Insight into Trophic Interactions of Spiders in Olive Groves with Integrated and Ecological Pest Management Using DNA Metabarcoding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Collection and Identification of Collected Samples
2.2. Isolation and Amplification of eDNA from Spiders’ Gut, Library Preparation and Next-Generation Sequencing
2.3. Bioinformatic Data Processing
2.4. Data Analyses
3. Results
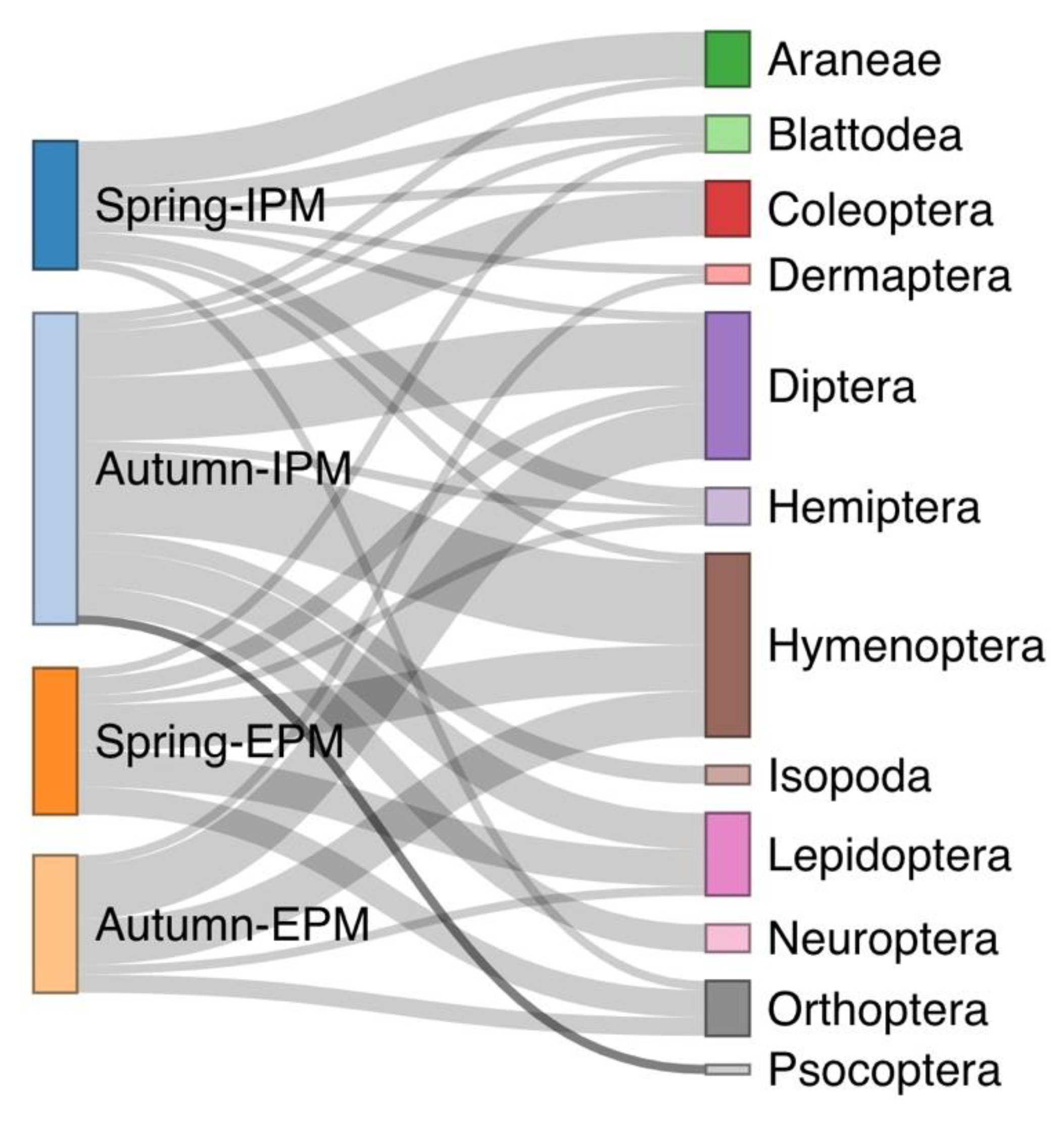
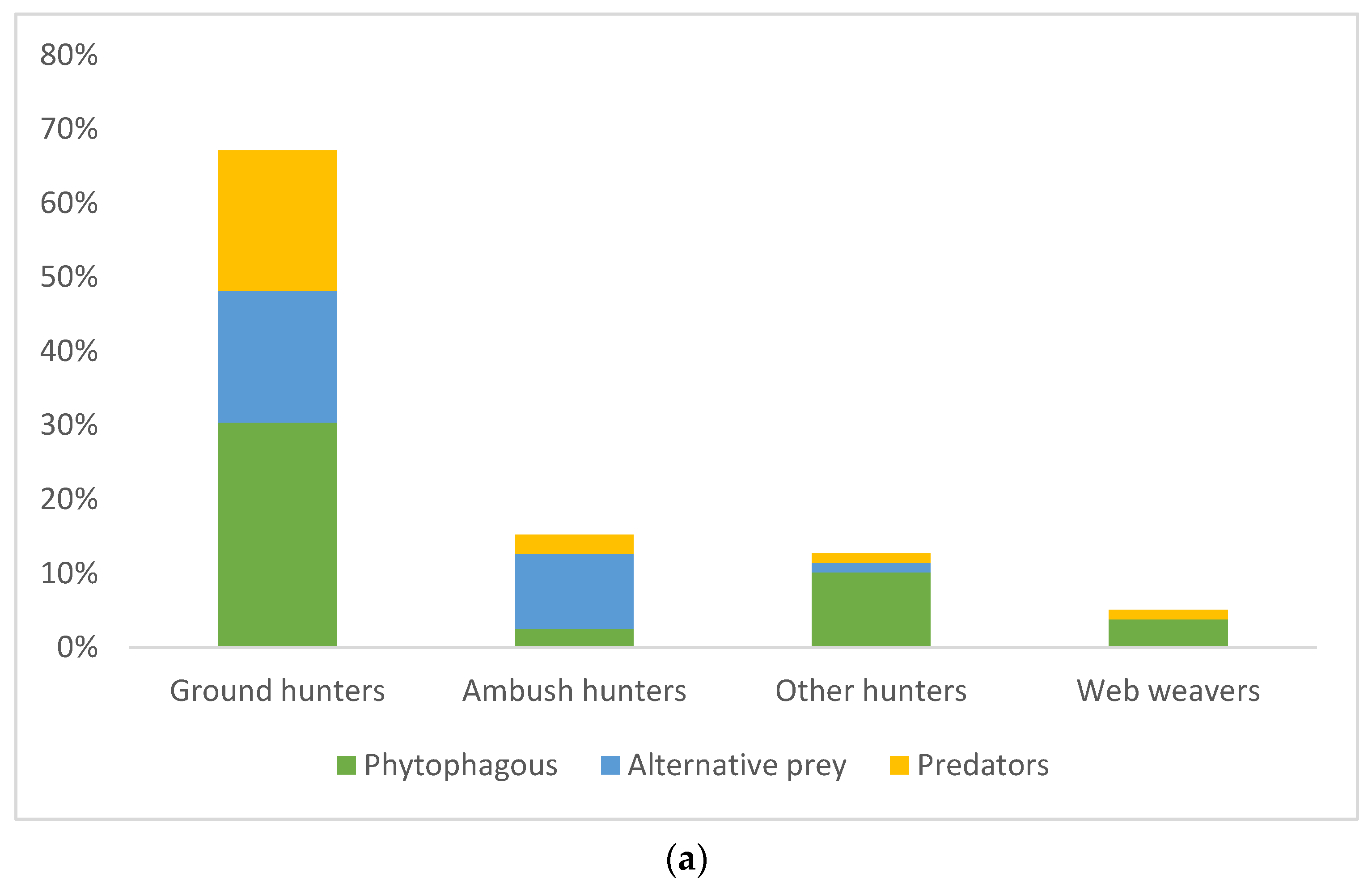
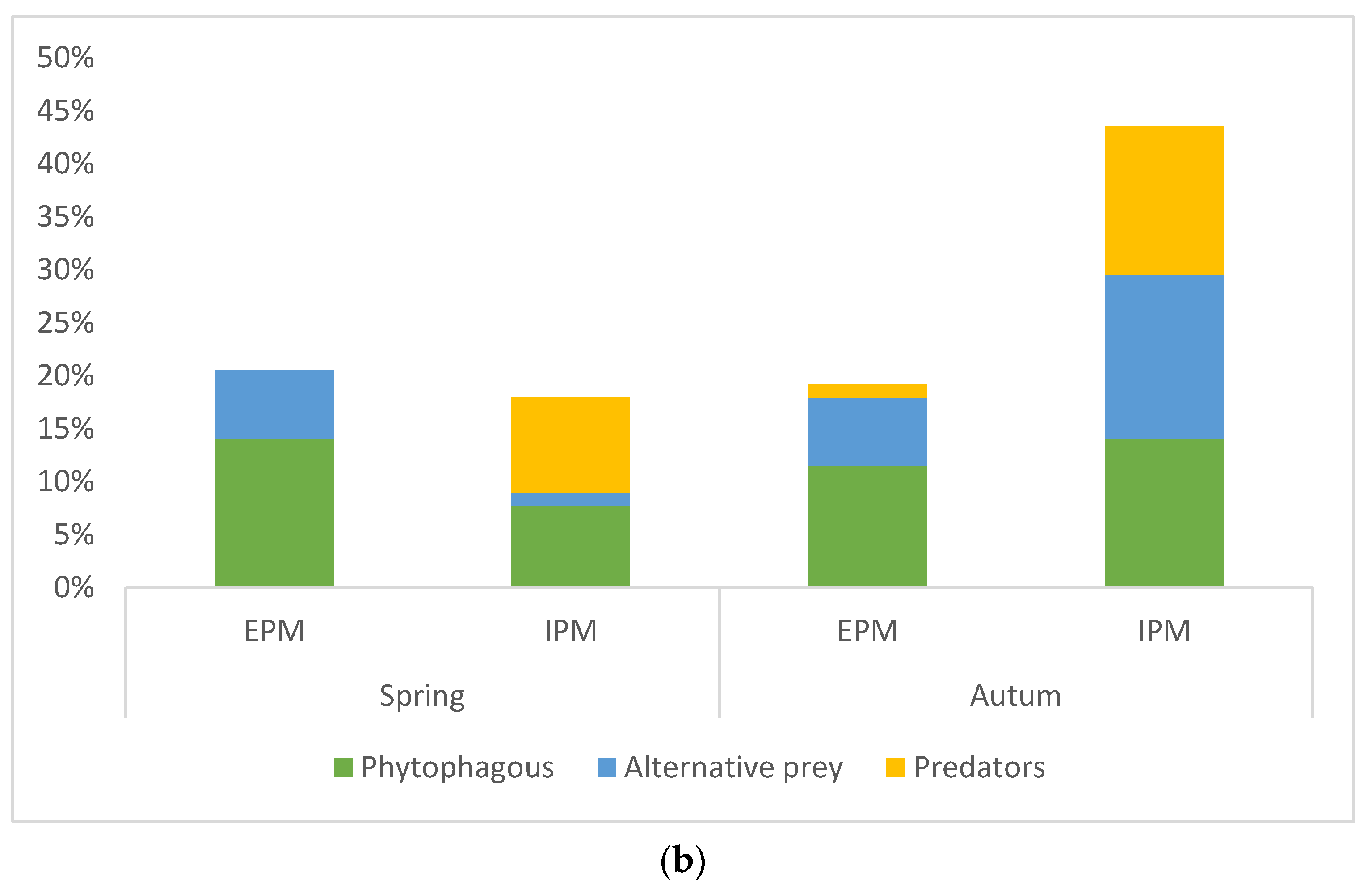
3.1. Spiders’ Dietary Data and Consumption of Different Prey Orders
3.2. Consumption of Invertebrates as an Indicator of Spiders’ Biocontrol Potential
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cuff, J.P.; Drake, L.E.; Tercel, M.P.; Stockdale, J.E.; Orozco-terWengel, P.; Bell, J.R.; Vaughan, I.P.; Müller, C.T.; Symondson, W.O. Money spider dietary choice in pre- and post-harvest cereal crops using metabarcoding. Ecol. Entomol. 2021, 46, 249–261. [Google Scholar] [CrossRef]
- Saqib, H.S.A.; Sun, L.; Pozsgai, G.; Liang, P.; You, M.; Gurr, G.M.; You, S. DNA metabarcoding of gut contents reveals key habitat and seasonal drivers of trophic networks involving generalist predators in agricultural landscapes. Pest Manag. Sci. 2022, 78, 5390–5401. [Google Scholar] [CrossRef]
- Mezőfi, L.; Markó, G.; Nagy, C.; Korányi, D.; Markó, V. Beyond polyphagy and opportunism: Natural prey of hunting spiders in the canopy of apple trees. PeerJ 2020, 8, e9334. [Google Scholar] [CrossRef] [PubMed]
- Nyffeler, M.; Sunderland, K.D. Composition, abundance and pest control potential of spider communities in agroecosystems: A comparison of European and US studies. Agriculture. Ecosyst. Environ. 2003, 95, 579–612. [Google Scholar] [CrossRef]
- Živković, P.; Lemić, D.; Samu, F.; Kos, T.; Barić, B. Spider communities affected by exclusion nets. Appl. Ecol. Environ. Res. 2019, 17, 879–887. [Google Scholar] [CrossRef]
- Sanders, D.; Vogel, E.; Knop, E. Individual and species-specific traits explain niche size and functional role in spiders as generalist predators. J. Anim. Ecol. 2015, 84, 134–142. [Google Scholar] [CrossRef]
- Cuff, J.P.; Tercel, M.P.T.G.; Drake, L.E.; Vaughan, I.P.; Bell, J.R.; Orozco-terWengel, P.; Müller, C.T.; Symondson, W.O.C. Density-independent prey choice, taxonomy, life history, and web characteristics determine the diet and biocontrol potential of spiders (Linyphiidae and Lycosidae) in cereal crops. Environ. DNA 2021, 4, 549–564. [Google Scholar] [CrossRef]
- Chapman, E.G.; Schmidt, J.M.; Welch, K.D.; Harwood, J.D. Molecular evidence for dietary selectivity and pest suppression potential in an epigeal spider community in winter wheat. Biol. Control 2013, 65, 72–86. [Google Scholar] [CrossRef]
- Gomez-Polo, P.; Alomar, O.; Castañé, C.; Agustí, N. Molecular tracking of arthropod predator–prey interactions in Mediterranean lettuce crops. Food Webs 2016, 9, 18–24. [Google Scholar] [CrossRef]
- Wagan, T.A.; Li, X.; Hua, H.; Cai, W. Starvation time and predatory efficiency of spider species on Bemisia tabaci (Homoptera: Aleyrodidae). Fla. Entomol. 2019, 102, 684–690. [Google Scholar] [CrossRef]
- Samiayyan, K. Spiders—The Generalist Super Predators in Agro-Ecosystems, Integrated Pest Management; Academic Press: Cambridge, MA, USA, 2014; pp. 283–310. [Google Scholar]
- Mestre, L.; Piñol, J.; Barrientos, J.A.; Espadaler, X.; Brewitt, K.; Werner, C.; Platner, C. Trophic structure of the spider community of a Mediterranean citrus grove: A stable isotope analysis. Basic Appl. Ecol. 2013, 14, 413–422. [Google Scholar] [CrossRef]
- Sow, A.; Haran, J.; Benoit, L.; Galan, M.; Brévault, T. DNA Metabarcoding as a Tool for Disentangling Food Webs in Agroecosystems. Insects 2020, 11, 294. [Google Scholar] [CrossRef] [PubMed]
- Hambäck, P.A.; Cirtwill, A.R.; García, D.; Grudzinska-Sterno, M.; Miñarro, M.; Tasin, M.; Yang, X.; Samnegård, U. More intraguild prey than pest species in arachnid diets may compromise biological control in apple orchards. Basic Appl. Ecol. 2021, 57, 1–13. [Google Scholar] [CrossRef]
- Agusti, N.; Shayler, S.P.; Harwood, J.D.; Vaughan, I.P.; Sunderland, K.D.; Symondson, W.O.C. Collembola as alternative prey sustaining spiders in arable ecosystems: Prey detection within predators using molecular markers. Mol. Ecol. 2003, 12, 3467–3475. [Google Scholar] [CrossRef]
- Harwood, J.D.; Sunderland, K.D.; Symondson, W.O. Prey selection by linyphiid spiders: Molecular tracking of the effects of alternative prey on rates of aphid consumption in the field. Mol. Ecol. 2004, 13, 3549–3560. [Google Scholar] [CrossRef]
- Kuusk, A.K.; Ekbom, B. Lycosid spiders and alternative food: Feeding behavior and implications for biological control. Biol. Control. 2010, 55, 20–26. [Google Scholar] [CrossRef]
- Axelsen, J.A.; Ruggle, P.; Holst, N.; Toft, S. Modelling natural control of cereal aphids. III. Linyphiid spiders and coccinellids. Acta. Jutl. 1997, 72, 221–231. [Google Scholar]
- Michalko, R.; Pekár, S.; Entling, M.H. An updated perspective on spiders as generalist predators in biological control. Oecologia 2019, 189, 21–36. [Google Scholar] [CrossRef] [PubMed]
- Newbold, T.; Oppenheimer, P.; Etard, A.; Williams, J.J. Tropical and Mediterranean biodiversity is disproportionately sensi-tive to land-use and climate change. Nat. Ecol. Evol. 2020, 4, 1630–1638. [Google Scholar] [CrossRef] [PubMed]
- Šerić Jelaska, L.; Franjević, D.; Jelaska, S.D.; Symondson, W.O.C. Prey detection in carabid beetles in woodland ecosystems by PCR analyses of gut content. E. Jour. Ent. 2014, 111, 631–638. [Google Scholar]
- European Commission. A Farm to Fork Strategy for a Fair, Healthy and Environmentally-Friendly Food System; COM (2020) 381 Final; European Commission: Brussels, Belgium, 2020. [Google Scholar]
- European Commission (EC 889/2008) Commission Regulation (EC) No 889/2008 of 5 September 2008 Laying down Detailed Rules for the Implementation of Council Regulation (EC) No 834/2007 on Organic Production and Labelling of Organic Products with Regard to Organic Production, Labelling and Control, Special edition in Croatian: Chapter 15 Volume 008 P. 173–256; Latest consolidated version: 01/01/2022; European Commission: Brussels, Belgium, 2008.
- Uhl, P.; Bucher, R.; Schäfer, R.B.; Entling, M.H. Sublethal effects of imidacloprid on interactions in a tritrophic system of non-target species. Chemosphere 2015, 132, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Petráková, L.; Michalko, R.; Loverre, P.; Sentenská, L.; Korenko, S.; Pekár, S. Intraguild predation among spiders and their effect on the pear psylla during winter. Agric. Ecosyst. Environ. 2016, 233, 67–74. [Google Scholar] [CrossRef]
- Šerić Jelaska, L.; Jurasović, J.; Brown, S.D.; Vaughan, P.I.; Symondson, W.O.C. Molecular field analysis of trophic relationships in soil-dwelling invertebrates to identify mercury, lead and cadmium transmission through forest ecosystems. Mol. Ecol. 2014, 23, 3755–3766. [Google Scholar] [CrossRef]
- Šerić Jelaska, L.; Symondson, O. Predation on epigeic, endogeic and anecic earthworms by carabids active in spring and autumn. Periodicum. Biologorum. 2016, 118, 281–289. [Google Scholar] [CrossRef]
- Menalled, F.D.; Smith, R.G.; Dauer, J.T.; Fox, T.B. Impact of agricultural management on carabid communities and weed seed predation. Agric. Ecosyst. Environ. 2007, 118, 49–54. [Google Scholar] [CrossRef]
- Caprio, E.; Nervo, B.; Isaia, M.; Allegro, G.; Rolando, A. Organic versus conventional systems in viticulture: Comparative effects on spiders and carabids in vineyards and adjacent forests. Agric. Syst. 2015, 136, 61–69. [Google Scholar] [CrossRef]
- Pekár, S. Spiders (Araneae) in the pesticide world: An ecotoxicological review. Pest Manag. Sci. 2012, 68, 1438–1446. [Google Scholar] [CrossRef] [PubMed]
- Benamú, M.; Schneider, M.; González, A.; Sánchez, N. Short and long-term effects of three neurotoxic insecticides on biological and behavioural attributes of the orb-web spider Alpaida veniliae (Araneae, Araneidae): Implications for IPM programs. Ecotoxicology 2013, 22, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Šimunović, V. Stanje maslinarstva i uljarstva u Republici Hrvatskoj. Pomologia Croatica 2005, 11, 69–78. [Google Scholar]
- Concepción, E.D.; Díaz, M.; Baquero, R.A. Effects of landscape complexity on the ecological effectiveness of agri-environment schemes. Landscape Ecol. 2008, 23, 135–148. [Google Scholar] [CrossRef]
- Ando, H.; Mukai, H.; Komura, T.; Dewi, T.; Ando, M.; Isagi, Y. Methodological trends and perspectives of animal dietary studies by noninvasive fecal DNA metabarcoding. Environ. DNA 2020, 2, 391–406. [Google Scholar] [CrossRef]
- Pompanon, F.; Deagle, B.E.; Symondson, W.O.C.; Brown, D.S.; Jarman, S.N.; Tabarlet, P. Who is eating what: Diet assessment using next generation sequencing? Mol. Ecol. 2012, 21, 1931–1950. [Google Scholar] [CrossRef]
- Kennedy, S.R.; Prost, S.; Overcast, I.; Rominger, A.J.; Gillespie, R.G.; Krehenwinkel, H. High-throughput sequencing for community analysis: The promise of DNA barcoding to uncover diversity, relatedness, abundances and interactions in spider communities. Dev. Genes Evol. 2020, 230, 185–201. [Google Scholar] [CrossRef] [PubMed]
- Drake, L.E.; Cuff, J.P.; Young, R.E.; Marchbank, A.; Chadwick, E.A.; Symondson, W.O.C. Post-bioinformatic methods to identify and reduce the prevalence of artefacts in metabarcoding data. Authorea 2021. [Google Scholar] [CrossRef]
- Clare, E.L.; Chain, F.J.; Littlefair, J.E.; Cristescu, M.E. The effects of parameter choice on defining molecular operational taxonomic units and resulting ecological analyses of metabarcoding data. Genome 2016, 59, 981–990. [Google Scholar] [CrossRef]
- Dušátková, L.P.; Pekár, S.; Michálek, O.; Líznarová, E.; Symondson, W.O.C. Estimation of trophic niches in myrmecophagous spider predators. Sci. Rep. 2020, 10, 8683. [Google Scholar] [CrossRef]
- Šerić Jelaska, L.; Ivanković Tatalović, L.; Kostanjšek, F.; Kos, T. Ground beetle assemblages and distribution of functional traits in olive orchards and vineyards depend on the agricultural management practice. BioControl 2022, 67, 275–286. [Google Scholar] [CrossRef]
- Nentwig, W.; Blick, T.; Bosmans, R.; Gloor, D.; Hänggi, A.; Kropf, C. Spiders of Europe. Version 3. 2022. Available online: https://www.araneae.nmbe.ch. (accessed on 20 February 2023).
- Macías-Hernández, N.; Athey, K.; Tonzo, V.; Wangensteen, O.S.; Arnedo, M.; Harwood, J.D. Molecular gut content analysis of different spider body parts. PLoS ONE 2018, 13, e0196589. [Google Scholar] [CrossRef] [PubMed]
- Elbrecht, V.; Braukmann, T.W.A.; Ivanova, N.V.; Prosser, S.W.J.; Hajibabaei, M.; Wright, M.; Zakharov, E.V.; Hebert, P.D.N.; Steinke, D. Validation of COI metabarcoding primers for terrestrial arthropods. PeerJ 2019, 7, e7745. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Alberdi, A.; Aizpurua, O.; Gilbert, M.T.P.; Bohmann, K. Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods Ecol. Evol. 2017, 9, 134–147. [Google Scholar] [CrossRef]
- Cardoso, P.; Pekár, S.; Jocqué, R.; Coddington, J.A. Global patterns of guild composition and functional diversity of spiders. PLoS ONE 2011, 6, e21710. [Google Scholar] [CrossRef] [PubMed]
- Deagle, B.E.; Thomas, A.C.; McInnes, J.C.; Clarke, L.J.; Vesterinen, E.J.; Clare, E.L.; Kartzinel, T.R.; Eveson, J.P. Counting with DNA in metabarcoding studies: How should we convert sequence reads to dietary data? Mol. Ecol. 2019, 28, 391–406. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R—A Language and Environment for Statistical Computing. Available: R Foundation for Statistical Computing, Vienna. Available online: http://www.R-project.org/ (accessed on 2 September 2022).
- Pekár, S.; Brabec, M. Generalized estimating equations: A pragmatic and flexible approach to the marginal GLM modelling of correlated data in the behavioural sciences. Ethology 2018, 124, 86–93. [Google Scholar] [CrossRef]
- Allaire, J.J.; Gandrud, C.; Russell, K.; Yetman, C.J. NetworkD3: D3 JavaScript Network Graphs from R. R Package Version 0.4. Available online: https://CRAN.R-project.org/package=networkD3. (accessed on 2 September 2022).
- TIBCO Software Inc. Data Science Workbench. 2020. Available online: http://tibco.com (accessed on 1 May 2023).
- Diehl, E.; Mader, V.L.; Wolters, V.; Birkhofer, K. Management intensity and vegetation complexity affect web-building spiders and their prey. Oecologia 2013, 173, 579–589. [Google Scholar] [CrossRef]
- Landis, D.A.; Wratten, S.D.; Gurr, G.M. Habitat Management to conserve natural enemies of arthropod pests in agriculture. Annu. Rev. Entomol. 2003, 45, 175–201. [Google Scholar] [CrossRef]
- Symondson, W.O.C.; Sunderland, K.D.; Greenstone, M.H. Can generalist predators be effective biocontrol agents? Annu. Rev. Entomol. 2002, 47, 561–594. [Google Scholar] [CrossRef]
- Anđelić Dmitrović, B.; Jelić, M.; Rota, E.; Šerić Jelaska, L. DNA Barcoding of Invertebrates Inhabiting Olive Orchards and Vineyards Accelerates Understudied Mediterranean Biodiversity Assessment. Diversity 2022, 14, 182. [Google Scholar] [CrossRef]
- Kubiak, K.L.; Pereira, J.A.; Tessaro, D.; Santos, S.A.P.; Benhadi-Marín, J. Functional diversity of epigeal spiders in the olive grove agroecosystem in northeastern Portugal: A comparison between crop and surrounding semi-natural habitats. Entomol. Exp. Appl. 2022, 170, 449–458. [Google Scholar] [CrossRef]
- Saqib, H.S.A.; Liang, P.; You, M.; Gurr, G.M. Molecular gut content analysis indicates the inter- and intraguild predation patterns of spiders in conventionally managed vegetable fields. Ecol. Evol. 2021, 11, 9543–9552. [Google Scholar] [CrossRef]
- Birkhofer, K.; Djoudi, E.A.; Schnerch, B.; Michalko, R. Climatic conditions and functional traits affect spider diets in agricultural and non-agricultural habitats worldwide. Ecography 2022, 3, e06090. [Google Scholar] [CrossRef]
- Aguilera, G.; Roslin, T.; Tamburini, G.; Birkhofer, K.; Caballero-Lopez, B.; Lindstrom, S.A.M.; Bommarco, R. Crop diversity benefits carabid and pollinator communities in landscapes with semi-natural habitats. J. Appl. Eco. 2000, 57, 2170–2179. [Google Scholar] [CrossRef]
- Gajski, D.; Mifková, T.; Košulič, O.; Michalek, O.; Štarhova-Serbina, L.; Michalko, R.; Pekar, S. Brace yourselves, winter is coming: The winter activity, natural diet, and prey preference of winter-active spiders on pear trees. J. Pest Sci. 2023. In press. [Google Scholar] [CrossRef]
| Predator | Pest Management | Season | Hunting Strategies | Prey Order | Prey |
|---|---|---|---|---|---|
| Agalenatea redii (Scopoli, 1763) | Integrated | Spring | Web weavers | Coleoptera | Oxytelus (Epomotylus) sculptus Gravenhorst, 1806 |
| Agalenatea Archer, 1951 spp | Ecological | Spring | Web weavers | Lepidoptera | Prays oleae (Bernard, 1788) |
| Prays fraxinella (Bjerkander, 1784) | |||||
| Alopecosa accentuata (Latreille, 1817) | Ecological | Spring | Ground hunters | Orthoptera | Aiolopus strepens (Latreille, 1804) |
| Integrated | Autumn | Ground hunters | Isopoda | Armadillidium vulgare (Latreille, 1804) | |
| Alopecosa albofasciata (Brullé, 1832) | Integrated | Autumn | Ground hunters | Coleoptera | Cantharis (Cantharis) livida Linnaeus, 1758 |
| Diptera | Sarcophaga (Myorhina) nigriventris Meigen, 1826 | ||||
| Bradysia brevispina Tuomikoski, 1960 | |||||
| Hymenoptera | Messor ibericus Santschi, 1931 | ||||
| Lepidoptera | Cadra figulilella (Gregson, 1871) | ||||
| Neuroptera | Chrysoperla carnea (Stephens, 1836) | ||||
| Alopecosa farinosa (Herman, 1879) | Integrated | Autumn | Ground hunters | Hymenoptera | Formica (Serviformica) glauca Ruzsky, 1896 |
| Alopecosa Simon, 1885 spp | Ecological | Spring | Ground hunters | Lepidoptera | Eriocottis fuscanella Zeller, 1847 |
| Integrated | Autumn | Ground hunters | Diptera | Aedes (Stegomyia) albopictus (Skuse, 1894) | |
| Neuroptera | Chrysoperla carnea (Stephens, 1836) | ||||
| * Araneidae Sundevall, 1833 | Ecological | Spring | Web weavers | Hemiptera | Euscelis Brullé, 1832 sp. |
| Arctosa C.L. Koch, 1847 spp | Integrated | Autumn | Ground hunters | Coleoptera | Poecilus (Macropoecilus) sericeus Fischer von Waldheim, 1824 |
| Diptera | Diptera (OTU1) | ||||
| Forcipomyia sp. Meigen, 1818 | |||||
| Bassaniodes caperatus (Simon, 1875) | Integrated | Autumn | Ambush hunters | Hymenoptera | Plagiolepis pygmaea (Latreille, 1798) |
| Tetramorium semilaeve Andre, 1883 | |||||
| Coleoptera | Poecilus sp. Bonelli, 1810 | ||||
| * Gnaphosidae Pocock, 1898 | Ecological | Autumn | Ground hunters | Hymenoptera | Tetramorium caespitum (Linnaeus, 1758) |
| Haplodrassus dalmatensis (L. Koch, 1866) | Ecological | Spring | Ground hunters | Orthoptera | Aiolopus strepens (Latreille, 1804) |
| Hogna radiata (Latreille, 1817) | Ecological | Autumn | Ground hunters | Diptera | Cystiphora sonchi (Vallot, 1827) |
| Oscinella sp. Becker, 1910 | |||||
| Philosepedon humeralis (Meigen, 1818) | |||||
| Integrated | Autumn | Ground hunters | Psocoptera | Ectopsocus McLachlan, 1899 sp. | |
| Araneae | Sitticus penicillatus (Simon, 1875) | ||||
| Hogna radiata (Latreille, 1817) | Ecological | Spring | Ground hunters | Blattodea | Loboptera decipiens (Germar, 1817) |
| Diptera | * Phoridae Curtis, 1833sp. | ||||
| Hymenoptera | Tetramorium semilaeve Andre, 1883 | ||||
| Orthoptera | Aiolopus strepens (Latreille, 1804) | ||||
| Araneae | Diplostyla concolor (Wider, 1834) | ||||
| Agyneta pseudorurestris Wunderlich, 1980 | |||||
| Hogna radiata (Latreille, 1817) | Integrated | Spring | Ground hunters | Blattodea | Loboptera decipiens (Germar, 1817) |
| Diptera | Bradysia brevispina Tuomikoski, 1960 | ||||
| Hemiptera | Nysius sp. Dallas, 1852 | ||||
| Hymenoptera | Aphaenogaster balcanica (Emery, 1898) | ||||
| Orthoptera | Tettigonia viridissima (Linnaeus, 1758) | ||||
| Araneae | Cheiracanthium mildei L. Koch, 1864 | ||||
| Sitticus penicillatus (Simon, 1875) | |||||
| Tegenaria hasperi Chyzer, 1897 | |||||
| * Lycosidae Sundevall, 1833 | Ecological | Autumn | Ground hunters | Isopoda | Armadillidium vulgare (Latreille, 1804) |
| Integrated | Autumn | Ground hunters | Blattodea | Loboptera decipiens (Germar, 1817) | |
| Hymenoptera | Aphaenogaster balcanica (Emery, 1898) | ||||
| Solenopsis Westwood, 1840 spp | |||||
| * Formicidae Latreille 1809 sp. | |||||
| Lepidoptera | Plutella (Plutella) xylostella (Linnaeus, 1758) | ||||
| Nomisia exornata (C.L. Koch, 1839) | Ecological | Autumn | Ground hunters | Lepidoptera | Stigmella freyella (Heyden, 1858) |
| Oxyopes Latreille, 1804 sp | Ecological | Autumn | Other hunters | Orthoptera | Aiolopus strepens (Latreille, 1804) |
| Oxyopes heterophthalmus (Latreille, 1804) | Ecological | Autumn | Other hunters | Dermaptera | Forficula auricularia Linnaeus, 1758 |
| Diptera | Chromatomyia horticola (Goureau, 1851) | ||||
| Cystiphora sonchi (Vallot, 1827) | |||||
| Philosepedon humeralis (Meigen, 1818) | |||||
| Orthoptera | Aiolopus strepens (Latreille, 1804) | ||||
| * Oxyopidae Thorell, 1870 | Integrated | Autumn | Other hunters | Hemiptera | * Ninidae Barber, 1956 |
| Hymenoptera | Tetramorium semilaeve Andre, 1883 | ||||
| Pardosa hortensis (Thorell, 1872) | Integrated | Autumn | Ground hunters | Coleoptera | Harmonia axyridis Pallas, 1773 |
| Hymenoptera | Solenopsis sp. Westwood, 1840 | ||||
| Lepidoptera | Dryobotodes (Dryobotodes) eremita (Fabricius, 1775) | ||||
| * Thomisidae Sundevall, 1833 | Integrated | Autumn | Ambush hunters | Diptera | Corynoptera perpusilla Winnertz, 1867 |
| Tibellus macellus Simon, 1875 | Ecological | Spring | Other hunters | Diptera | Cystiphora sonchi (Vallot, 1827) |
| Lepidoptera | Eriocottis fuscanella Zeller, 1847 | ||||
| Xerolycosa nemoralis (Westring, 1861) | Integrated | Spring | Ground hunters | Hemiptera | Nysius graminicola (Kolenati, 1845) |
| Xerolycosa Dahl, 1908 sp. | Ecological | Autumn | Ground hunters | Hymenoptera | * Formicidae Latreille 1809 |
| Xysticus acerbus Thorell, 1872 | Integrated | Autumn | Ambush hunters | Diptera | Corynoptera perpusilla Winnertz, 1867 |
| Xysticus acerbus Thorell, 1872 | Ecological | Autumn | Ambush hunters | Hymenoptera | Aphaenogaster balcanica (Emery, 1898) |
| Xysticus kochi Thorell, 1872 | Integrated | Spring | Ambush hunters | Dermaptera | * Forficulidae Latreille 1810 |
| Xysticus marmoratus Thorell, 1875 | Ecological | Spring | Ambush hunters | Hymenoptera | Lasioglossum glabriusculum (Morawitz, 1853) |
| Tetramorium semilaeve Andre, 1883 | |||||
| Aphaenogaster balcanica (Emery, 1898) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anđelić Dmitrović, B.; Gajski, D.; Kos, T.; Jelić, M.; Šerić Jelaska, L. Insight into Trophic Interactions of Spiders in Olive Groves with Integrated and Ecological Pest Management Using DNA Metabarcoding. Diversity 2023, 15, 976. https://doi.org/10.3390/d15090976
Anđelić Dmitrović B, Gajski D, Kos T, Jelić M, Šerić Jelaska L. Insight into Trophic Interactions of Spiders in Olive Groves with Integrated and Ecological Pest Management Using DNA Metabarcoding. Diversity. 2023; 15(9):976. https://doi.org/10.3390/d15090976
Chicago/Turabian StyleAnđelić Dmitrović, Barbara, Domagoj Gajski, Tomislav Kos, Mišel Jelić, and Lucija Šerić Jelaska. 2023. "Insight into Trophic Interactions of Spiders in Olive Groves with Integrated and Ecological Pest Management Using DNA Metabarcoding" Diversity 15, no. 9: 976. https://doi.org/10.3390/d15090976
APA StyleAnđelić Dmitrović, B., Gajski, D., Kos, T., Jelić, M., & Šerić Jelaska, L. (2023). Insight into Trophic Interactions of Spiders in Olive Groves with Integrated and Ecological Pest Management Using DNA Metabarcoding. Diversity, 15(9), 976. https://doi.org/10.3390/d15090976









