Fragilaria shirshovii sp. nov.—A New Species of Araphid Diatoms (Bacillariophyta, Fragilariophyceae) from the Gulf of Ob (Kara Sea, Arctic)
Abstract
1. Introduction
2. Materials and Methods
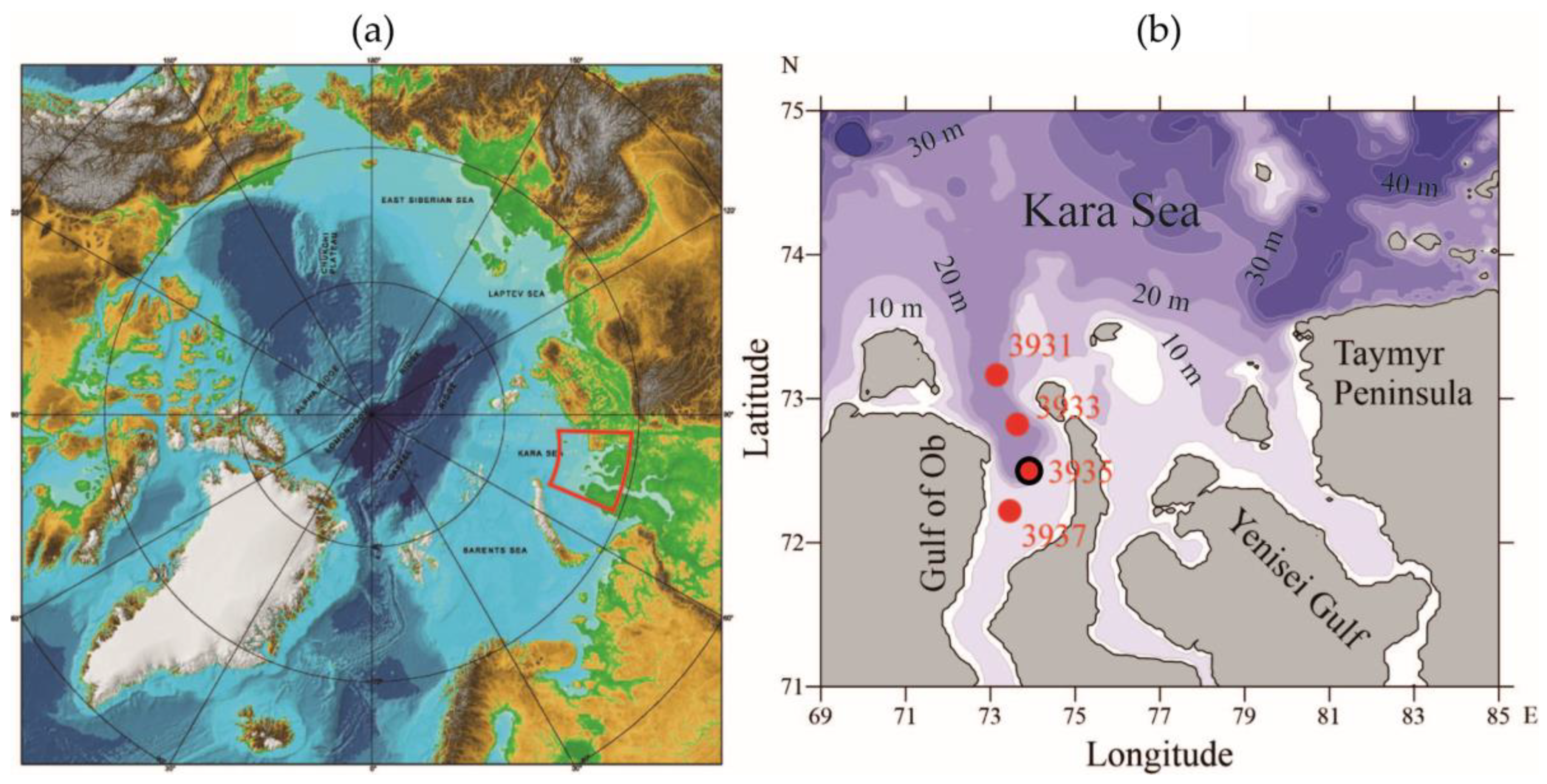
2.1. Study Area and Environmental Conditions
2.2. Sample Collection
2.3. Diatom Strain Isolation and Culturing
2.4. Preparation of Slides and Microscope Investigation
2.5. Molecular Analyses
3. Results
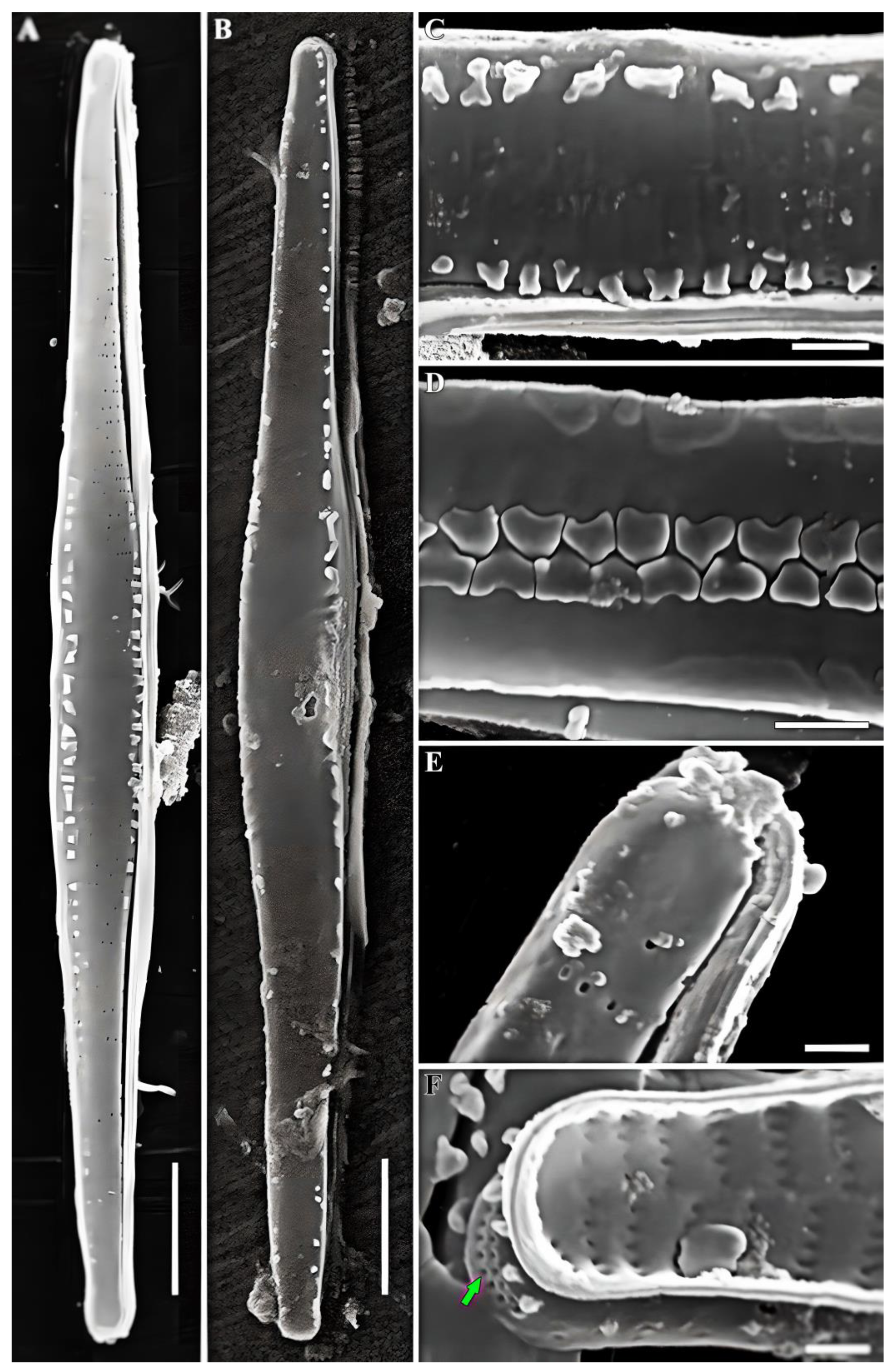
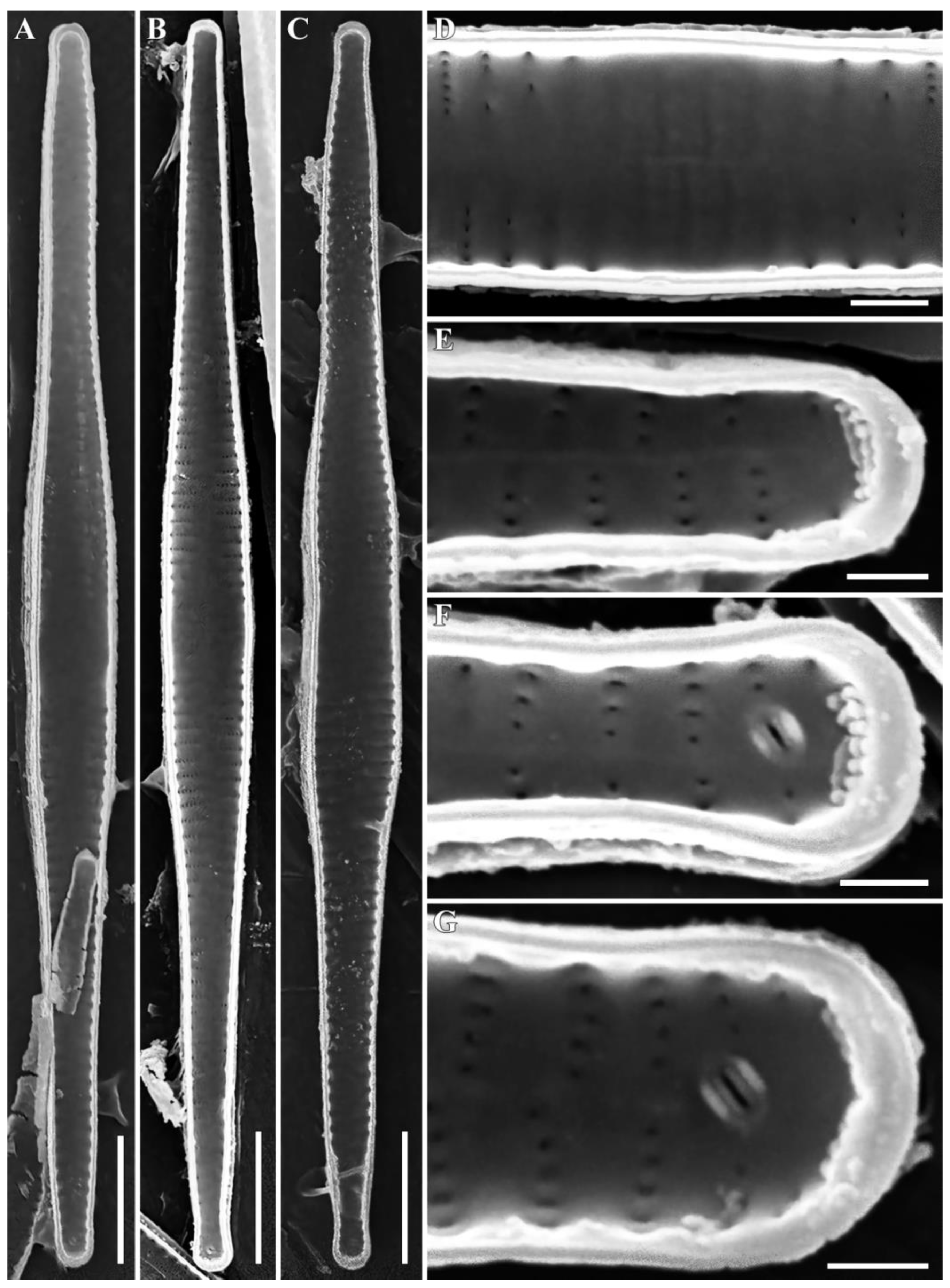
3.1. Morphological Analysis
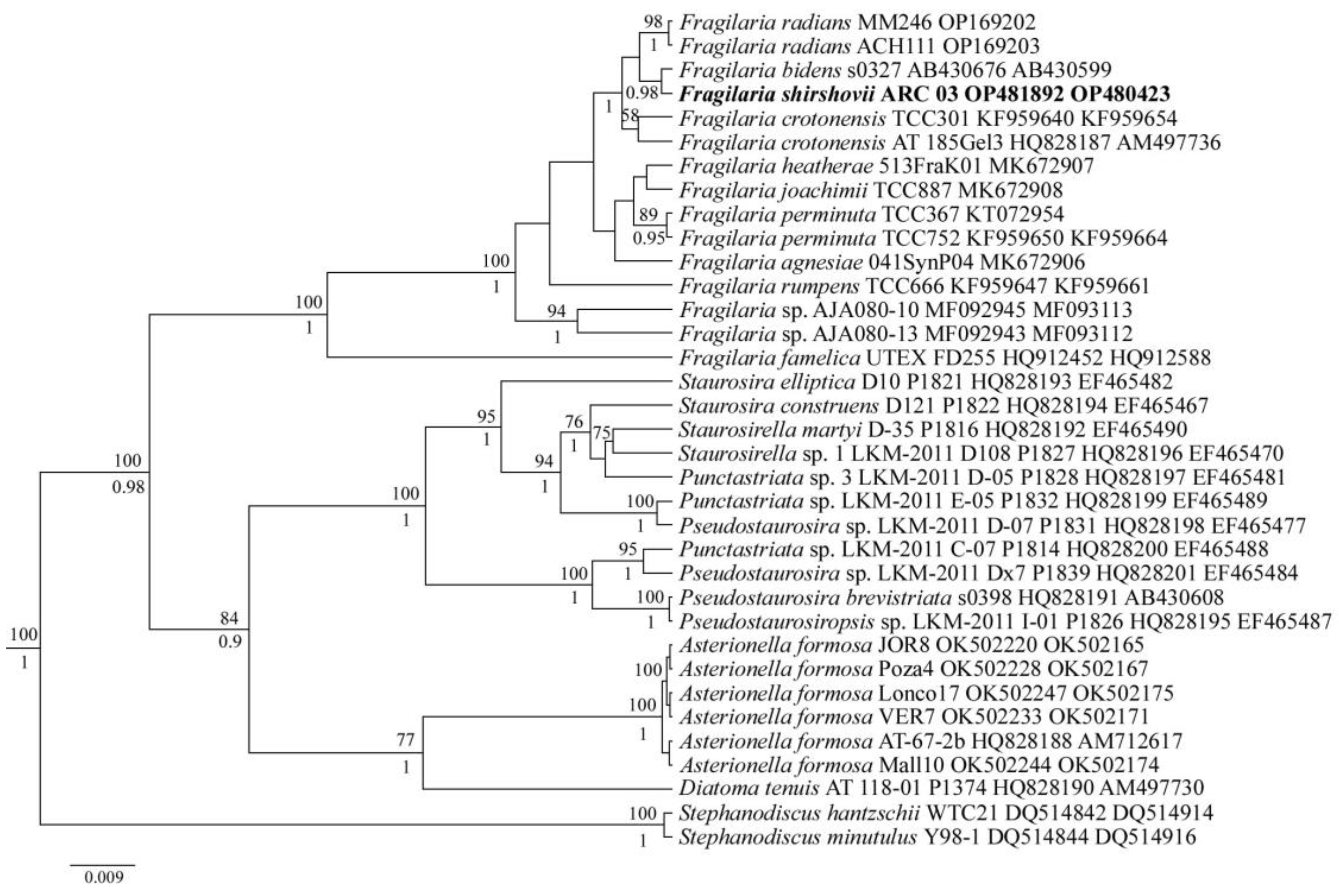
3.2. Molecular Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carmack, E.C.; Yamamoto-Kawai, M.; Haine, T.W.N.; Bacon, S.; Bluhm, B.A.; Lique, C.; Melling, H.; Polyakov, I.V.; Straneo, F.; Timmermans, M.L. Freshwater and its role in the Arctic Marine System: Sources, disposition, storage, export, and physical and biogeochemical consequences in the Arctic and global oceans. J. Geophys. Res. Biogeosci. 2016, 121, 675–717. [Google Scholar] [CrossRef]
- Flint, M.V.; Poyarkov, S.G.; Rimsky-Korsakov, N.A. Ecosystems of the Russian Arctic-2015 (63rd Cruise of the research vessel Akademik Mstislav Keldysh). Oceanology 2016, 56, 459–461. [Google Scholar] [CrossRef]
- PAME. Working Group on the Protection of the Arctic Marine Environment. Report to the Third Ministerial Conference on the Protection of the Arctic Environment, Norwegian Ministry of Environment; PAME: Oslo, Norway, 1996. [Google Scholar]
- Flint, M.V. Cruise 54th of the research vessel Akademik Mstislav Keldysh in the Kara Sea. Oceanology 2010, 50, 637–642. [Google Scholar] [CrossRef]
- Polyakov, I.; Walsh, D.; Dmitrenko, I.; Colony, R.L.; Timokhov, L.A. Arctic ocean variability derived from historical observations. Geophys. Res. Lett. 2003, 30, 1298. [Google Scholar] [CrossRef]
- Polyakov, I.V.; Alexeev, V.A.; Belchansky, G.I.; Dmitrenko, I.A.; Ivanov, V.V.; Kirillov, S.A.; Korablev, A.A.; Steele, M.; Timokhov, L.A.; Yashayaev, I. Arctic Ocean Freshwater Changes over the Past 100 Years and Their Causes. J. Clim. 2008, 21, 364–384. [Google Scholar] [CrossRef]
- Dittmar, T.; Kattner, G. The biogeochemistry of the river and shelf ecosystem of the Arctic Ocean: A review. Mar. Chem. 2003, 83, 103–120. [Google Scholar] [CrossRef]
- Kaika, M. The Water Framework Directive: A New Directive for a Changing Social, Political and Economic European Framework. Eur. Plan. Stud. 2003, 11, 299–316. [Google Scholar] [CrossRef]
- Lobus, N.V.; Kulikovskiy, M.S. The Co-Evolution Aspects of the Biogeochemical Role of Phytoplankton in Aquatic Ecosystems: A Review. Biology 2023, 12, 92. [Google Scholar] [CrossRef]
- Kelly, M.; Boonham, N.; Juggins, S.; Kille, P.; Mann, D.; Pass, D.; Sapp, M.; Sato, S.; Glover, R.; Walsh, K. A DNA Based Diatom Metabarcoding Approach for Water Framework Directive Classification of Rivers; Environment Agency: Bristol, UK, 2018; ISBN 978-1-84911-406-6.
- Wang, X.; Zheng, B.; Liu, L.; Li, L. Use of Diatoms in River Health Assessment. Annu. Res. Rev. Biol. 2014, 4, 4054–4074. [Google Scholar] [CrossRef]
- Lange-Bertalot, H.; Hofmann, G.; Werum, M.; Cantonati, M. Freshwater Benthic Diatoms of Central Europe: Over 800 Common Species Used in Ecological Assessment; Koeltz Botanical Books: Stuttgart, Germany, 2017. [Google Scholar]
- Su, Y.; Lundholm, N.; Ellegaard, M. Effects of abiotic factors on the nanostructure of diatom frustules—Ranges and variability. Appl. Microbiol. Biotechnol. 2018, 102, 5889–5899. [Google Scholar] [CrossRef]
- Kulikovskiy, M.S.; Glushchenko, A.M.; Genkal, S.I.; Kuznetsova, I.V. Identification Book of Diatoms from Russia; Filigran: Yaroslavl, Russia, 2016; ISBN 978-5-906682-72-7. [Google Scholar]
- Kahlert, M.; Kelly, M.G.; Mann, D.G.; Rimet, F.; Sato, S.; Bouchez, A.; Keck, F. Connecting the morphological and molecular species concepts to facilitate species identification within the genus Fragilaria (Bacillariophyta). J. Phycol. 2019, 55, 948–970. [Google Scholar] [CrossRef]
- Zakharova, Y.; Marchenkov, A.; Petrova, D.; Bukin, Y.; Morozov, A.; Bedoshvili, Y.; Podunay, Y.; Davidovich, O.; Davidovich, N.; Bondar, A.; et al. Delimitation of Some Taxa of Ulnaria and Fragilaria (Bacillariophyceae) Based on Genetic, Morphological Data and Mating Compatibility. Diversity 2023, 15, 271. [Google Scholar] [CrossRef]
- Sukhanova, I.N.; Flint, M.V.; Mosharov, S.A.; Sergeeva, V.M. Structure of the phytoplankton communities and primary production in the Ob River estuary and over the adjacent Kara Sea shelf. Oceanology 2010, 50, 743–758. [Google Scholar] [CrossRef]
- Lange-Bertalot, H.; Ulrich, S. Contributions to the taxonomy of needle-shaped Fragilaria and Ulnaria species. Lauterbornia 2014, 78, 1–73. [Google Scholar]
- Krammer, K.; Lange-Bertalot, H. Bacillariophyceae 3. Teil: Centrales, Fragilariaceae, Eunotiaceae. In Suβwasserflora von Mitteleuropa; Ett, H., Gerloff, J., Heynig, H., Mollenhauer, D., Eds.; Gustav Fischer: Stuttgart, Germany, 1991; Volume 2, p. 598. [Google Scholar]
- Pavlov, V.K.; Timokhov, L.A.; Baskakov, G.A.; Kulakov, M.Y.; Kurazhov, V.K.; Pavlov, P.V.; Pivovarov, S.V.; Stanovoy, V.V. Hydrometeorological Regime of the Kara, Laptev, and East-Siberian Seas; Washington University Seattle Applied Physics Lab, USA: Seattle, WA, USA, 1996. [Google Scholar]
- Osadchiev, A.; Konovalova, O.; Gordey, A. Water Exchange between the Gulf of Ob and the Kara Sea during Ice-Free Seasons: The Roles of River Discharge and Wind Forcing. Front. Mar. Sci. 2021, 8, 741143. [Google Scholar] [CrossRef]
- Lobus, N.V.; Glushchenko, A.M.; Osadchiev, A.A.; Maltsev, Y.I.; Kapustin, D.A.; Konovalova, O.P.; Kulikovskiy, M.S.; Krylov, I.N.; Drozdova, A.N. Production of Fluorescent Dissolved Organic Matter by Microalgae Strains from the Ob and Yenisei Gulfs (Siberia). Plants 2022, 11, 3361. [Google Scholar] [CrossRef] [PubMed]
- Sazhin, A.F.; Romanova, N.D.; Mosharov, S.A. Bacterial and primary production in the pelagic zone of the Kara Sea. Oceanology 2010, 50, 759–765. [Google Scholar] [CrossRef]
- Guillard, R.R.L.; Lorenzen, C.J. Yellow-green algae with chlorophyllide c1,2. J. Phycol. 1972, 8, 10–14. [Google Scholar] [CrossRef]
- Andersen, R.A.; Masanobu, K. Microalgae Isolation Techniques. In Algal Culturing Techniques; Andersen, R.A., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2005; p. 578. ISBN 0-12-088426-7. [Google Scholar]
- Zimmermann, J.; Jahn, R.; Gemeinholzer, B. Barcoding diatoms: Evaluation of the V4 subregion on the 18S rRNA gene, including new primers and protocols. Org. Divers. Evol. 2011, 11, 173–192. [Google Scholar] [CrossRef]
- Ruck, E.C.; Theriot, E.C. Origin and Evolution of the Canal Raphe System in Diatoms. Protist 2011, 162, 723–737. [Google Scholar] [CrossRef]
- Daugbjerg, N.; Andersen, R.A. A molecular phylogeny of the heterokont algae based on analyses of chloroplast encoded rbcL sequence data. J. Phycol. 1997, 33, 1031–1041. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 2010, 26, 1899–1900. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Delgado, C.; Novais, M.H.; Blanco, S.; de Almeida, S.F.P. Examination and comparison of Fragilaria candidagilae sp. nov. with type material of Fragilaria recapitellata, F. capucina, F. perminuta, F. intermedia and F. neointermedia (Fragilariales, Bacillariophyceae). Phytotaxa 2015, 231, 1–18. [Google Scholar] [CrossRef]
- Heudre, D.; Wetzel, C.E.; Moreau, L.; Van de Vijver, B.; Ector, L. On the identity of the rare Fragilaria subconstricta (Fragilariaceae), with Fragilaria species forming ribbon-like colonies shortly reconsidered. Plant Ecol. Evol. 2019, 152, 327–339. [Google Scholar] [CrossRef]
- Van de Vijver, B.; Schuster, T.M.; Williams, D.M.; Kusber, W.H. Was Fragilaria pararumpens Lange-Bertalot, G. Hofmann & Werum 2011 new to science? Not. Algarum 2021, 180, 1–4. [Google Scholar]
- Medlin, L.; Yang, I.; Sato, S. Evolution of the Diatoms. VII. Four gene Phylogeny assesses the validity of selected araphid genera. Nov. Hedwigia 2012, 141, 505–513. [Google Scholar]
- Alexson, E.E.; Reavie, E.D.; Van de Vijver, B.; Wetzel, C.E.; Ector, L.; Wellard Kelly, H.A.; Aliff, M.N.; Estepp, L.R. Revision of the needle-shaped Fragilaria species (Fragilariaceae, Bacillariophyta) in the Laurentian Great Lakes (United States of America, Canada). J. Great Lakes Res. 2022, 48, 999–1020. [Google Scholar] [CrossRef]
- Kulikovskiy, M.S.; Genkal, S.I.; Mikheyeva, T.M. New Data for the Bacillariophyta Flora of Belarus. 1. Family Naviculaceae Kütz. Int. J. Algae 2010, 12, 37–56. [Google Scholar] [CrossRef]
- Bąk, M.; Witkowski, A.; Żelazna-Wieczorek, J.; Wojtal, A.Z.; Szczepocka, E.; Szulc, K.; Szulc, B. Klucz do Oznaczania Okrzemek w Fitobentosie na Potrzeby Oceny Stanu Ekologicznego Wód Powierzchniowych w Polsce; Biblioteka Monitoringu Środowiska: Warszawa, Poland, 2012. [Google Scholar]
- Chudaev, D.A.; Gololobova, M.A. Diatoms of the Glubokoe Lake (Moscow Region); KMK: Moscow, Russia, 2016. [Google Scholar]
- Bazhenova, O.P.; Glushchenko, A.M.; Igoshkina, I.Y.; Shkilev, T.E.; Kulikovskiy, M.S. Plankton diatoms (Fragilariophyceae, Bacillariophyceae) from the middle part of Irtysh River. Nov. Sist. Nizshikh Rastenii 2019, 53, 207–240. [Google Scholar] [CrossRef]



| F. shirshovii sp. nov. | F. crotonensis (Type Material) | F. crotonensis (Sensu Auct.) | F. pararumpens | F. bidens | F. perminuta | |
|---|---|---|---|---|---|---|
| Colony formation | double comb-shaped | ribbon-like | ribbon- or comb-shaped | band-like aggregates, double comb-shaped | band-like | none |
| Outline | fusiform (in large specimens) to almost linear (in smaller specimens) | n.d. | narrowly lanceolate with proximal inflations, which can sometimes be constricted | rather narrow, lanceolate | linear | lanceolate or rhombic |
| Spines | spatula-shaped at the central part of the valve, small conical near the valve ends | n.d. | spatula-shaped at the central part of the valve, small conical near the valve ends | conical near the apex to spatulate in the middle | no data | absent |
| Valve ends | capitate in large specimens to subcapitate in smaller specimens | n.d. | subcapitate | subcapitate | wedge-shaped, often weakly protracted, bluntly to widely rounded | slightly rostrate |
| Axial area | narrow lanceolate | n.d. | in the form of very narrow sternum | absent | absent | absent |
| Central area | rounded or rectangular fascia | n.d. | rectangular hyaline zone | elliptic | rounded or rectangular fascia | strongly unilateral |
| Length, μm | 30.4–47.8 | 45 | 40–170 | 25–50 | 10–50 | 7–40 |
| Breadth, μm | 3.1–3.7 | n.d. | 2–4(5) | 2.5–3.5 | (2)3–4 | 3–4 |
| Striae in 10 μm | 17–18 | 15–16 | 14–19 | 16–18 | (11)15–18 | 17–21 |
| Areolae per 10 μm | 55–60 | n.d. | composed of up to six apically elongated to almost rounded areolae | ≈50 | no data | 60–65 1 |
| Position of slit of rimoportula | about under 45° to striae | n.d. | oriented perpendicular to the striae, located in valve face or face/mantle junction | oriented almost parallel to the striae | no data | oriented almost parallel to the striae |
| Habitat and ecology | This species is known from the type locality with the environmental conditions described in this study | n.d. | Despite intensive investigations, ecology amplitude is still not well defined, as differentiation from closely related forms in not straightforward. Mainly observed in mesotrophic, alkaline freshwater habitats with different electrolyte content. Eurythermic species. | Small, oligosaprobic running water and lakes on siliceous substrata | Alkaline meso- and strongly eutrophic waters | Calcium-bicarbonate-rich, meso- to moderately eutrophic lakes. Usually absent from more strongly eutrophic and saprobically impacted freshwater habitats. |
| References | This study | [18] | [14,18,38] | [12,35,36] | [39,40,41,42] | [12,15,34] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lobus, N.V.; Glushchenko, A.M.; Genkal, S.I.; Maltsev, Y.I.; Kulikovskiy, M.S. Fragilaria shirshovii sp. nov.—A New Species of Araphid Diatoms (Bacillariophyta, Fragilariophyceae) from the Gulf of Ob (Kara Sea, Arctic). Diversity 2023, 15, 916. https://doi.org/10.3390/d15080916
Lobus NV, Glushchenko AM, Genkal SI, Maltsev YI, Kulikovskiy MS. Fragilaria shirshovii sp. nov.—A New Species of Araphid Diatoms (Bacillariophyta, Fragilariophyceae) from the Gulf of Ob (Kara Sea, Arctic). Diversity. 2023; 15(8):916. https://doi.org/10.3390/d15080916
Chicago/Turabian StyleLobus, Nikolay V., Anton M. Glushchenko, Sergei I. Genkal, Yevhen I. Maltsev, and Maxim S. Kulikovskiy. 2023. "Fragilaria shirshovii sp. nov.—A New Species of Araphid Diatoms (Bacillariophyta, Fragilariophyceae) from the Gulf of Ob (Kara Sea, Arctic)" Diversity 15, no. 8: 916. https://doi.org/10.3390/d15080916
APA StyleLobus, N. V., Glushchenko, A. M., Genkal, S. I., Maltsev, Y. I., & Kulikovskiy, M. S. (2023). Fragilaria shirshovii sp. nov.—A New Species of Araphid Diatoms (Bacillariophyta, Fragilariophyceae) from the Gulf of Ob (Kara Sea, Arctic). Diversity, 15(8), 916. https://doi.org/10.3390/d15080916








