Genome-Wide Analysis of SPL Gene Families Illuminate the Evolution Patterns in Three Rubber-Producing Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. SPL Identification across the Genome and Phylogenetic Analysis
2.2. Physicochemical Characterization and Structural Analysis of the Protein
2.3. Analysis of Protein Domains and Conserved Motifs
2.4. Collinearity Analysis
2.5. Analysis of Expression Patterns across Several Developmental Stages and Tissue Types
3. Results
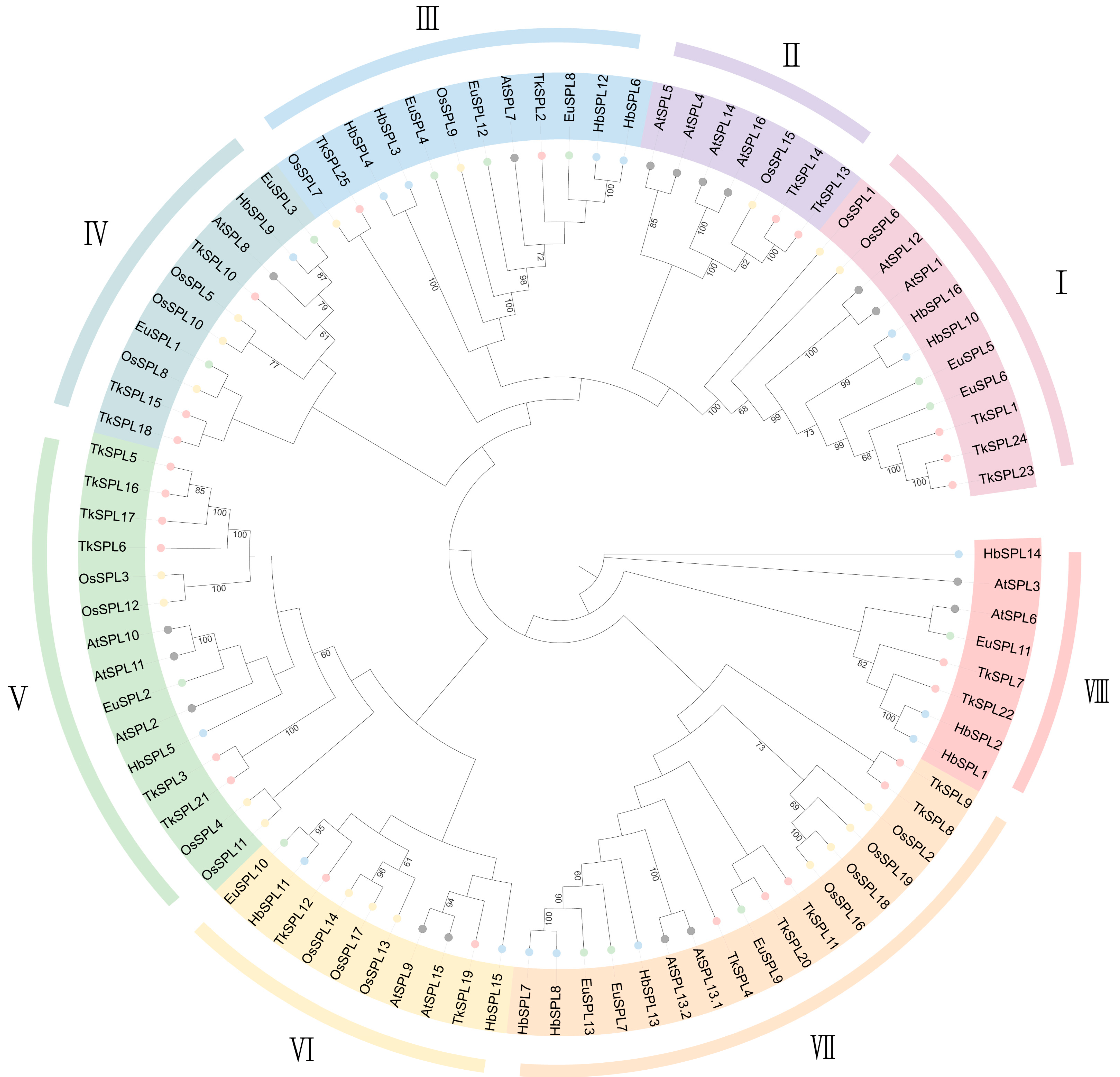
3.1. SPL Identification across the Genome and Phylogenetic Analysis
3.2. Physicochemical Properties and Secondary and Three-Dimensional Structural Analysis
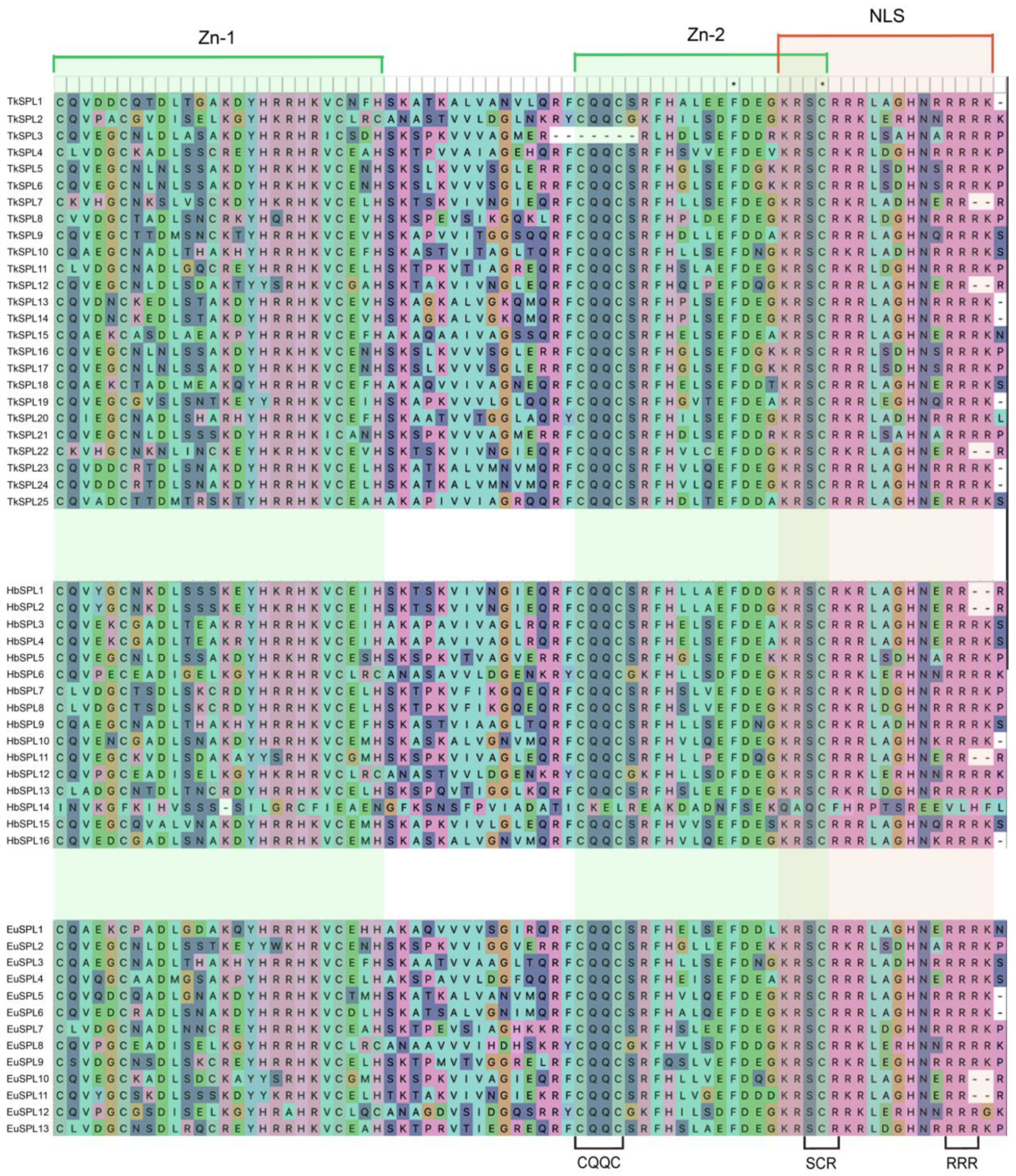
3.3. Analysis of Protein Domains and Conserved Motifs
3.4. Cis-Acting Elements and Collinearity Analysis
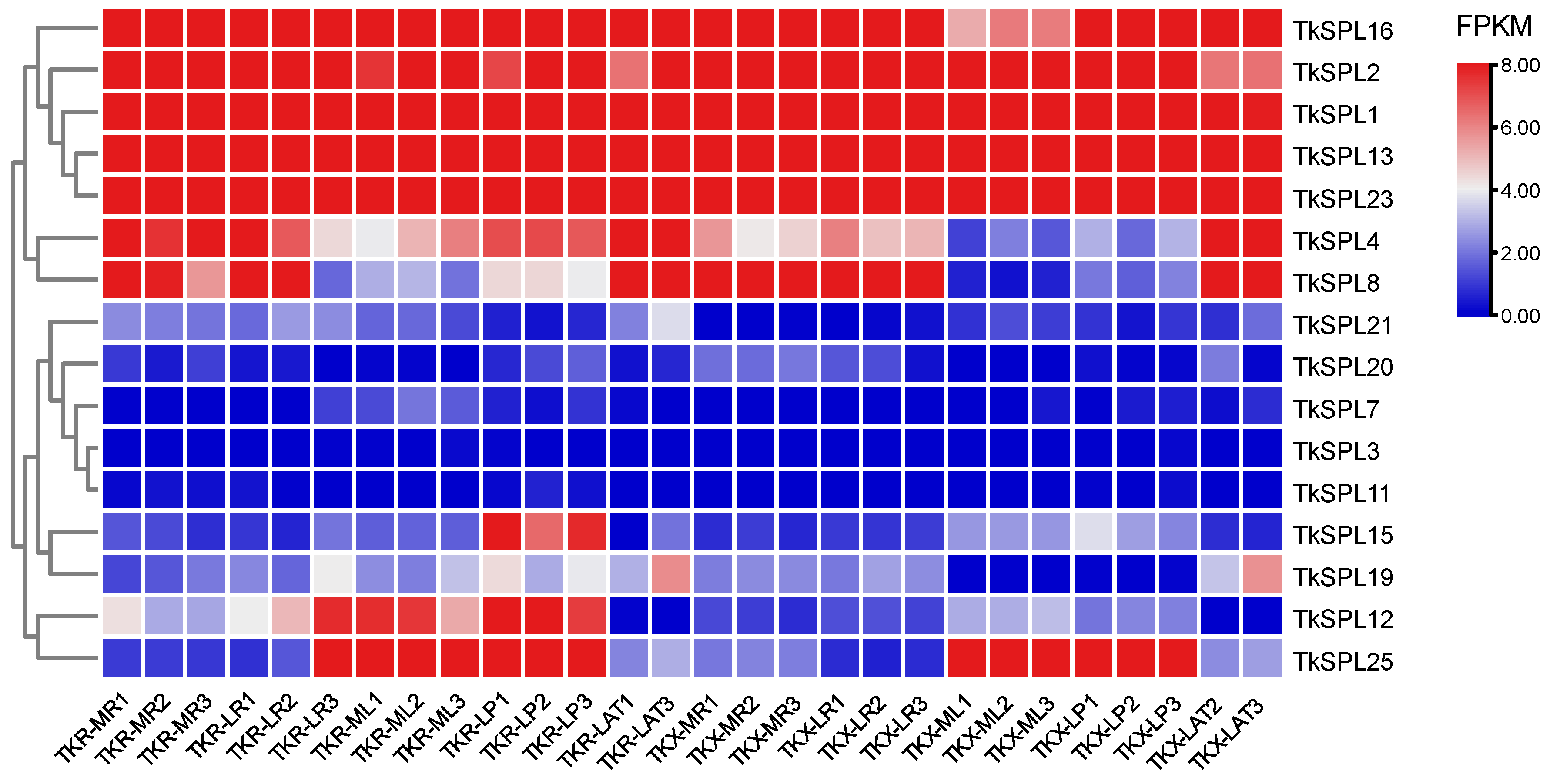
3.5. Analysis of Expression Patterns across Several Developmental Stages and Tissue Types
4. Discussion
4.1. SPL Identification across the Genome and Phylogenetic Analysis
4.2. Analysis of Protein Domains and Conserved Motifs
4.3. Analysis of Responses and Expression Patterns
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E.; et al. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Y.; Xiao, Y.; Zhang, X.; Du, B.; Turupu, M.; Wang, C.; Yao, Q.; Gai, S.; Huang, J.; et al. Genome-Wide Identification of the SQUAMOSA Promoter-Binding Protein-like (SPL) Transcription Factor Family in Sweet Cherry Fruit. Int. J. Mol. Sci. 2023, 24, 2880. [Google Scholar] [CrossRef]
- Zhao, H.; Cao, H.; Zhang, M.; Deng, S.; Li, T.; Xing, S. Genome-Wide Identification and Characterization of SPL Family Genes in Chenopodium quinoa. Genes 2022, 13, 1455. [Google Scholar] [CrossRef]
- Lai, D.; Fan, Y.; Xue, G.; He, A.; Yang, H.; He, C.; Li, Y.; Ruan, J.; Yan, J.; Cheng, J. Genome-wide identification and characterization of the SPL gene family and its expression in the various developmental stages and stress conditions in foxtail millet (Setaria italica). BMC Genom. 2022, 23, 389. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Wu, M.F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 2009, 17, 268–278. [Google Scholar] [CrossRef]
- Gandikota, M.; Birkenbihl, R.P.; Hohmann, S.; Cardon, G.H.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007, 49, 683–693. [Google Scholar] [CrossRef]
- Zhang, Y.; Schwarz, S.; Saedler, H.; Huijser, P. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 2007, 63, 429–439. [Google Scholar] [CrossRef]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA Promoter Binding Protein-Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef]
- Kropat, J.; Tottey, S.; Birkenbihl, R.P.; Depege, N.; Huijser, P.; Merchant, S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 2005, 102, 18730–18735. [Google Scholar] [CrossRef]
- Chen, W.W.; Jin, J.F.; Lou, H.Q.; Liu, L.; Kochian, L.V.; Yang, J.L. LeSPL-CNR negatively regulates Cd acquisition through repressing nitrate reductase-mediated nitric oxide production in tomato. Planta 2018, 248, 893–907. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ding, H.; Jiang, H.; Wang, H.; Chen, K.; Duan, J.; Feng, S.; Wu, G. Regulation of cadmium tolerance and accumulation by miR156 in Arabidopsis. Chemosphere 2020, 242, 125168. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef]
- Wang, J.; Ye, Y.; Xu, M.; Feng, L.; Xu, L.A. Roles of the SPL gene family and miR156 in the salt stress responses of tamarisk (Tamarix chinensis). BMC Plant Biol. 2019, 19, 370. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Yan, S.; Jing, Y.; Yang, R.; Zhang, Y.; Zhou, Y.; Zhu, Y.; Sun, J. MIR156-Targeted SPL9 Is Phosphorylated by SnRK2s and Interacts With ABI5 to Enhance ABA Responses in Arabidopsis. Front. Plant Sci. 2021, 12, 708573. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Xue, H.; Zhang, F.; Jiang, Q.; Yang, S.; Yue, P.; Wang, F.; Zhang, Y.; Li, L.; He, P.; et al. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. 2021, 19, 311–323. [Google Scholar] [CrossRef]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008, 67, 183–195. [Google Scholar] [CrossRef]
- Wang, J.W.; Schwab, R.; Czech, B.; Mica, E.; Weigel, D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 2008, 20, 1231–1243. [Google Scholar] [CrossRef]
- Wang, L.; Yu, P.; Lyu, J.; Hu, Y.; Han, C.; Bai, M.Y.; Fan, M. BZR1 physically interacts with SPL9 to regulate the vegetative phase change and cell elongation in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 10415. [Google Scholar] [CrossRef]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Niu, Q.W.; Ng, K.H.; Chua, N.H. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 2015, 83, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Cai, W.J.; Wang, S.; Shan, C.M.; Wang, L.J.; Chen, X.Y. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 2010, 22, 2322–2335. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.Y.; Zhao, B.; Chao, L.M.; Chen, D.Y.; Cui, W.R.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Interaction between two timing microRNAs controls trichome distribution in Arabidopsis. PLoS Genet. 2014, 10, e1004266. [Google Scholar] [CrossRef]
- Ioannidi, E.; Rigas, S.; Tsitsekian, D.; Daras, G.; Alatzas, A.; Makris, A.; Tanou, G.; Argiriou, A.; Alexandrou, D.; Poethig, S.; et al. Trichome patterning control involves TTG1 interaction with SPL transcription factors. Plant Mol. Biol. 2016, 92, 675–687. [Google Scholar] [CrossRef]
- Nodine, M.D.; Bartel, D.P. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010, 24, 2678–2692. [Google Scholar] [CrossRef]
- Martin, R.C.; Asahina, M.; Liu, P.-P.; Kristof, J.R.; Coppersmith, J.L.; Pluskota, W.E.; Bassel, G.W.; Goloviznina, N.A.; Nguyen, T.T.; Martínez-Andújar, C.; et al. The regulation of post-germinative transition from the cotyledon- to vegetative-leaf stages by microRNA-targeted Squamosa Promoter-Binding Protein LIKE13 in Arabidopsis. Seed Sci. Res. 2010, 20, 89–96. [Google Scholar] [CrossRef]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C.; et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Backhaus, R.A. Rubber formation in plants. Isr. J. Bot. 1985, 34, 283–293. [Google Scholar]
- van Beilen, J.B.; Poirier, Y. Establishment of new crops for the production of natural rubber. Trends Biotechnol. 2007, 25, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Cornish, K. Alternative natural rubber crops: Why should we care? Technol. Innov. 2017, 18, 244–255. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, D.; Li, H.L.; Peng, S.Q. Characterization of HbWRKY1, a WRKY transcription factor from Hevea brasiliensis that negatively regulates HbSRPP. Plant Physiol. Biochem. 2013, 71, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Wei, L.R.; Guo, D.; Wang, Y.; Zhu, J.H.; Chen, X.T.; Peng, S.Q. HbMADS4, a MADS-box transcription factor from Hevea brasiliensis, negatively regulates HbSRPP. Front. Plant Sci. 2016, 7, 1709. [Google Scholar] [CrossRef]
- Wang, Y.; Zhan, D.F.; Li, H.L.; Guo, D.; Zhu, J.H.; Peng, S.Q. Transcriptome-wide identification and characterization of MYB transcription factor genes in the laticifer cells of Hevea brasiliensis. Front. Plant Sci. 2017, 8, 1974. [Google Scholar] [CrossRef]
- Qu, L.; Li, H.L.; Guo, D.; Zhu, J.H.; Wang, Y.; Yin, L.Y.; Peng, S.Q. HbWRKY27, a group IIe WRKY transcription factor, positively regulates HbFPS1 expression in Hevea brasiliensis. Sci. Rep. 2020, 10, 20639. [Google Scholar] [CrossRef]
- Guo, D.; Yang, Z.P.; Li, H.L.; Wang, Y.; Zhu, J.H.; Peng, S.Q. The 14-3-3 protein HbGF14a interacts with a RING zinc finger protein to regulate expression of the rubber transferase gene in Hevea brasiliensis. J. Exp. Bot. 2018, 69, 1903–1912. [Google Scholar] [CrossRef]
- Guo, D.; Li, H.L.; Wang, Y.; Zhu, J.H.; Peng, S.Q. A myelocytomatosis transcription factor from Hevea brasiliensis positively regulates the expression of the small rubber particle protein gene. Ind. Crops Prod. 2019, 133, 90–97. [Google Scholar] [CrossRef]
- Cheng, Y.; Luo, J.; Li, H.; Wei, F.; Zhang, Y.; Jiang, H.; Peng, X. Identification of the WRKY Gene Family and Characterization of Stress-Responsive Genes in Taraxacum kok-saghyz Rodin. Int. J. Mol. Sci. 2022, 23, 10270. [Google Scholar] [CrossRef]
- Zhang, H.; Gong, Y.; Sun, P.; Chen, S.; Ma, C. Genome-wide identification of CBF genes and their responses to cold acclimation in Taraxacum kok-saghyz. PeerJ 2022, 10, e13429. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X.; Chen, Y.; Liu, Y.; Wu, Y.; Ren, S.; Li, L. Identification, evolution and expression analysis of WRKY gene family in Eucommia ulmoides. Genomics 2021, 113, 3294–3309. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, X.; Pan, L.; Guo, W.; Li, Y.; Wang, W. Genome-wide identification and expression analysis of MADS-box transcription factors reveal their involvement in sex determination of hardy rubber tree (Eucommia ulmoides oliv). Front. Genet. 2023, 14, 1138703. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Birkenbihl, R.P.; Jach, G.; Saedler, H.; Huijser, P. Functional Dissection of the Plant-specific SBP-Domain: Overlap of the DNA-binding and Nuclear Localization Domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, X.; Gu, S.; Hu, Z.; Xu, H.; Xu, C. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 2008, 407, 1–11. [Google Scholar] [CrossRef]
- Hyun, Y.; Richter, R.; Vincent, C.; Martinez-Gallegos, R.; Porri, A.; Coupland, G. Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev. Cell 2015, 37, 254–266. [Google Scholar] [CrossRef]
- Gao, R.; Wang, Y.; Gruber, M.Y.; Hannoufa, A. miR156/SPL10 modulates lateral root development, branching and leaf mor-phology in Arabidopsis by silencing AGAMOUS-LIKE 79. Front. Plant Sci. 2017, 8, 2226. [Google Scholar] [CrossRef]
- Kabin, X.; Congqing, W.; Lizhong, X. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef]
- Zhang, D.; Han, Z.; Li, J.; Qin, H.; Zhou, L.; Wang, Y.; Zhu, X.; Ma, Y.; Fang, W. Genome-wide analysis of the SBP-box gene family transcription factors and their responses to abiotic stresses in tea (Camellia sinensis). Genomics 2020, 112, 2194–2202. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Ma, D.; Yin, J.; Yang, L.; Gao, C. Genome-wide characterization and expression profiling of squamosa promoter binding protein-like (SBP) transcription factors in wheat (Triticum aestivum L). Agronomy 2019, 9, 527. [Google Scholar] [CrossRef]
- Cai, C.; Guo, W.; Zhang, B. Genome-wide identification and characterization of SPL transcription factor family and their evolution and expression profiling analysis in cotton. Sci. Rep. 2018, 8, 762. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Calderon-Villalobos, L.I.; Estelle, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Leng, P.; Yuan, B.; Guo, Y. The role of abscisic acid in fruit ripening and responses to abiotic stress. J. Exp. Bot. 2014, 65, 4577–4588. [Google Scholar] [CrossRef]
- Guo, J.; Wang, S.; Yu, X.; Dong, R.; Li, Y.; Mei, X.; Shen, Y. Polyamines regulate strawberry fruit ripening by abscisic acid, auxin, and ethylene. Plant. Physiol. 2018, 177, 339–351. [Google Scholar] [CrossRef]
- Duan, Q.; Li, G.-R.; Qu, Y.-P.; Yin, D.-X.; Zhang, C.-L.; Chen, Y.-S. Genome-Wide Identification, Evolution and Expression Analysis of the Glutathione S-Transferase Supergene Family in Euphorbiaceae. Front. Plant Sci. 2022, 13, 808279. [Google Scholar] [CrossRef]
- Jaiswal, P.; Cheruku, J.R.; Kumar, K.; Yadav, S.; Singh, A.; Kumari, P.; Dube, S.C.; Upadhyaya, K.C.; Verma, P.K. Differential transcript accumulation in chickpea during early phases of compatible interaction with a necrotrophic fungus Ascochyta rabiei. Mol. Biol. Rep. 2012, 39, 4635–4646. [Google Scholar] [CrossRef]




| Gene | gene ID | Chr ID | Gene Range | Exon | Intron | CDS | UTR | Size/aa | MW/kD | PI | Instability Index |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TkSPL1 | GWHPBCHF001261 | GWHBCHF00000001 | 14822969:14828316 | 12 | 11 | 11 | 3 | 927 | 103,394.48 | 6.2 | 53.89 |
| TkSPL2 | GWHPBCHF002675 | GWHBCHF00000002 | 2073280:2078116 | 11 | 10 | 10 | 3 | 748 | 83,959.66 | 5.95 | 58.08 |
| TkSPL3 | GWHPBCHF006473 | GWHBCHF00000002 | 100814431:100818025 | 4 | 3 | 4 | 0 | 261 | 29,108.21 | 7.73 | 74.85 |
| TkSPL4 | GWHPBCHF019170 | GWHBCHF00000004 | 830539:832919 | 3 | 2 | 2 | 3 | 302 | 34,271.32 | 9.08 | 68.14 |
| TkSPL5 | GWHPBCHF020579 | GWHBCHF00000004 | 30786520:30796342 | 5 | 4 | 5 | 2 | 357 | 39,818.25 | 9 | 51.09 |
| TkSPL6 | GWHPBCHF020580 | GWHBCHF00000004 | 30786520:30796342 | 5 | 4 | 4 | 3 | 349 | 38,902.15 | 9.1 | 51.1 |
| TkSPL7 | GWHPBCHF022201 | GWHBCHF00000004 | 84269226:84270914 | 4 | 3 | 4 | 2 | 298 | 33,372.53 | 8.41 | 56.09 |
| TkSPL8 | GWHPBCHF023484 | GWHBCHF00000004 | 107063807:107065169 | 2 | 1 | 2 | 2 | 198 | 22,079.54 | 9.24 | 65.76 |
| TkSPL9 | GWHPBCHF024887 | GWHBCHF00000004 | 129182786:129185573 | 2 | 1 | 2 | 2 | 192 | 22,005.9 | 8.36 | 70.5 |
| TkSPL10 | GWHPBCHF025127 | GWHBCHF00000004 | 132569671:132571972 | 3 | 2 | 3 | 2 | 298 | 33,497.02 | 8.76 | 53.76 |
| TkSPL11 | GWHPBCHF026791 | GWHBCHF00000004 | 155862873:155865148 | 4 | 3 | 3 | 3 | 279 | 30,897.47 | 9.74 | 67.09 |
| TkSPL12 | GWHPBCHF028134 | GWHBCHF00000005 | 14701253:14704746 | 4 | 3 | 3 | 3 | 351 | 37,175.07 | 7.61 | 52.58 |
| TkSPL13 | GWHPBCHF029402 | GWHBCHF00000005 | 33473738:33478697 | 11 | 10 | 10 | 3 | 1042 | 115,521.09 | 8.36 | 56.28 |
| TkSPL14 | GWHPBCHF029413 | GWHBCHF00000005 | 33656280:33661229 | 11 | 10 | 10 | 3 | 1039 | 115,165.69 | 8.36 | 57.09 |
| TkSPL15 | GWHPBCHF035319 | GWHBCHF00000006 | 28696570:28698241 | 2 | 1 | 2 | 2 | 183 | 20,547.1 | 9.49 | 44.27 |
| TkSPL16 | GWHPBCHF039801 | GWHBCHF00000007 | 11469584:11479658 | 5 | 4 | 5 | 2 | 357 | 39,883.37 | 9.1 | 53.96 |
| TkSPL17 | GWHPBCHF039802 | GWHBCHF00000007 | 11469584:11479658 | 5 | 4 | 4 | 3 | 349 | 38,967.27 | 9.19 | 54.04 |
| TkSPL18 | GWHPBCHF040059 | GWHBCHF00000007 | 13895402:13897881 | 3 | 2 | 2 | 3 | 171 | 19,026.37 | 8.94 | 60.4 |
| TkSPL19 | GWHPBCHF044300 | GWHBCHF00000008 | 4487440:4488774 | 2 | 1 | 2 | 2 | 296 | 32,163.23 | 9.48 | 57.94 |
| TkSPL20 | GWHPBCHF044302 | GWHBCHF00000008 | 4501786:4503817 | 3 | 2 | 2 | 3 | 261 | 29,845.17 | 7.03 | 73.8 |
| TkSPL21 | GWHPBCHF050461 | GWHBCHF00000009 | 9840573:9843749 | 5 | 4 | 4 | 2 | 275 | 30,724.02 | 9.55 | 73.95 |
| TkSPL22 | GWHPBCHF052188 | GWHBCHF00000009 | 28195236:28196956 | 3 | 2 | 3 | 2 | 316 | 35,456.72 | 9.36 | 68.57 |
| TkSPL23 | GWHPBCHF053518 | GWHBCHF00000009 | 42852554:42856744 | 10 | 9 | 9 | 3 | 928 | 104,212.96 | 6.82 | 46.08 |
| TkSPL24 | GWHPBCHF053519 | GWHBCHF00000009 | 42852554:42856744 | 9 | 8 | 9 | 2 | 924 | 103,871.69 | 6.95 | 45.77 |
| TkSPL25 | GWHPBCHF056162 | GWHBCHF00000009 | 103280194:103281999 | 2 | 1 | 2 | 2 | 137 | 15,812.68 | 8.56 | 63.94 |
| EuSPL1 | GWHPAAAL001077 | GWHAAAL00000030 | 988480:992976 | 11 | 10 | 11 | 0 | 178 | 20,179.44 | 9.03 | 55.87 |
| EuSPL2 | GWHPAAAL003913 | GWHAAAL00000058 | 2037987:2040046 | 3 | 2 | 3 | 0 | 547 | 60,636.72 | 6.91 | 54.8 |
| EuSPL3 | GWHPAAAL004171 | GWHAAAL00000080 | 203789:215644 | 2 | 1 | 2 | 0 | 369 | 41,404.83 | 8.07 | 72.6 |
| EuSPL4 | GWHPAAAL007895 | GWHAAAL00000112 | 735958:741319 | 5 | 4 | 5 | 0 | 133 | 15,322.89 | 7.69 | 84.16 |
| EuSPL5 | GWHPAAAL008461 | GWHAAAL00000115 | 1029716:1033122 | 5 | 4 | 5 | 0 | 968 | 107,979.44 | 7.33 | 57.38 |
| EuSPL6 | GWHPAAAL008647 | GWHAAAL00000172 | 475295:476266 | 2 | 1 | 2 | 0 | 978 | 109,120.78 | 8.36 | 51.07 |
| EuSPL7 | GWHPAAAL012748 | GWHAAAL00000184 | 1290342:1300388 | 11 | 10 | 11 | 0 | 347 | 37,321.69 | 8.1 | 64.57 |
| EuSPL8 | GWHPAAAL012866 | GWHAAAL00000249 | 1634971:1649669 | 12 | 11 | 12 | 0 | 875 | 98,038.82 | 6.26 | 58.57 |
| EuSPL9 | GWHPAAAL016673 | GWHAAAL00006095 | 779825:852698 | 9 | 8 | 9 | 0 | 259 | 28,558.46 | 8.79 | 61.98 |
| EuSPL10 | GWHPAAAL020125 | GWHAAAL00014124 | 1676:4668 | 3 | 2 | 3 | 0 | 363 | 38,927.58 | 9.01 | 64.09 |
| EuSPL11 | GWHPAAAL025240 | GWHAAAL00017881 | 529023:533014 | 3 | 2 | 3 | 0 | 354 | 40,444.43 | 8.94 | 48.71 |
| EuSPL12 | GWHPAAAL026381 | GWHAAAL00020293 | 536331:540436 | 3 | 2 | 3 | 0 | 696 | 77,869.21 | 8.88 | 53.91 |
| EuSPL13 | GWHPAAAL026606 | GWHAAAL00026424 | 227394:229146 | 3 | 2 | 3 | 0 | 131 | 15,166.23 | 10.47 | 72.74 |
| HbSPL1 | GT021460.t1 | CM021228.1 | 13875757:13877545 | 2 | 1 | 2 | 0 | 531 | 57,107.57 | 7.27 | 46.45 |
| HbSPL2 | GT021495.t1 | CM021228.1 | 13970213:13972002 | 2 | 1 | 2 | 0 | 442 | 48,140.64 | 8.2 | 48.31 |
| HbSPL3 | GT010583.t1 | CM021228.1 | 86118185:86120916 | 4 | 3 | 4 | 0 | 194 | 21,651.15 | 9.06 | 64.25 |
| HbSPL4 | GT010615.t1 | CM021233.1 | 98164232:98172531 | 10 | 9 | 10 | 0 | 194 | 21,651.15 | 9.06 | 64.25 |
| HbSPL5 | GT017984.t1 | CM021234.1 | 94639220:94645994 | 4 | 3 | 4 | 0 | 458 | 49,852.23 | 8.87 | 56.52 |
| HbSPL6 | GT003116.t1 | CM021238.1 | 65041899:65062959 | 24 | 23 | 24 | 0 | 707 | 79,038.55 | 7.23 | 55.88 |
| HbSPL7 | GT030712.t1 | CM021238.1 | 65225362:65248838 | 24 | 23 | 24 | 0 | 1065 | 117,893.71 | 8.75 | 46.86 |
| HbSPL8 | GT030809.t1 | CM021239.1 | 12363240:12365845 | 3 | 2 | 3 | 0 | 1005 | 110,811.41 | 8.33 | 48.37 |
| HbSPL9 | GT019690.t1 | CM021239.1 | 23400072:23405078 | 10 | 9 | 10 | 0 | 293 | 32,788.22 | 9.21 | 64.77 |
| HbSPL10 | GT039310.t1 | CM021239.1 | 26622170:26632180 | 12 | 11 | 12 | 0 | 995 | 110,256.79 | 6.4 | 43.85 |
| HbSPL11 | GT033052.t1 | CM021239.1 | 39504948:39506565 | 3 | 2 | 3 | 0 | 339 | 37001.37 | 9.5 | 59.73 |
| HbSPL12 | GT019861.t1 | CM021240.1 | 72675955:72677659 | 3 | 2 | 3 | 0 | 686 | 77,405.9 | 5.86 | 55.24 |
| HbSPL13 | GT025439.t1 | CM021241.1 | 2028142:2034204 | 10 | 9 | 10 | 0 | 368 | 41,070.02 | 9.44 | 51.55 |
| HbSPL14 | GT042310.t1 | CM021241.1 | 27863168:27864903 | 3 | 2 | 3 | 0 | 862 | 97,241.22 | 6.29 | 47.94 |
| HbSPL15 | GT040479.t1 | JAAGAX010000040.1 | 559530:565521 | 4 | 3 | 4 | 0 | 398 | 44,823.71 | 8.64 | 49.13 |
| HbSPL16 | GT028737.t1 | JAAGAX010000040.1 | 620658:626675 | 4 | 3 | 4 | 0 | 404 | 44,567.07 | 8.41 | 55.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, R.; Yuan, B.; Yang, Y.; Ao, G.; Wang, J. Genome-Wide Analysis of SPL Gene Families Illuminate the Evolution Patterns in Three Rubber-Producing Plants. Diversity 2023, 15, 983. https://doi.org/10.3390/d15090983
Su R, Yuan B, Yang Y, Ao G, Wang J. Genome-Wide Analysis of SPL Gene Families Illuminate the Evolution Patterns in Three Rubber-Producing Plants. Diversity. 2023; 15(9):983. https://doi.org/10.3390/d15090983
Chicago/Turabian StyleSu, Renping, Boxuan Yuan, Yang Yang, Guoen Ao, and Juanying Wang. 2023. "Genome-Wide Analysis of SPL Gene Families Illuminate the Evolution Patterns in Three Rubber-Producing Plants" Diversity 15, no. 9: 983. https://doi.org/10.3390/d15090983
APA StyleSu, R., Yuan, B., Yang, Y., Ao, G., & Wang, J. (2023). Genome-Wide Analysis of SPL Gene Families Illuminate the Evolution Patterns in Three Rubber-Producing Plants. Diversity, 15(9), 983. https://doi.org/10.3390/d15090983







