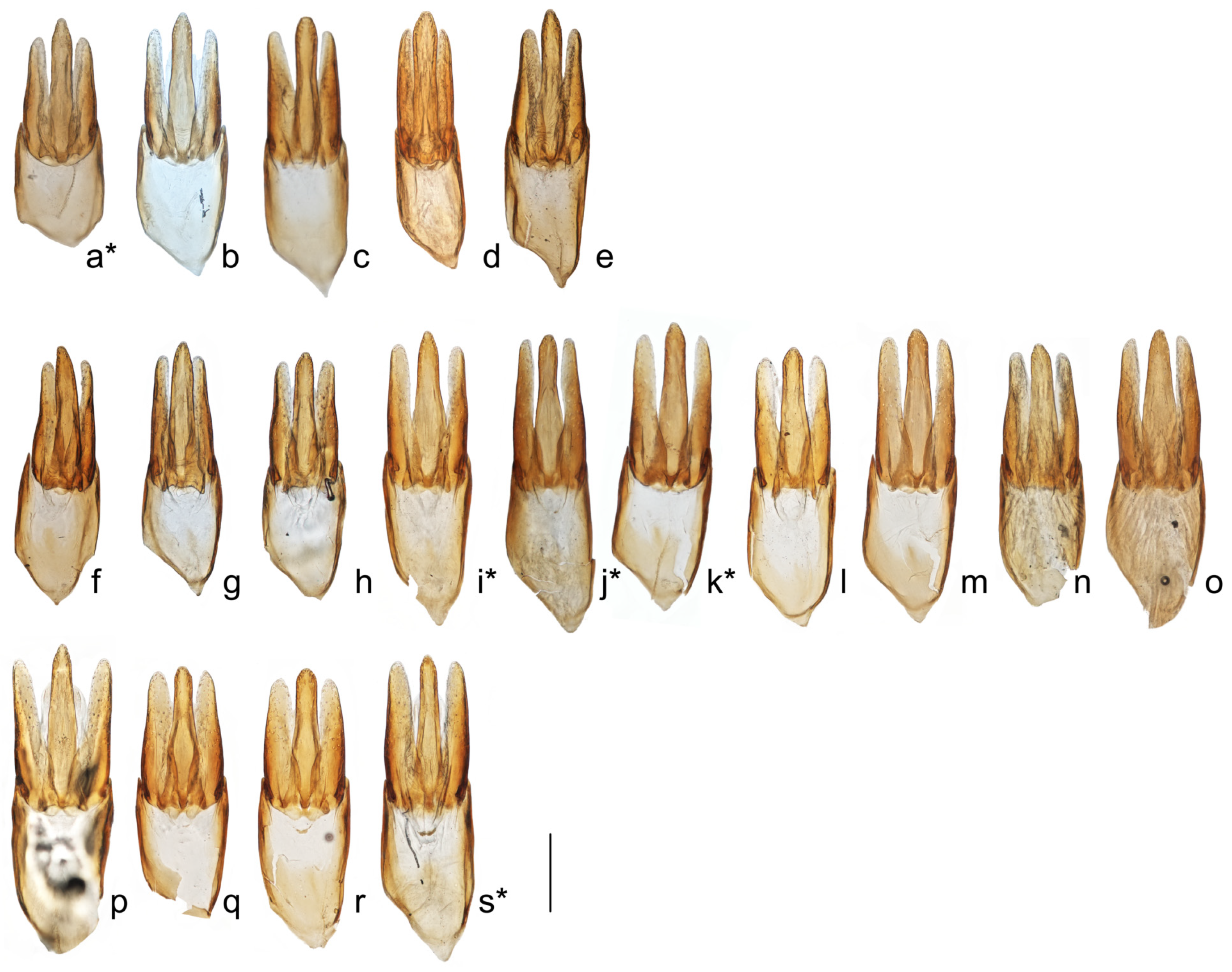
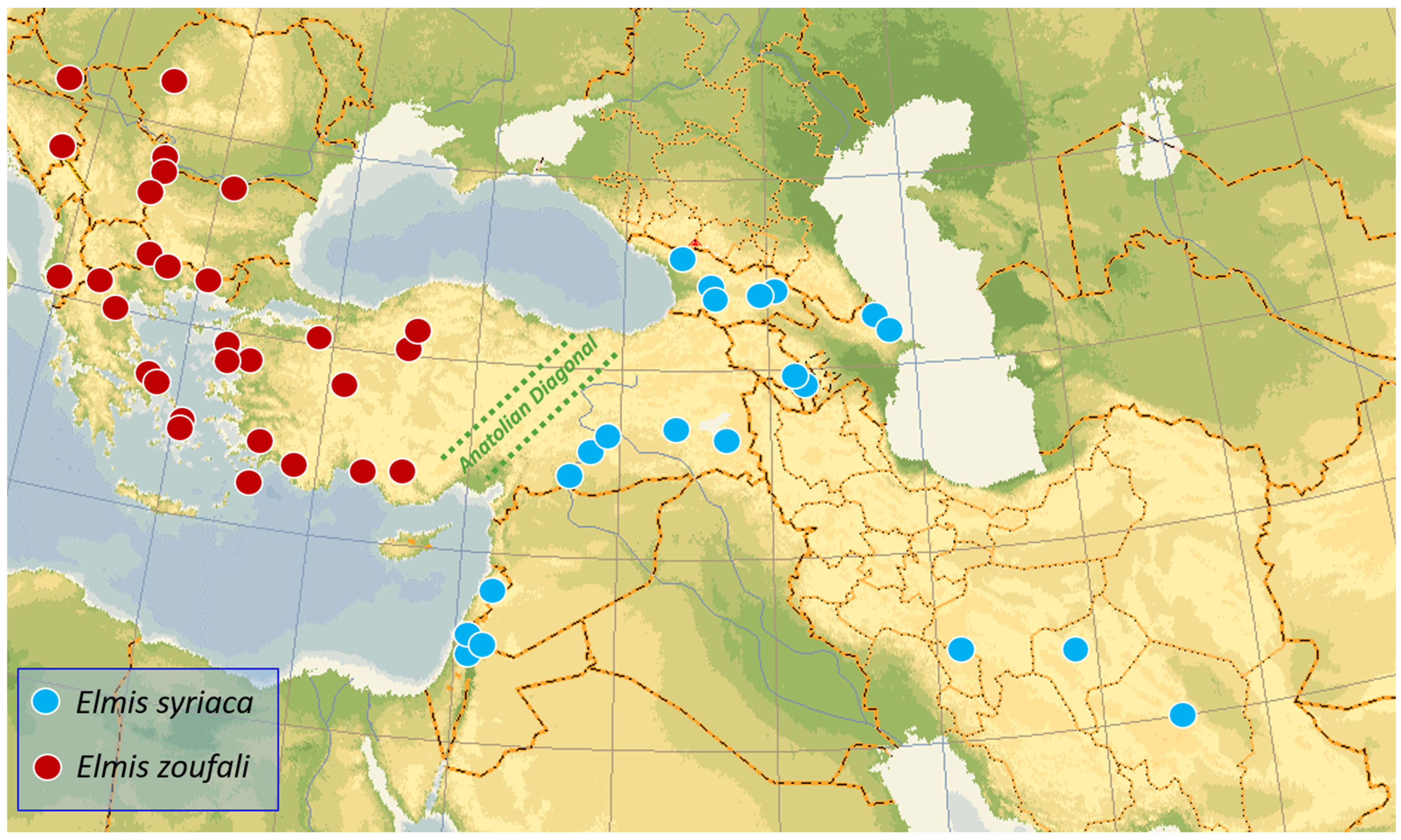
3.1. Morphology and Distribution
3.2. Identification, Delimitation and Divergence Time Calibration Based on Molecular Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B Biol. Sci. 2003, 270(1512), 313–321. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7(3), 355–364. [Google Scholar] [CrossRef] [PubMed]
- Trizzino, M.; Jäch, M.A.; Audisio, P.; Ribera, I. Molecular and morphological analyses confirm two new species of the Hydraena emarginata–saga clade (Coleoptera, Hydraenidae) from Spain and France. Zootaxa 2011, 2760(1), 29–38. [Google Scholar] [CrossRef]
- Villastrigo, A.; Jäch, M.A.; Cardoso, A.; Valladares, L.F.; Ribera, I. A molecular phylogeny of the tribe Ochthebiini (Coleoptera, Hydraenidae, Ochthebiinae). Syst. Entomol. 2019, 44(2), 273–288. [Google Scholar] [CrossRef]
- Kobayashi, T.; Hayashi, M.; Kamite, Y.; Sota, T. Molecular phylogeny of Elmidae (Coleoptera: Byrrhoidea) with a focus on Japanese species: implications for intrafamilial classification. Syst. Entomol. 2021, 46(4), 870–886. [Google Scholar] [CrossRef]
- Čiampor, F.; Kodada, J. Taxonomy of the Oulimnius tuberculatus species group (Coleoptera: Elmidae) based on molecular and morphological data. Zootaxa 2010, 2670(1), 59–68. [Google Scholar] [CrossRef]
- Čiampor, F.; Laššová, K.; Maier, C.A.; Čiamporová-Zaťovičová, Z.; Kodada, J. Phanoceroides Hinton, 1939: Description of new species, morphology of larvae, and revised taxonomic position of the genus (Coleoptera: Elmidae) based on molecular evidence. Zootaxa 2016, 4117(2), 277–288. [Google Scholar] [CrossRef]
- Linský, M.; Čiamporová-Zaťovičová, Z.; Čiampor, F. A revision of Onychelmis Hinton, 1941 (Coleoptera: Elmidae), with description of new species, DNA barcoding and notes on the geography of the genus. Eur. J. Taxon. 2021, 739(1), 1–35. [Google Scholar] [CrossRef]
- Linský, M.; Čiamporová-Zaťovičová, Z.; Laššová, K.; Čiampor, F. Molecular phylogeny of the riffle beetle genus Hexanchorus revealed a presence of a new genus (Coleoptera: Elmidae). Arthropod. Syst. Phylogeny 2022, 80, 575–602. [Google Scholar] [CrossRef]
- Múrria, C.; Bonada, N.; Vellend, M.; Zamora-Muñoz, C.; Alba-Tercedor, J.; Sainz-Cantero, C.E.; Garrido, J.; Acosta, R.; El Alami, M.; Barquín, J.; et al. Local environment rather than past climate determines community composition of mountain stream macroinvertebrates across Europe. Mol. Ecol. 2017, 26(21), 6085–6099. [Google Scholar] [CrossRef]
- Bozáňová, J.; Čiamporová Zat’ovičová, Z.; Čiampor, F.; Mamos, T.; Grabowski, M. The tale of springs and streams: how different aquatic ecosystems impacted the mtDNA population structure of two riffle beetles in the Western Carpathians. PeerJ 2020, 8, e10039. [Google Scholar] [CrossRef] [PubMed]
- Novaković, B.B.; Raković, M.B.; Čiampor, F.; Teofilova, T.M.; Živić, I.M. Genetic variability of the riffle beetle Elmis maugetii Latreille, 1802 (Coleoptera: Elmidae) in Europe and North Africa. Biologia 2022, 77(11), 3173–3183. [Google Scholar] [CrossRef]
- Litovkin, S.V.; Bruvo-Mađarić, B.; Jäch, M.A.; Jung, S.W.; Efimov, D.A. Stenelmis koreana Satô, 1978 (Coleoptera: Elmidae): confirmed as a wide-spread species by DNA-sequencing. Zootaxa 2019, 4651(3), 596–600. [Google Scholar] [CrossRef] [PubMed]
- Mičetić Stanković, V.; Bruvo Mađarić, B.; Jäch, M.A.; Kučinić, M. Elmis rietscheli Steffan, 1958 (Insecta: Coleoptera: Elmidae) in Croatia: first record and DNA barcoding. Nat. Croat. Period. Musei Hist. Nat. Croat. 2018, 27(1), 185–194. [Google Scholar] [CrossRef]
- Mičetić Stanković, V.; Bruvo Mađarić, B.; Kučinić, M. Ubiquitous but ignored? A case of water beetle in southeastern Europe. Diversity 2022, 14(1), 26. [Google Scholar] [CrossRef]
- Kodada, J.; Jäch, M.A.; Freitag, H.; Čiamporová-Zaťovičová, Z.; Goffová, K.; Selnekovič, D.; Čiampor, F. Ancyronyx clisteri, a new spider riffle beetle species from Borneo, redescription of A. sarawacensis Jäch including a description of the larva and new distribution data for A. procerus Jäch using DNA barcodes (Coleoptera, Elmidae). ZooKeys 2020, 912, 25–64. [Google Scholar] [CrossRef] [PubMed]
- Kodada, J.; Bennas, N.; Goffová, K.; Čiampor, F. Potamophilus acuminatus (Fabricius, 1792): distribution update in North Africa confirmed by COI barcoding sequencing (Coleoptera, Elmidae). Zootaxa 2022, 5200(6), 565–575. [Google Scholar] [CrossRef] [PubMed]
- Linský, M.; Čiamporová-Zaťovičová, Z.; Čiampor, F. Four new species of Hexanchorus Sharp from Ecuador (Coleoptera, Elmidae) with DNA barcoding and notes on the distribution of the genus. ZooKeys 2019, 838, 85–109. [Google Scholar] [CrossRef]
- Seno, C.B.N.; Delocado, E.D.; Freitag, H. Three new species of Ancyronyx Erichson, 1847 (Coleoptera, Elmidae) from Mindanao, Philippines. Tijdschr. Voor Entomol. 2022, 165(1–3), 61–80. [Google Scholar] [CrossRef]
- Novaković, B.B.; Teofilova, T.M.; Pandakov, P.G.; Živić, I.M. New distributional records of rare riffle beetles (Coleoptera: Elmidae) from the Balkan Peninsula. Arch. Biol. Sci. 2020, 72(1), 129–135. [Google Scholar] [CrossRef]
- Raupach, M.J.; Astrin, J.J.; Hannig, K.; Peters, M.K.; Stoeckle, M.Y.; Wägele, J.-W. Molecular species identification of Central European ground beetles (Coleoptera: Carabidae) using nuclear rDNA expansion segments and DNA barcodes. Front. Zool. 2010, 7(1), 26. [Google Scholar] [CrossRef] [PubMed]
- Gorring, P.S.; Cognato, A.I. The case for a nuclear barcode: using the CAD CPS region for species and genus level discrimination in beetles. Diversity 2023, 15(7), 847. [Google Scholar] [CrossRef]
- Huang, W.; Xie, X.; Peng, F.; Liang, X.; Wang, X.; Chen, X. Optimizing the widely used nuclear protein-coding gene primers in beetle phylogenies and their application in the genus Sasajiscymnus Vandenberg (Coleoptera: Coccinellidae). Ecol. Evol. 2020, 10(14), 7731–7738. [Google Scholar] [CrossRef] [PubMed]
- Che, L.-H.; Zhang, S.-Q.; Li, Y.; Liang, D.; Pang, H.; Ślipiński, A.; Zhang, P. Genome-wide survey of nuclear protein-coding markers for beetle phylogenetics and their application in resolving both deep and shallow-level divergences. Mol. Ecol. Resour. 2017, 17(6), 1342–1358. [Google Scholar] [CrossRef] [PubMed]
- Reitter, E. Elmis zoufali n. sp. Wien. Entomol. Ztg. 1910, 29, 36. [Google Scholar]
- Berthélemy, C. Elmidae de la région paléarctique occidentale: systématique et répartition [Coleoptera Dryopoidea]. Ann. Limnol. 1979, 15(1), 1–102. [Google Scholar] [CrossRef]
- Jäch, M.A.; Kodada, J.; Brojer, M.; Shepard, W.D.; Čiampor, F. Coleoptera: World Catalogue of Insects, Volume 14: Elmidae and Protelmidae; Brill: Leiden, The Netherlands, 2016; XXI + 318 pp. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3(5), 294–299. [Google Scholar] [PubMed]
- Hurtado, N.S.; Pérez-García, C.; Morán, P.; Pasantes, J.J. Genetic and cytological evidence of hybridization between native Ruditapes decussatus and introduced Ruditapes philippinarum (Mollusca, Bivalvia, Veneridae) in NW Spain. Aquaculture 2011, 311(1–4), 123–128. [Google Scholar] [CrossRef]
- Ogden, T.H.; Whiting, M.F. The problem with “the Paleoptera Problem:” sense and sensitivity. Cladistics 2003, 19(5), 432–442. [Google Scholar] [CrossRef]
- Wahlberg, N.; Wheat, C.W. Genomic outposts serve the phylogenomic pioneers: designing novel nuclear markers for genomic DNA extractions of Lepidoptera. Syst. Biol. 2008, 57(2), 231–242. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20(4), 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37(5), 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44 (W1), W232–W235. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14(6), 587–589. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35(2), 518–522. [Google Scholar] [CrossRef] [PubMed]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65(6), 997–1008. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32(1), 268–274. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLoS ONE 2013, 8(7), e66213. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33(7), 1870–1874. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29(22), 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble Species by Automatic Partitioning. Mol. Ecol. Resour. 2021, 21(2), 609–620. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29(8), 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Brower, A.V. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl. Acad. Sci. USA 1994, 91(14), 6491–6495. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67(5), 901–904. [Google Scholar] [CrossRef] [PubMed]
- ICZN [International Commission on Zoological Nomenclature]. International Code of Zoological Nomenclature, 4th ed.; International Trust for Zoological Nomenclature: London, UK, 1999; 306 pp. [Google Scholar]
- Kuwert, A. Bestimmungstabelle der Parniden Europas, der Mittelmeerfauna, sowie der angrenzenden Gebiete. Verhandlungen der Zool.-Bot. Ges. Wien 1890, 40, 15–54. [Google Scholar]
- Eberle, J.; Warnock, R.C.M.; Ahrens, D. Bayesian species delimitation in Pleophylla chafers (Coleoptera) – the importance of prior choice and morphology. BMC Evol. Biol. 2016, 16(1), 94. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Szalanski, A.L.; Roehrdanz, R.L.; Taylor, D.B. Genetic relationship among Diabrotica species (Coleoptera: Chrysomelidae) based on rDNA and mtDNA sequences. Fla. Entomol. 2000, 83(3), 262–267. [Google Scholar] [CrossRef]
- Cognato, A.I.; Sari, G.; Smith, S.M.; Beaver, R.A.; Li, Y.; Hulcr, J.; Jordal, B.H.; Kajimura, H.; Lin, C.-S.; Pham, T.H.; et al. The essential role of taxonomic expertise in the creation of DNA databases for the identification and delimitation of Southeast Asian ambrosia beetle species (Curculionidae: Scolytinae: Xyleborini). Front. Ecol. Evol. 2020, 8, 27. [Google Scholar] [CrossRef]
- Karpiński, L.; Gorring, P.; Hilszczański, J.; Szczepański, W.T.; Plewa, R.; Łoś, K.; Cognato, A.I. Integrative taxonomy tests possible hybridisation between Central Asian cerambycids (Coleoptera). Zool. Scr. 2023, 52(1), 70–85. [Google Scholar] [CrossRef]
- Bilton, D.T.; Jäch, M.A.; Ribera, I.; Toussaint, E.F.A. Minute moss beetles in the Southern Hemisphere: Molecular phylogeny, historical biogeography and habitat shifts (Coleoptera: Hydraenidae). Syst. Entomol. 2023, 48(1), 142–162. [Google Scholar] [CrossRef]
- Fossen, E.; Ekrem, T.; Nilsson, A.; Bergsten, J. Species delimitation in Northern European water scavenger beetles of the genus Hydrobius (Coleoptera, Hydrophilidae). ZooKeys 2016, 564, 71–120. [Google Scholar] [CrossRef] [PubMed]
- García-Vázquez, D.; Bilton, D.T.; Alonso, R.; Benetti, C.J.; Garrido, J.; Valladares, L.F.; Ribera, I. Reconstructing ancient Mediterranean crossroads in Deronectes diving beetles. J. Biogeogr. 2016, 43(8), 1533–1545. [Google Scholar] [CrossRef]
- Hidalgo-Galiana, A.; Ribera, I. Late Miocene diversification of the genus Hydrochus (Coleoptera, Hydrochidae) in the West Mediterranean area. Mol. Phyl. Evol. 2011, 59(2), 377–385. [Google Scholar] [CrossRef] [PubMed]
- Ribera, I.; Vogler, A.P. Speciation of Iberian diving beetles in Pleistocene refugia (Coleoptera, Dytiscidae). Mol. Ecol. 2004, 13(1), 179–193. [Google Scholar] [CrossRef] [PubMed]
- Ribera, I.; Fresneda, J.; Bucur, R.; Izquierdo, A.; Vogler, A.P.; Salgado, J.M.; Cieslak, A. Ancient origin of a Western Mediterranean radiation of subterranean beetles. BMC Evol. Biol. 2010, 10(1), 29. [Google Scholar] [CrossRef] [PubMed]
- Villastrigo, A.; Hernando, C.; Millán, A.; Ribera, I. The neglected diversity of the Ochthebius fauna from Eastern Atlantic and Central and Western Mediterranean coastal rockpools (Coleoptera, Hydraenidae). Org. Divers. Evol. 2020, 20(4), 785–801. [Google Scholar] [CrossRef]
- Sabatelli, S.; Ruspantini, P.; Cardoli, P.; Audisio, P. Underestimated diversity: Cryptic species and phylogenetic relationships in the subgenus Cobalius (Coleoptera: Hydraenidae) from marine rockpools. Mol. Phyl. Evol. 2021, 163, 107243. [Google Scholar] [CrossRef]
- Davis, P.H. Distribution patterns in Anatolia with particular reference to endemism. In Plant Life of South-West Asia; Davis, P.H., Harper, P.C., Hedge, I.C., Eds.; The Botanical Society of Edinburgh: Edinburgh, UK, 1971; pp. 15–27. [Google Scholar]
- Gür, H. The Anatolian diagonal revisited: Testing the ecological basis of a biogeographic boundary. Zool. Middle East 2016, 62(3), 189–199. [Google Scholar] [CrossRef]
- Mutun, S. Review of oak gall wasps phylogeographic patterns in Turkey suggests a main role of the Anatolian diagonal. Turk. J. For. 2016, 17, 1–6. [Google Scholar] [CrossRef]
- Kuzucuoğlu, C.; Çiner, A.; Kazancı, N. The geomorphological regions of Turkey. In Landscapes and Landforms of Turkey; Kuzucuoğlu, C., Çiner, A., Kazancı, N., Eds.; World Geomorphological Landscapes; Springer International Publishing: Cham, Switzerland, 2019; pp. 41–178. [Google Scholar]
- Seyi̇toğlu, G.; Tunçel, E.; Kaypak, B.; Esat, K.; Gökkaya, E. The Anatolian Diagonal: A broad left-lateral shear zone between the North Anatolian Fault Zone and the Aegean/Cyprus Arcs. Geol. Bull. Turk. 2022, 65(2), 93–116. [Google Scholar] [CrossRef]
- Rögl, V.F. Palaeogeographic considerations for Mediterranean and Paratethys seaways (Oligocene to Miocene). Ann. Naturhistorischen Mus. Wien (Ser. A) 1998, 99, 279–310. [Google Scholar]
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 2001, 292(5517), 686–693. [Google Scholar] [CrossRef] [PubMed]
- Lisiecki, L.E.; Raymo, M.E. Plio-Pleistocene climate evolution: trends and transitions in glacial cycle dynamics. Quat. Sci. Rev. 2007, 26(1), 56–69. [Google Scholar] [CrossRef]
- Martinetto, E.; Bertini, A.; Bhandari, S.; Bruch, A.A.; Cerilli, E.; Cherin, M.; Field, J.H.; Gabrielyan, I.; Gianotti, F.; Kern, A.K.; et al. The last three millions of unequal spring thaws. In Nature through Time: Virtual field trips through the Nature of the past; Martinetto, E., Tschopp, E., Gastaldo, R.A., Eds.; Springer Textbooks in Earth Sciences, Geography and Environment; Springer International Publishing: Cham, Switzerland, 2020; 53 pp. [Google Scholar]
- Dröllner, M.; Barham, M.; Kirkland, C.L.; Danišík, M.; Bourdet, J.; Schulz, M.; Aspandiar, M. Directly dating Plio-Pleistocene climate change in the terrestrial record. Geophys. Res. Lett. 2023, 50(8), e2023GL102928. [Google Scholar] [CrossRef]
- Bilgin, R. Back to the suture: The distribution of intraspecific genetic diversity in and around Anatolia. Int. J. Mol. Sci. 2011, 12(6), 4080–4103. [Google Scholar] [CrossRef]
- Mutun, S. Intraspecific genetic variation and phylogeography of the oak gallwasp Andricus caputmedusae (Hymenoptera: Cynipidae): Effects of the Anatolian Diagonal. Acta Zool. Acad. Sci. Hung. 2010, 56(2), 153–172. [Google Scholar]
- Mutun, S.; Atay, G. Phylogeography of Trigonaspis synaspis (Hymenoptera: Cynipidae) from Anatolia based on mitochondrial and nuclear DNA sequences. Eur. J. Entomol. 2015, 112(2), 259–269. [Google Scholar] [CrossRef]
- Mutun, S.; Dinç, S. The Anatolian Diagonal and paleoclimatic changes shaped the phylogeography of Cynips quercus (Hymenoptera, Cynipidae). Ann. Zool. Fenn. 2019, 56(1–6), 65. [Google Scholar] [CrossRef]
- Challis, R.J.; Mutun, S.; Nieves-Aldrey, J.-L.; Preuss, S.; Rokas, A.; Aebi, A.; Sadeghi, E.; Tavakoli, M.; Stone, G.N. Longitudinal range expansion and cryptic eastern species in the Western Palaearctic oak gallwasp, Andricus coriarius. Mol. Ecol. 2007, 16(10), 2103–2114. [Google Scholar] [CrossRef]
- Perreau, M. The Near Eastern species of Leptodirini (Coleoptera: Leiodidae: Cholevinae). Ann. Soc. Entomol. Fr. (N.S.) 2021, 57(4), 289–312. [Google Scholar] [CrossRef]
- Şenol, Ö.; Görür, G.; Akyıldırım Beğen, H. Contributions of the Anatolian Diagonal effect on Turkish aphid diversity. Artvin Çoruh Üniv. Orman Fak. Derg. 2019, 20(1), 102–109. [Google Scholar] [CrossRef]
- Yazici, G. Overview of the zoogeographical distribution of aquatic and semi-aquatic Heteroptera (Hemiptera) in Turkey. J. Insect Biodivers. Syst. 2020, 6(2), 135–155. [Google Scholar] [CrossRef]
- García-Vázquez, D.; Bilton, D.T.; Foster, G.N.; Ribera, I. Pleistocene range shifts, refugia and the origin of widespread species in Western Palaearctic water beetles. Mol. Phyl. Evol. 2017, 114, 122–136. [Google Scholar] [CrossRef]
- Jäch, M.A. Beitrag zur Kenntnis der Elmidae der (asiatischen) Türkei (Col.). Entomol. Blätter 1984, 80(2–3), 136–142. [Google Scholar]
- Reitter, E. Neue Coleopteren aus Europa, den angrenzenden Ländern und Sibirien, mit Bemerkungen über bekannte Arten. Dritter Theil. Dtsch. Entomol. Z. 1887, 31(1), 241–288. [Google Scholar]
- Olmi, M. Fauna d’Italia, Volume 12: Coleoptera Dryopidae, Elminthidae; Edizioni Calderini: Bologna, Italy, 1976; 280 pp. [Google Scholar]




| Species | Country | BIN | BOLD Process ID/GenBank acc. nr. for COI 1,2 | ITS1 | H3 | CAD |
|---|---|---|---|---|---|---|
| Elmis zoufali | Croatia Serbia Serbia Serbia Albania Bulgaria Bulgaria | BOLD:ADP5230 | ELMEU105-18/- ELMEU101-18/- CROEL003-20/OR138318 CROEL019-20/OR138316 NMWEL003-23/OR138311 NMWEL004-23/OR138310 ELMEU103-18/- | - - OR142274 OR142273 OR142271 OR142272 - | - - OR146742 OR146741 OR146739 OR146740 - | - - OR146727 OR146726 OR146724 OR146725 - |
| Elmis syriaca | Israel Israel Israel Israel Israel Israel Georgia Georgia Georgia Armenia Azerbaijan Azerbaijan | BOLD:AFC9293 | -/OR187603 -/OR187604 -/OR187605 -/OR187606 NMWEL007-23/OR138313 NMWEL008-23/OR138308 NMWEL005-23/OR138305 NMWEL006-23/OR138309 NMWEL010-23/OR138306 NMWEL013-23/OR138312 NMWEL009-23/OR138307 NMWEL011-23/OR138315 | - - - - OR142269 OR142270 - OR142267 OR142268 - OR142266 OR142265 | - - - - OR146737 OR146738 OR146734 OR146735 OR146736 OR146733 OR146732 OR146731 | - - - - OR146722 OR146723 OR146719 OR146720 OR146721 OR146718 OR146717 OR146716 |
| Elmis ? quadricollis | China China | BOLD:ADL0116 | NMWEL012-23/OR138314 XJDQD253-18/- | OR142275 - | - - | - - |
| Elmis aenea | Croatia | BOLD:AAF0074 | CROBF005-19/OR138317 | OR142277 | OR146743 | OR146729 |
| Elmis rioloides | Croatia | BOLD:ABY6700 | CROBF017-19/OR138319 | OR142278 | OR146744 | OR146730 |
| Elmis bosnica | Croatia | BOLD:ADR7537 | CROBF008-19/OL874462 | OR142276 | - | OR146728 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jäch, M.A.; Brojer, M.; Mičetić Stanković, V.; Bošnjak, M.; Luz, D.; Dorchin, N.; Hershkovitz, Y.; Novaković, B.; Živić, I.; Dorfer, W.; et al. Elmis syriaca (Kuwert, 1890) and E. zoufali (Reitter, 1910) (Coleoptera: Elmidae) confirmed as distinct species based on molecular data, morphology and geographical distribution. Diversity 2023, 15, 994. https://doi.org/10.3390/d15090994
Jäch MA, Brojer M, Mičetić Stanković V, Bošnjak M, Luz D, Dorchin N, Hershkovitz Y, Novaković B, Živić I, Dorfer W, et al. Elmis syriaca (Kuwert, 1890) and E. zoufali (Reitter, 1910) (Coleoptera: Elmidae) confirmed as distinct species based on molecular data, morphology and geographical distribution. Diversity. 2023; 15(9):994. https://doi.org/10.3390/d15090994
Chicago/Turabian StyleJäch, Manfred A., Michaela Brojer, Vlatka Mičetić Stanković, Marija Bošnjak, Dafna Luz, Netta Dorchin, Yaron Hershkovitz, Boris Novaković, Ivana Živić, Wolfgang Dorfer, and et al. 2023. "Elmis syriaca (Kuwert, 1890) and E. zoufali (Reitter, 1910) (Coleoptera: Elmidae) confirmed as distinct species based on molecular data, morphology and geographical distribution" Diversity 15, no. 9: 994. https://doi.org/10.3390/d15090994
APA StyleJäch, M. A., Brojer, M., Mičetić Stanković, V., Bošnjak, M., Luz, D., Dorchin, N., Hershkovitz, Y., Novaković, B., Živić, I., Dorfer, W., & Bruvo Mađarić, B. (2023). Elmis syriaca (Kuwert, 1890) and E. zoufali (Reitter, 1910) (Coleoptera: Elmidae) confirmed as distinct species based on molecular data, morphology and geographical distribution. Diversity, 15(9), 994. https://doi.org/10.3390/d15090994








