Phylogenetic Position of the Genus Manunema (Nematoda, Plectida, Leptolaimidae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Morphological Analysis
2.2. Molecular Analysis
2.2.1. Sample Preparation and Species Verification
2.2.2. DNA Extraction and Amplification
2.2.3. Data Analysis
3. Results
3.1. Systematics
3.2. Diagnosis (Amended and Updated after Gerlach 1957, Vitiello and De Coninck 1968, Lorenzen 1996, and Holovachov 1914)
3.3. Annotated List of Manunema Species
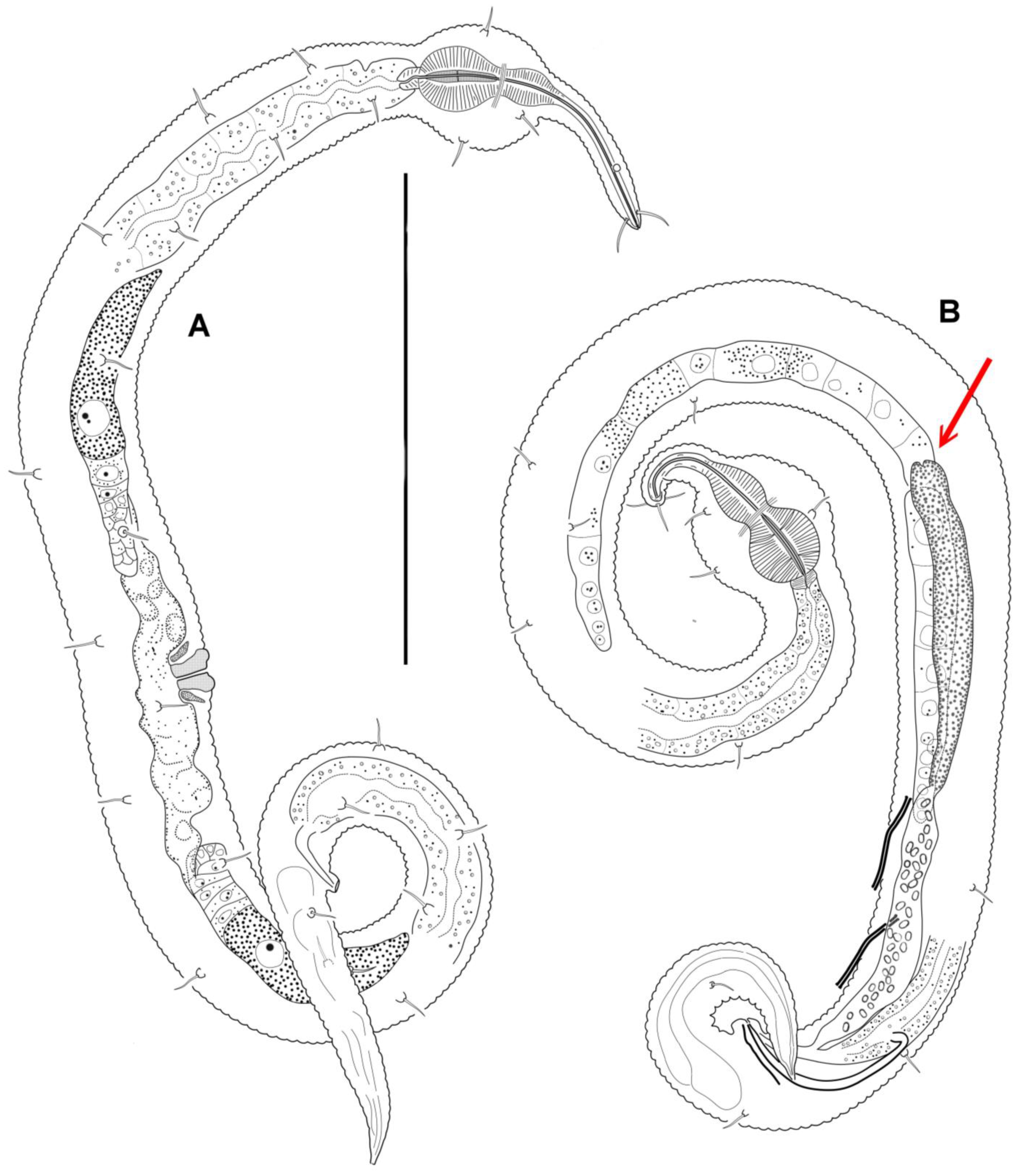
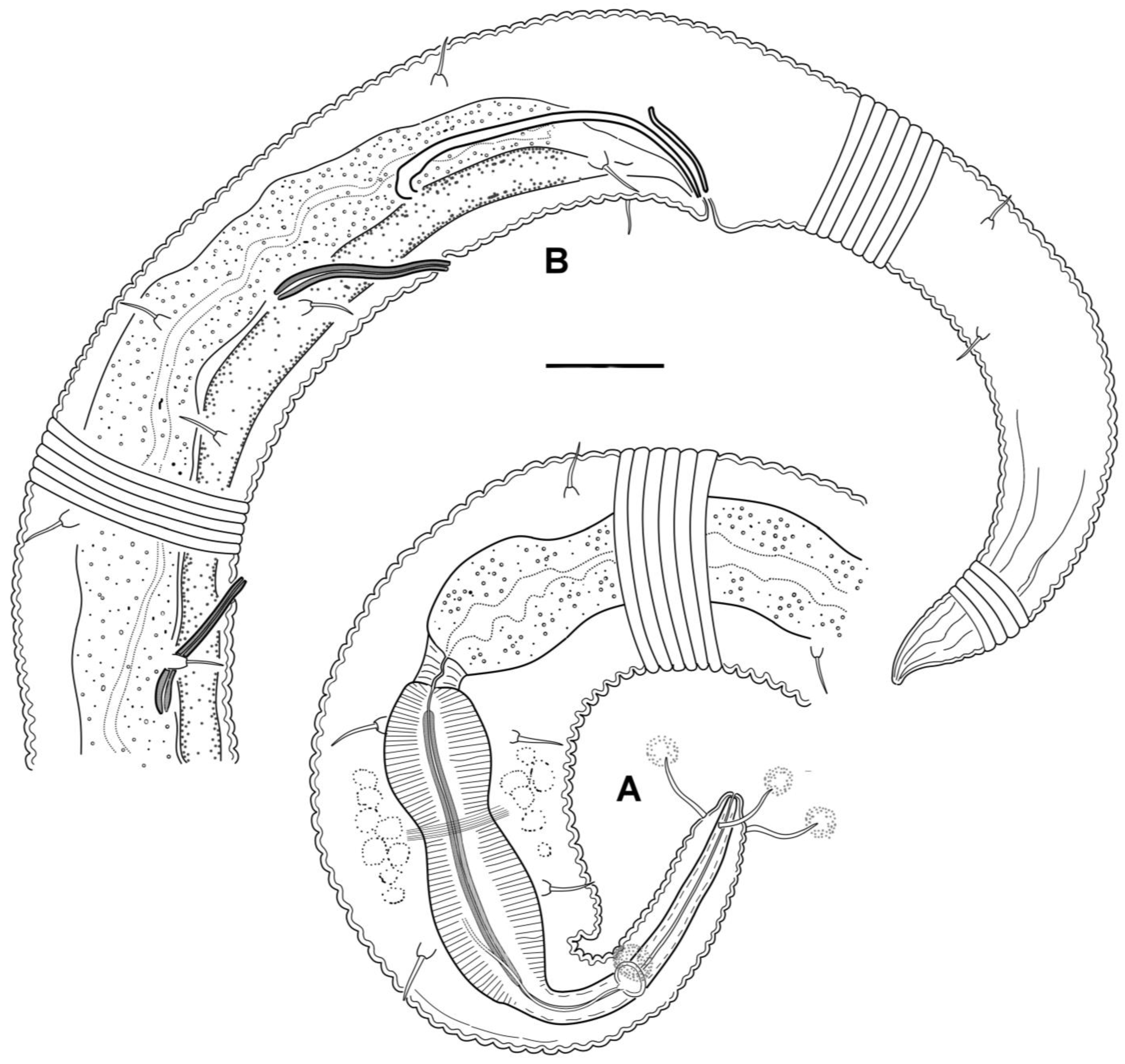
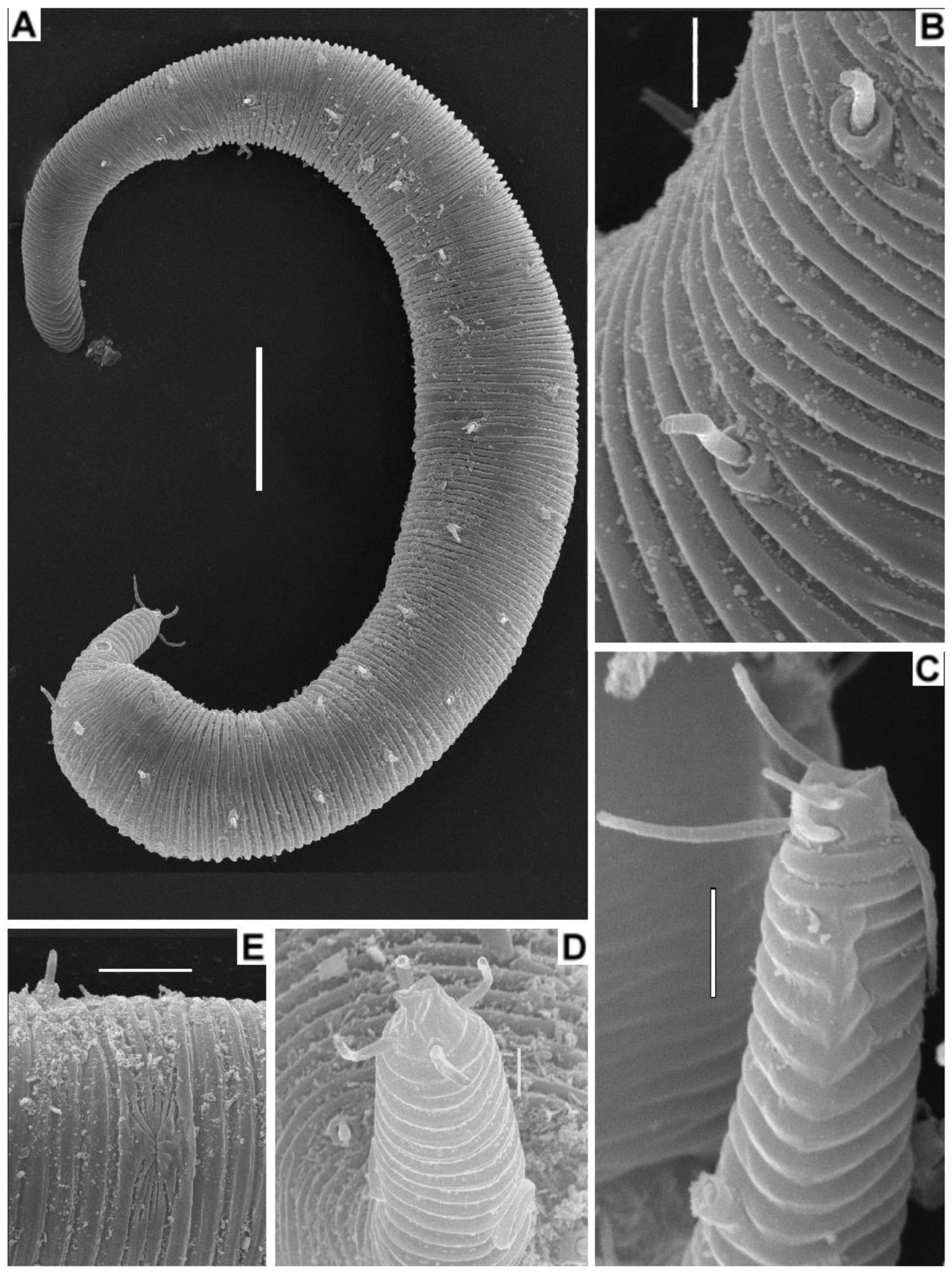
3.4. Description
3.5. Molecular Phylogenetics
3.5.1. Pairwise Distance
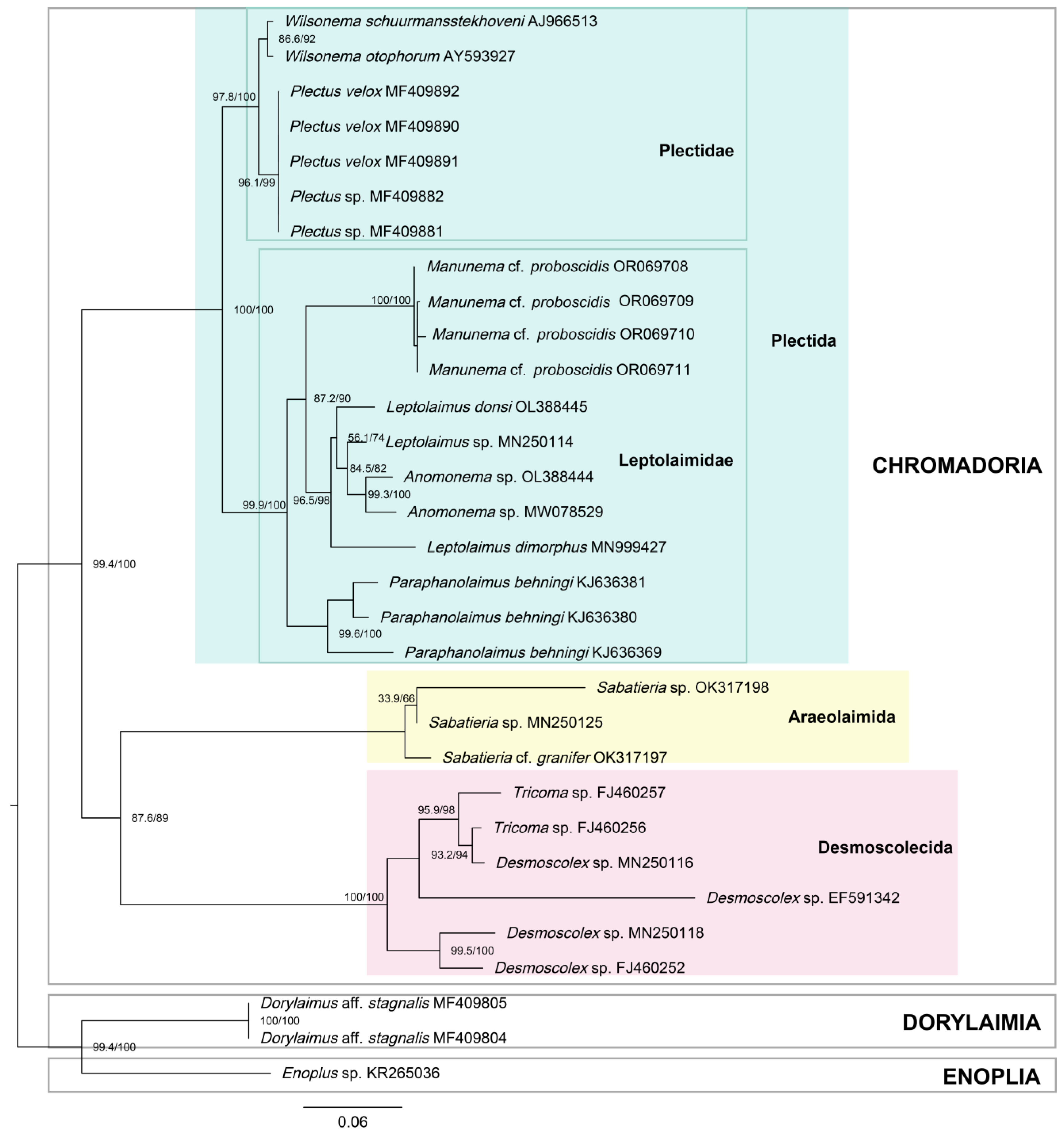
3.5.2. Phylogenetic Tree
4. Discussion
4.1. Female Reproductive System
4.2. Male Reproductive System
4.3. Phylogeny and Position in the Nematode System
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jeong, R.; Tchesunov, A.V.; Lee, W. Two species of Thoracostomopsidae (Nematoda: Enoplida) from Jeju Island, South Korea. PeerJ 2020, 8, e9037. [Google Scholar] [CrossRef]
- Jeong, R.; Tchesunov, A.V.; Lee, W. Bibliographic revision of Mesacanthion Filipjev, 1927 (Nematoda: Thoracostomopsidae) with description of a new species from Jeju Island, South Korea. PeerJ 2019, 7, e8023. [Google Scholar] [CrossRef]
- Tchesunov, A.; Jeong, R.; Lee, W. Two New Marine Free-Living Nematodes from Jeju Island Together with a Review of the Genus Gammanema Cobb 1920 (Nematoda, Chromadorida, Selachinematidae). Diversity 2020, 12, 19. [Google Scholar] [CrossRef]
- Tchesunov, A.V.; Jeong, R.; Lee, W. Onyx disparamphis sp. n. (Nematoda, Desmodorida) from South Korea with a taxonomic review of the genus. PeerJ 2022, 10, e13010. [Google Scholar] [CrossRef] [PubMed]
- Tchesunov, A.V.; Jeong, R.; Lee, W. A new genus and species of the family Microlaimidae (Nematoda: Chromadorea) from intertidal sand of the Jeju Island, South Korea. Zootaxa 2021, 5020, 130–140. [Google Scholar] [CrossRef]
- Gerlach, S.A. Die Nematodenfauna des Sandstrandes an der Küste von Mittelbrasilien (Brasilianische Meeres-Nematoden IV). Mitteilungen Aus Dem Zool. Mus. Berl. 1957, 33, 411–459. [Google Scholar] [CrossRef]
- Vitiello, P.; De Coninck, L. Peresiana annulata n. gen., n. sp., type intéressant de Desmoscolecida. Rapp. Comm. Int. Mer Médit. 1968, 19, 201–204. [Google Scholar]
- Andrássy, I. Nematoden aus Strand- und Hohlenbiotopen von Kuba. Acta Zool. Acad. Sci. Hung. 1973, 19, 233–270. [Google Scholar]
- Hopper, B.E. Marine nematodes from the coast line of the Gulf of Mexico. III. Additional species from Gulf shores, Alabama. Can. J. Zool. 1963, 41, 841–863. [Google Scholar] [CrossRef]
- Lorenzen, S. Entwurf eines phylogenetischen Systems der freilebenden Nematoden. In Veröffentlichungen des Instituts für Meeresforschung in Bremerhaven; Kommissionsverlag F. Leuwer: Bremerhaven, Germany, 1981; Volume 7, 472p. [Google Scholar]
- Riemann, F.; von Thun, W.; Lorenzen, S. Über den phylogenetischen Zusammenhang zwischen Desmoscolecidae und Leptolaimidae (freilebende Nematoden). In Veröffentlichungen des Instituts für Meeresforschung Bremerhaven; Kommissionsverlag F. Leuwer: Bremerhaven, Germany, 1971; Volume 13, pp. 147–152. [Google Scholar]
- Lorenzen, S. The Phylogenetic Systematics of Freeliving Nematodes (Translation of the 1981 Paper); Ray Society: Andover, UK, 1994; Volume 162, pp. 1–383. [Google Scholar]
- Holovachov, O.; Boström, S. Morphology and systematics of the superfamilies Leptolaimoidea Örley, 1880 and Camacolaimoidea Micoletzky, 1824 (Nematoda: Plectida). J. Nematode Morphol. Syst. 2004, 7, 1–49. [Google Scholar]
- Stewart, A.C.; Nicholas, W.L. Manunema pectenophora sp. nov. (Peresianidae, Leptolaimina), a nematode possessing unusual male supplementary organs. Trans. R. Soc. S. Aust. 1995, 119, 163–169. [Google Scholar]
- Barnes, N.; Ferrero, T.J. Two new species of Manunema (Plectida: Peresianidae) from the Arabian Gulf, with notes on the phylogeny of the genus. Zootaxa 2009, 2053, 43–58. [Google Scholar] [CrossRef]
- De Ley, P.; Blaxter, M.L. A new system for Nematoda: Combining morphological characters with molecular trees, and translating clades into ranks and taxa. In Proceedings of the Fourth International Congress of Nematology, Tenerife, Spain, 8–13 June 2002; Brill: Leiden, The Netherlands, 2004; Volume 2, pp. 633–653. [Google Scholar]
- Hodda, M. Phylum Nematoda: A classification, catalogue and index of valid genera, with a census of valid species. Zootaxa 2022, 5114, 1–289. [Google Scholar] [CrossRef]
- Holovachov, O. 7.16 Order Plectida Gadea, 1973. In Handbook of Zoology Gastrotricha, Cycloneuralia and Gnathifera. Volume 2: Nematoda; Schmidt-Rhaesa, A., Ed.; De Gruyter: Berlin, Germany; Boston, MA, USA, 2014; pp. 487–535. [Google Scholar] [CrossRef]
- Burgess, B. An improved protocol for separating meiofauna from sediments using colloidal silica sols. Mar. Ecol. Prog. Ser. 2001, 214, 161–165. [Google Scholar] [CrossRef]
- Seinhorst, J. A rapid method for the transfer of nematodes from fixative to anhydrous glycerin. Nematologica 1959, 4, 67–69. [Google Scholar] [CrossRef]
- Hopper, D. Drawing and Measuring Nematodes; Ministry of Agriculture, Fisheries and Food: Her Majesty’s Stationery Office: London, UK, 1970.
- Meeker, N.D.; Hutchinson, S.A.; Ho, L.; Trede, N.S. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques 2007, 43, 610–614. [Google Scholar] [CrossRef]
- Derycke, S.; Vanaverbeke, J.; Rigaux, A.; Backeljau, T.; Moens, T. Exploring the use of cytochrome oxidase c subunit 1 (COI) for DNA barcoding of free-living marine nematodes. PLoS ONE 2010, 5, e13716. [Google Scholar] [CrossRef]
- Bhadury, P.; Austen, M.C.; Bilton, D.T.; Lambshead, P.J.D.; Rogers, A.D.; Smerdon, G.R. Development and evaluation of a DNA-barcoding approach for the rapid identification of nematodes. Mar. Ecol. Prog. Ser. 2006, 320, 1–9. [Google Scholar] [CrossRef]
- Carta, L.K.; Li, S. Improved 18S small subunit rDNA primers for problematic nematode amplification. J. Nematol. 2018, 50, 533–542. [Google Scholar] [CrossRef]
- Carta, L.K.; Li, S. PCR amplification of a long rDNA segment with one primer pair in agriculturally important nematodes. J. Nematol. 2019, 51, 1–8. [Google Scholar] [CrossRef] [PubMed]
- De Ley, P.; De Ley, I.T.; Morris, K.; Abebe, E.; Mundo-Ocampo, M.; Yoder, M.; Heras, J.; Waumann, D.; Rocha-Olivares, A.; Jay Burr, A.H.; et al. An integrated approach to fast and informative morphological vouchering of nematodes for applications in molecular barcoding. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 1945–1958. [Google Scholar] [CrossRef] [PubMed]
- Bowles, J.; Blair, D.; Mcmanus, D.P. Genetic-Variants within the Genus Echinococcus Identified by Mitochondrial-DNA Sequencing. Mol. Biochem. Parasit. 1992, 54, 165–174. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal-W—Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide-Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tchesunov, A.V. Biology of Marine Nematodes; KMK Scientific Press: Moscow, Russia, 2006; p. 367. [Google Scholar]
- Lorenzen, S. New and known gonadal characters in free-living nematodes and the phylogenetic implications. Z. Syst. Zool. Evol. 1978, 16, 108–115. [Google Scholar] [CrossRef]
- Filipjev, I.N. The classification of the free-living nematodes and their relation to the parasitic nematodes. Smithson. Misc. Collect. 1934, 89, 1–63. [Google Scholar]
- Khan, S.H.; Coomans, A. Observations on the juvenile stages of Miconchus studeri (Nematoda: Mononchida). Biol. Jaarb. 1980, 48, 111–118. [Google Scholar]
- Vincx, M.; Sharma, J.; Smol, N. On the identity and nomenclature of Paracanthonchus caecus (Bastian, 1865), with a redefinition of the genus Paracanthonchus Micoletzky (Nematoda, Cyatholaimidae). Zool. Scr. 1982, 11, 243–263. [Google Scholar] [CrossRef]
- Coomans, A.; Jacobs, L.J. Halalaimus algeriensis n. sp. (Nematoda) from the Sahara. Hydrobiologia 1983, 102, 39–44. [Google Scholar] [CrossRef]
- Vincx, M. Redescription and ontogenetic of Desmodora schulzi Gerlach 1950. Biol. Jaarb. 1983, 51, 171–179. [Google Scholar]
- Coomans, A.; Heyns, J. Oncholaimus-Jessicae N-Sp (Nematoda, Oncholaimidae) from Fresh-Water in the Transvaal. S. Afr. J. Zool. 1986, 21, 197–201. [Google Scholar]
- Tchesunov, A.V.; Sturhan, D. Studies on Domorganus macronephriticus Goodey, 1947 (Nematoda: Ohridiidae). J. Nematode Morphol. Syst. 2004, 6, 139–150. [Google Scholar]
- Jensen, P. Free-living marine nematodes from a sublittoral station in the North Sea off the Belgian coast. Biol. Jaarb. 1976, 44, 231–255. [Google Scholar]
- Holovachov, O.; de Ley, P. Order Plectida. In Freshwater Nematodes: Ecology and Taxonomy; CABI Publishing: Wallingford, UK, 2006; pp. 611–647. [Google Scholar]
- Martinez-Arce, A.; De Jesus-Nayarrete, A.; Leasi, F. DNA Barcoding for Delimitation of Putative Mexican Marine Nematodes Species. Diversity 2020, 12, 107. [Google Scholar] [CrossRef]


| Marker | Primer (Direction) | Sequence 5′-3′ | Sequence Length (bp) | Reference |
|---|---|---|---|---|
| mtCOI | JB3 (f) | TTTTTTGGGCATCCTGAGGTTTAT | 350–395 | [23,28] |
| JB5 (r) | AGCACCTAAACTTAAAACATAATGAAAATG | |||
| 18S | MN18F (f) | CGCGAATRGCTCATTACAACAGC | 336–349 | [24] |
| 22R (r) | GCCTGCTGCCTTCCTTGGA | |||
| 18S-CL-F (f) | TCAAAGATTAAGCCATGCAT | 983–1723 | [25,26] | |
| 18S-CL-R2 (r) | GTTGAGTCAAATTAAGCCGCA | |||
| 1912R (r) | TTTACGGTCAGAACTAGGG | |||
| 550F (f) | GGCAAGTCTGGTGCCAGCAGCC | |||
| 530R (r) | GCGGCTGCTGGCACCACACTT | |||
| 18S-CL-F2 (f) | CTGTGATGCCCTTAGATGTCC | |||
| 18S-CL-R5 (r) | GCGGTGTGTACAAAGGGCAGGGAC | |||
| 18S-CL-R7 (r) | ACCTTGTTACGACTTTTGCCCGGTTCA | |||
| 28S | D2A (f) | ACAAGTACCGTGAGGGAAAGTTG | 780 bp | [27] |
| D3B (r) | TCGGAAGGAACCAGCTACTA |
| Specimen | Species Name | GenBank Accession Number | ||||
|---|---|---|---|---|---|---|
| mtCOI | 18S | 28S | ||||
| JB3 | 18S-CL-F | 18S-CL-F | MN18F | D2A | ||
| /JB5 | /18S-CL-R7 | /1912R | /22R | /D3B | ||
| (~390 bp) | (~1700 bp) | (~1100 bp) | (~300 bp) | (~730 bp) | ||
| 1 | Manunema cf. proboscidis | OR068235 | OR069708 | – | OR069703 | OR069692 |
| 2 | Manunema cf. proboscidis | OR068236 | – | OR069709 | OR069704 | OR069693 |
| 3 | Manunema cf. proboscidis | OR068237 | – | – | OR069705 | OR069694 |
| 4 | Manunema cf. proboscidis | OR068238 | – | OR069710 | OR069706 | OR069695 |
| 5 | Manunema cf. proboscidis | OR068239 | – | OR069711 | OR069707 | – |
| Character | Male (from Original Description of Gerlach, 1957) | Males (n = 9) mean ± sd (Range) | Females (n = 15) Mean ± sd (Range) |
|---|---|---|---|
| Body length | 353 | 354.4 ± 54.2 (301.0–477.0) | 415.5 ± 66.0 (330.0–533.0) |
| Pharynx length | 62 | 59.9 ± 5.6 (52.0–69.0) | 63.9 ± 3.9 (58.0–71.8) |
| Tail length | 58 | 63.5 ± 8.1 (52.2–76.0) | 65.2 ± 10.3 (50.0–85.5) |
| a | 16.1 | 17.4 ± 1.6 (14.3–19.4) | 14.6 ± 2.6 (12.2–18.7) |
| b | 5.7 | 5.7 ± 0.8 (5.0–7.6) | 6.5 ± 1.0 (5.4–8.3) |
| c | 6.1 | 5.6 ± 0.6 (4.5–6.6) | 6.2 ± 0.6 (4.7–6.9) |
| c′ | 2.9 | 3.9 ± 0.6 (3.0–4.7) | 4.1 ± 0.5 (3.2–4.6) |
| V | – | – | 49.7 ± 2.6 (43.0–53.3) |
| Body diameter at level of cephalic setae | 5 | 4.3 ± 0.4 (3.8–4.8) | 4.3 ± 0.7 (3.1–5.3) |
| Body diameter at level of amphid | – | 7.5 ± 0.4 (6.9–8.1) | 8.0 ± 0.9 (6.4–8.9) |
| Body diameter at level of nerve ring | – | 24.3 ± 1.8 (22.7–27.5) | 26.8 ± 1.7 (24.6–30.3) |
| Body diameter at level of cardia | – | 21.4 ± 2.8 (16.3–23.6) | 22.8 ± 4.5 (16.0–28.0) |
| Body diameter at level of midbody | 25 | 20.4 ± 2.2 (18.0–24.8) | 28.1 ± 1.5 (25.6–31.0) |
| Body diameter at level of cloaca/anus | 20 | 15.7 ± 1.4 (14.1–17.7) | 15.7 ± 1.4 (13.8–18.6) |
| Cephalic setae length | 5 | 6.7 ± 1.2 (4.7–8.0) | 6.4 ± 0.9 (4.8–8.0) |
| Amphid width | – | 2.7 ± 0.5 (2.0–3.4) | 2.9 ± 1.0 (1.5–4.0) |
| Distance head apex—amphid | – | 18.2 ± 1.3 (17.0–19.7) | 17.6 ± 1.7 (15.0–19.0) |
| Length of pharynx posterior muscular part | – | 30.4 ± 3.3 (24.0–34.2) | 32.9 ± 3.2 (28.2–37.9) |
| Pharynx diameter of preneural muscular swelling | 5–6 | 8.9 ± 0.5 (8.3–9.7) | 2.6 ± 0.5 (1.8–3.6) |
| Pharynx diameter at nerve ring | – | 7.0 ± 0.8 (5.2–7.8) | 8.1 ± 0.8 (6.9–9.3) |
| Pharynx diameter of postneural muscular swelling | 13 | 10.8 ± 1.8 (9.3–15.0) | 13.2 ± 2.1 (10.7–17.3) |
| Length of spicules along arc | 26 | 31.2 ± 4.6 (25.9–40.6) | – |
| Length of gubernaculum | 16 | 11.2 ± 2.3 (7.9–13.2) | – |
| Length of anterior supplement | 15 | 13.9 ± 4.1 (10.4–23.0) | – |
| Length of posterior supplement | 15 | 14.0 ± 3.4 (10.0–19.6) | – |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, R.; Tchesunov, A.V. Phylogenetic Position of the Genus Manunema (Nematoda, Plectida, Leptolaimidae). Diversity 2023, 15, 914. https://doi.org/10.3390/d15080914
Jeong R, Tchesunov AV. Phylogenetic Position of the Genus Manunema (Nematoda, Plectida, Leptolaimidae). Diversity. 2023; 15(8):914. https://doi.org/10.3390/d15080914
Chicago/Turabian StyleJeong, Raehyuk, and Alexei V. Tchesunov. 2023. "Phylogenetic Position of the Genus Manunema (Nematoda, Plectida, Leptolaimidae)" Diversity 15, no. 8: 914. https://doi.org/10.3390/d15080914
APA StyleJeong, R., & Tchesunov, A. V. (2023). Phylogenetic Position of the Genus Manunema (Nematoda, Plectida, Leptolaimidae). Diversity, 15(8), 914. https://doi.org/10.3390/d15080914






