Abstract
The sea star Patiria pectinifera (Asteroidea; Asterinidae; homotypic synonym: Asterina pectinifera; Muller & Troschel, 1842) is widely distributed in the coastal regions of the Seas of East Asia and the northern Pacific Ocean. Here, a de novo genome sequence of P. pectinifera as a reference for fundamental and applied research was constructed by employing a combination of long-read Oxford Nanopore Technology (ONT) PromethION, short-read Illumina platforms, and 10 × Genomics. The draft genome of P. pectinifera, containing 13,848,344 and 156,878,348 contigs from ONT and Illumina platforms, respectively, was obtained. Assembly with CANU resulted in 2262 contigs with an N50 length of 367 kb. Finally, ARCS + LINKS assembly combined these contigs into 328 scaffolds, totaling 499 Mb with an N50 length of 2 Mbp. The estimated genome size by GenomeScope analysis was 461 Mb. BUSCO analysis indicated that 930 (97.5%) of the expected genes were found in the assembly, with 889 (93.2%) being single-copy and 41 (4.3%) duplicated after searching against the metazoan database. Annotation, utilizing sequences obtained from Illumina RNA-Seq and Pacific Biosciences Iso-Seq, led to the identification of 22,367 protein-coding genes. When examining the orthologous relationship of P. pectinifera against the scaffolds of the common sea star Patiria miniata, high contiguity was observed. Annotation of repeat elements highlighted an enrichment of 1,121,079 transposable elements, constituting 47% of the genome, suggesting their potential role in shaping the genome structure of P. pectinifera. This de novo genome assembly is expected to be a valuable resource for future studies, providing insight into the developmental, environmental, and ecological aspects of P. pectinifera biology.
1. Introduction
The Asteroidea, commonly known as sea stars or starfishes, constitutes one of the largest and most distinctive classes in the phylum Echinodermata, which has four additional other well-defined clades: Crinoidea (sea lilies and feather stars), Echinoidea (sea urchins, sand dollars, and sea biscuits), Holothuroidea (sea cucumbers), and Ophiuroidea (basket stars and brittle stars) [1,2]. Sea stars are widely distributed throughout the oceans, occupying various depths, and play a crucial role as highly predatory scavengers of benthic macrofauna, engaging in grazing activities. They exhibit remarkable diversity in terms of phenotypes, displaying extraordinary coloration, varying sizes, and responses to environmental conditions. The life cycle of sea stars involves a unique metamorphosis during growth, transitioning from a bilateral symmetric planktonic larval stage to settling as radial symmetric adults. Noteworthy for their longevity, ubiquity, prolificacy, prevalence, dominancy, possession of calcified skin for protection, and phenotypic plasticity, changes in the population dynamics of sea stars can significantly impact marine communities. Sea stars often function as keystone species, influencing the balance of ecosystems, and occasionally, their population fluctuations can have detrimental effects on commercially valuable marine resources [3,4,5]. As a result, sea stars are acknowledged as one of the most successful life forms within the animal kingdom.
Sea stars have been extensively employed in various realms of biological and ecological research. Beyond their distinct phenotypes, sea stars have long been renowned for their remarkable ability to regenerate not only their limbs but also their central bodies and, in some instances, their entire bodies [6,7]. They have been instrumental in embryonic developmental research [8,9]. The ecological and environmental significance of sea stars has been consistently emphasized due to their wide geographic habitats and their adaptive capacity to environmental fluctuations. Endowed with unique evolutionary characteristics, such as being one of three deuterostome phyla (along with Chordata and Hemichordata), the genomic information of echinoderms, such as the sea stars studied here, holds the potential to provide insights into the evolutionary transition to vertebrates. In the class Asteroidea, the genome of the crown-of-thorns sea star, Acanthaster planci, was the first to be sequenced and assembled, shedding light on its chemical signaling method for colonizing from signal individuals to large numbers [10]. This discovery suggests the potential for controlling outbreaks and preventing the substantial loss of coral reefs [10]. However, large-scale genome analysis has not been extensively characterized in other sea stars, although genome assemblies of Asterias rubens, Patiria miniata, Patiriella regularis, and Pisaster ochraceus are registered in the NCBI genome database. To date, several studies have aimed at annotating potentially functional transcripts [11,12,13] and profiling the transcriptome in tissues [14,15,16,17,18,19] and developmental stages [20,21]. To fully leverage the sea star model system and comprehend the intricate mechanisms behind regeneration, unique metamorphosis, and molecular evolution with chordates, improving the quality of nucleotide sequences and employing advanced bioinformatics for orthology assignment and enhancing annotations are imperative on the genome resource.
The sea star Patiria pectinifera (Asteroidea; Asterinidae), formerly known as Asterina pectinifera, is widely distributed in the Yellow Sea, the East Sea of Korea, and coastal regions of Japan and the Russian Federation, serving as a keystone species in the northern Pacific and Atlantic Oceans. Three species, P. pectinifera, P. miniata (North America), and P. chilensis (South America), are registered in the genus Patiria. They occasionally form dense aggregations, leading to detrimental consequences for mussel beds. Although the high quality of genome assembly has been reported for the genus Patiria, fundamentally, interspecific variations such as nucleotide polymorphism, sequence inversions, and/or chromosomal inversions are observable in different species, even within the same genus. These variations have the potential to influence distinct population structures and geographical distributions due to varying susceptibility to environmental factors and adaptive capacities. Conducting a comparative genome analysis across multiple species can yield crucial insights into speciation, adaptability to environmental changes, ecological and evolutionary patterns, genetic diversity, and the future organization of populations. The diploid chromosome number of P. pectinifera was determined to be 44 [22], but genome size estimation has not been conducted yet. In this study, we established a reference genome for P. pectinifera using long-read Oxford Nanopore Technology (ONT) PromethION. The genome of P. pectinifera was statistically compared to the available sea star genomes to understand Asteroidea-specific genomic structures. This genomic assembly will be an essential resource for genetic and genomic studies, as well as research on the ecological and evolutionary aspects of P. pectinifera.
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
Individuals of P. pectinifera were collected from Banam-ri, Goseong-gun and Gangwon-do, South Korea (38°25′37.1″ N 128°27′38.0″ E). A single specimen was used for DNA extraction and total RNA preparation [23]. For DNA isolation, fresh gonad tissue from one specimen was used, employing the classical extraction method with saturated phenol (25:24:1) and chloroform–octanol (24:1). The quantity and quality of the isolated DNA were measured using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and the integrity of the DNA was confirmed through electrophoresis on 1% agarose gel. Total RNA was also extracted from the same tissues using TRI Reagent® (Sigma–Aldrich, Inc., St. Louis, MO, USA) following the manufacturer’s instructions. The integrity, purity, and concentration of total RNA were measured using a NanoDrop® 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA), with absorbance values recorded at 230, 260, and 280 nm (A230/260, A260/280).
2.2. Oxford Nanopore Technology (ONT) PromethION Sequencing
For ONT sequencing, 1.5 μg of genomic DNA underwent a 40 kb size selection and shearing process using the BluePippin system (Sage Science, Beverly, MA, USA). Libraries were constructed using a Ligation Sequencing Kit (SQK–LSK109; Oxford Nanopore Technologies, Oxford, UK), following the manufacturer’s instructions. The libraries were sequenced with the PromethION Flow Cell Priming kit (EXP–FLP001.PRO.6) on the Flowcell (FLO–PRO002) with pore ver. R9.4. All sequencing runs were performed on the PromethION instrument (Oxford Nanopore Technologies).
2.3. 10 × Chromium Genome Library Sequencing
Genomic DNA was prepared for 10 × Genomics (Pleasanton, CA, USA) Chromium sequencing (Chromium Genome Library & Gel Bead Kit v2, PN–120 258; Chromium i7 Multiplex Kit, PN–120 262). The quality of the DNA samples was verified by pulsed-field gel electrophoresis and a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). Gel beads-in-emulsion (GEMs) were generated by combining a library of genome gel beads with 1.5 ng of genomic DNA in a master mix and partitioning oil. This process was performed using a 10 × Genomics Chromium Controller instrument along with a microfluidic Genome chip (PN-120257). BluePippin sample size selection was conducted to remove fragments shorter than 40 kb. The Chromium Controller was used, and GEM preparation was performed as instructed by the manufacturer. Barcoded DNA fragments were extracted and subjected to Illumina library construction following the Chromium Genome Reagent Kits Version 2 User Guide (PN-120258). The library yield was measured with a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA), and library fragment size and distribution were determined using an Agilent 2100 Bioanalyzer High Sensitivity DNA chip (Santa Clara, CA, USA). The library was constructed by end repairing, A-tailing, adapter ligation, and PCR amplification, and sequenced with paired-end 150 bp runs on the Illumina Novaseq 6000 platform (Illumina Inc., San Diego, CA, USA).
2.4. Illumina Sequencing
The genomic DNA library was prepared following the protocol of the Illumina TruSeq Nano DNA Library preparation kit (Illumina Inc.). For sample library preparation, 0.1 μg of high molecular weight genomic DNA with an insert size of 550 bp was randomly sheared to yield DNA fragments using the Covaris S2 instrument (Covaris Inc., Woburn, MA, USA). The fragmented DNA samples were blunt-ended and phosphorylated, and a single ‘A’ nucleotide was added to the 3′ ends of the fragments in preparation for ligation to an adapter that has a single-base ‘T’ overhang. Adapter ligation at both ends of the genomic DNA fragment conferred different sequences at the 5′ and 3′ ends of each strand in the genomic fragment. Ligated DNA was amplified with PCR to enrich fragments that have adapters on both ends. The quality of the amplified libraries was verified by capillary electrophoresis (Bioanalyzer, Agilent Technologies, Waldbronn, Germany). After quantitative PCR using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), we combined libraries that were index-tagged in equimolar amounts in the pool. High-throughput sequencing was performed using an Illumina NovaSeq 6000 platform (Illumina Inc.) following provided protocols for 2 × 150 paired-end sequencing.
2.5. Assembly
Genome size and heterozygosity were estimated using Jellyfish 2.1.4 [24] with K–values of 29. De novo assembly was conducted using CANU ver. 1.8 (RRID:SCR 015880) [25] with the ONT sequences. Error correction was performed using Pilon ver. 1.2.3 (RRID:SCR 014731) [26] with the Illumina whole genome sequences to improve the base-pair-level quality of the genome assembly. To purge duplicated haplotype contigs, the ONT sequences were screened using Purge_haplotigs ver. 1.1.1 (RRID:SCR 017616) [27] with default parameters. Finally, to obtain long continuous sequences, the ARCS ver. 1.2.4 [28] + LINKS ver. 1.8 [29] pipeline was used with the 10× Chromium barcoded paired-end reads that were created by Longranger software (version 2.2; https://support.10xgenomics.com; accessed on 16 March 2021).
We compared genome sequences between P. pectinifera scaffolds at a 25 Mb resolution and the scaffolds of P. miniata using MUMmer (version 4.02b, RRID:SCR_018171) [30]. Raw sequence hits were computed with a minimum alignment length of 300 bp. Circos (RRID:SCR_011798) [31] was employed to visualize and compare genome sequences based on the homogeneous coordinates identified using MUMmer.
2.6. Transcriptome Sequencing
Transcriptome data were obtained with Illumina paired-end sequencing (150–bp; Illumina Novaseq platform). The complementary DNA (cDNA) library was prepared using a TruSeq Sample Preparation Kit (Illumina) according to the manufacturer’s instructions. Two micrograms of total RNA were used from each sample and pooled for RNA sequencing. The pooled samples were sequenced using one SMRT cell v3 based on P6–C4 chemistry after standard full-length cDNA (1–3 kb) library preparation, and a total of two SMRT cells were sequenced on a PacBio Sequel system (Pacific Biosciences, Menlo Park, CA, USA). To obtain clean data, raw reads were filtered out by removing low-quality reads and reads containing adapters and poly–N. Demultiplexing, filtering, quality control, clustering, and polishing of the Iso-Seq sequencing data were performed using SMRT Link (ver. 6.0.0).
2.7. Completeness Assessment
To evaluate the completeness of the P. pectinifera assembly, the assembled scaffolds were subjected to Benchmarking Universal Single-Copy Orthologs (BUSCO) ver. 3.0 (RRID:SCR 015008) with default parameters, using the conservation of a core set of genes from the metazoan database (metazoa_odb10) [32].
2.8. Genome Annotation and Repeat Analysis
A de novo repeat library was constructed using RepeatModeler ver. 1.0.3 (RRID:SCR 015027) [33], which includes RECON ver. 1.08 [29] and RepeatScout ver. 1.0.5 (RRID:SCR 014653) [34], with default parameters. Tandem Repeats Finder ver. 4.09 was employed to predict consensus sequences, classification information for each repeat, and tandem repeats, including simple repeats, satellites, and low-complexity repeats [35].
Genome annotation was performed using MAKER ver. 2.31.8 (RRID:SCR 005309) [36], a portable and easily configurable genome annotation pipeline. Subsequently, the repeat-masked genomes were used for ab initio gene prediction with SNAP v2006–07–28 (RRID:SCR 002127) [37] and Augustus ver. 3.2.3 (RRID:SCR 008417) [38]. MAKER was initially run in the est2genome mode, based on Iso-Seq data full-length transcripts and transcriptome assemblies from RNA-Seq using Trinity ver. 2.5.1 (RRID:SCR_013048) [39]. Exonerate software, ver. 2.2, providing integrated information for the SNAP software program, was used to polish MAKER alignments. MAKER was used to select and revise the final gene model considering all available information. Other non-coding RNAs were identified using the Basic Rapid Ribosomal RNA Predictor ver. 0.9 (Barrnap, RRID:SCR_015995). The putative tRNA genes were identified using tRNAscan-SE ver. 2.0.5 (RRID:SCR 010835) [40].
The predicted genes were annotated by aligning them to the NCBI non-redundant protein (nr) databases [41] using BLAST ver. 2.4.0 with a maximum E-value cut-off of 1 × 10−5. To obtain protein domain information, InterProScan ver. 5.44.79 (RRID:SCR 005829) [42] was employed for a protein sequence translated from a transcript. Gene Ontology (GO) terms (RRID:SCR 002811) [43] were assigned to the genes using the BLAST2GO ver. 4.0 pipeline (RRID:SCR 005828) [44]. Pathway annotation analysis utilized the Kyoto Encyclopedia of Genes and Genomes (KEGG) Automatic Annotation Server.
A comparative Venn diagram of the paralogous and orthologous groups among the four sea star genomes (A. planci, A. rubens, P. miniata, and P. pectinifera) was generated using OrthoFinder [45]. Orthologous gene clusters of the four genomes were classified using the OrthoMCL pipeline [46] with the Markov clustering algorithm [47] and default parameters.
3. Results and Discussion
3.1. Genome Sequencing and Assembly
Genomic information and knowledge on molecular evolution are scarce in sea stars. In this study, leveraging the advantages of long-read sequencing technology, we sequenced 60 Gb and 24 Gb of genomic data using ONT and Illumina platforms, respectively (Table S1). The average coverage of the raw sequences on the P. pectinifera genome was 120-fold and 48-fold, with N50 lengths of reads sequenced from ONT reaching 9.6 kb. The genome was assembled, yielding a reference genome size of 499 Mb with an N50 length of about 2 Mb and a longest length of 7.5 Mb (Figure 1A; Table 1). The genome size and N50 value of P. pectinifera are comparable to those of the bat star P. miniata (Asteroidea; Asterinidae; ASM1570657v1; 608 Mb; N50 2 Mb; unpublished), crown-of-thorns starfish A. planci (Asteroidea; Acanthasteridae; OKI–Apl_1.0; 384 Mb; N50 5 Kb; H; [7]) and European starfish A. rubens (Asteroidea; Asteriidae; eAstRub1.3; 418 Mb; N50 1.4 Mb; unpublished).
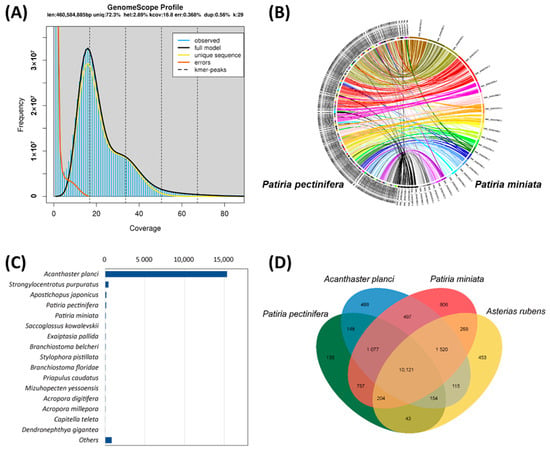
Figure 1.
(A) Results of genome size estimation of Patiria pectinifera assembly obtained by GenomeScope analysis. Diagram shortcuts are as follows: len, inferred total genome length; uniq, percent of the genome that is unique (not repetitive); het, overall rate of heterozygosity; kcov, mean k-mer coverage for heterozygous bases; err, error rate of the reads; dup, average rate of read duplications; k, k-mer size; observed, the observed k-mer profile; full model, estimated GenomeScope model; unique sequence, line representing unique sequences (k-mers below the line are treated as unique); errors, line representing sequencing errors (k-mers below the line are treated as incorrect); k-mer peaks, increased number of k-mers compared to the number of k-mers with lower and higher coverage. (B) Analysis of contiguity of Patiria pectinifera scaffolds at 25 Mb resolution against that of Patiria miniata. (C) Numbers of major BLAST hits matched to Patiria pectinifera transcripts at the species level. (D) Venn diagram of orthologous gene families. Four sea star genomes (Acanthaster planci, Asterias rubens, Patiria miniata, and Patiria pectinifera) were used to generate the Venn diagram based on the gene family cluster analysis.

Table 1.
Patiria pectinifera genome assembly statistics.
When the scaffolds of P. pectinifera were compared to the largest 30 pseudochromosomes of P. miniata, all the scaffolds were directly aligned (Figure 1B), suggesting that our assembly contained crucial synteny with contiguity. The completeness of the P. pectinifera genome assembly was assessed using BUSCO against the metazoan database. Of the 954 total BUSCO groups aligned, 930 and 10 BUSCO core genes were completed and fragmented, respectively (Table 2), resulting in a total of 98.5% of core metazoan genes being annotated in the P. pectinifera genome assembly. This result suggests that the assembled P. pectinifera genome is intact for completing the annotation of protein-coding sequences.

Table 2.
Benchmarking Universal Single-Copy Orthologs (BUSCO) evaluated for the completeness of the Patiria pectinifera genome assembly.
3.2. Gene Annotation and Comparison with Sea Star Genomes
A total of 17,334 P. pectinifera genes were annotated through bioinformatics, and 99.4% of these genes aligned with known proteins in public databases (Table S2). Principal BLAST hits revealed that about 15,343 P. pectinifera contigs exhibited sequence similarities to transcripts of the crown-of-thorns starfish A. planci (Figure 1C). However, the number of hits to the genes of the bat star P. miniata was very low. BLAST hit analysis was not performed on the bat star P. miniata or European starfish A. rubens due to the absence of their transcript information in the NR database. Orthologous analysis identified a core set of 10,121 genes shared among the four sea star genomes, with 135 genes being P. pectinifera-specific (Figure 1D). Approximately 12,698 and 7880 genes were functionally annotated by GO and KEGG orthology prediction, respectively.
The P. pectinifera genome contained 47.43% repetitive sequences, with 7.31%, 4.62%, 1.94%, and 1.38% of repetitive sequences attributed to DNA transposons, long terminal repeats (LTRs), long interspersed elements (LINEs), and short interspersed elements (SINEs), respectively (Table S3). The amount of transposable elements (TEs) in the genome is comparable, with 51.65%, 28.48%, and 45.87% TEs occupying the genomes of P. miniata, A. planci, and A. rubens, respectively (Table S3). Approximately 31% of TEs were specific unknown repeats, a figure comparable to the results for P. miniata (34%), A. planci (21%), and A. rubens (32%). Although only four genomes, including P. pectinifera, are available in sea stars, and thus generalization is limited, a positive correlation between TE content and genome size was observed in sea star genomes. The largest genome of P. miniata (608 Mb) had the largest TE content, while the smallest genome of A. planci (384 Mb) possessed the smallest TE content (Table S3). Overall, the composition of TEs was similar between Patiria species (P. pectinifera and P. miniata) (Figure 2), but the ratio was quite different from those of A. planci and A. rubens (Figure 3; Table S3). A relatively higher number of LTR members was observed in Patiria species than in A. planci and A. rubens.
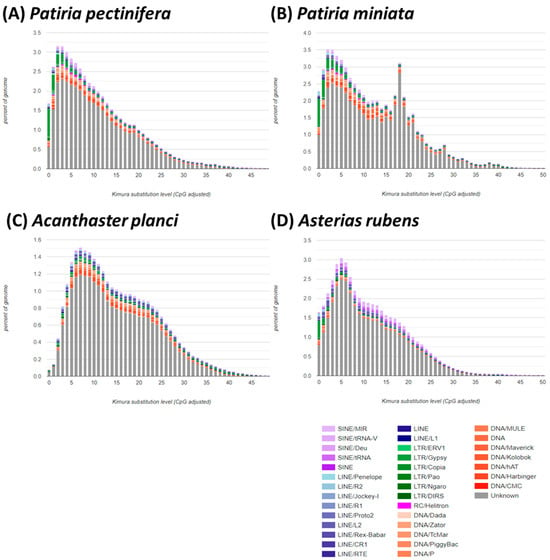
Figure 2.
Comparison of repetitive components and orthologous in sea star genomes: (A) Patiria pectinifera, (B) Patiria miniata, (C) Acanthaster planci, and (D) Asterias rubens. Kimura distance-based copy divergence analysis of transposable elements in four sea star genomes. Graphs represent genome coverage (Y-axis) for each type of TE (DNA transposons, SINE, LINE, and LTR retrotransposons) in the different genomes analyzed, clustered to their corresponding consensus sequence according to Kimura distances (X-axis, K-value from 0 to 50).

Figure 3.
Comparison of enrichment of transposable elements in sea star genomes, Patiria pectinifera, Patiria miniata, Acanthaster planci, and Asterias rubens. The numbers of each transposable element are represented by the intensity of the boxes.
3.3. Comparison of Transposable Elements
Approximately half of the genome in the Patiria species is comprised of transposable elements (TEs). TEs are major contributors to genome rearrangement and expansion due to their replicative nature [48]. Across vertebrates and invertebrates, the numbers and compositions of TEs vary greatly [49,50,51,52,53], indicating their crucial role in genome structure and evolution. While there is limited information on the positive correlation between TE content and genome size in invertebrates, recent studies have provided evidence for this relationship. For instance, larvaceans, with ~12-fold variation in genome size, show multiple independent genome expansions driven by TEs [54]. TEs are also recognized as important drivers of large genome sizes in the order Trichoptera, which exhibits ~14-fold variation in genome size [55]. Thus, the relatively higher composition of TEs in the genomes of Patiria species, with their larger genome sizes than those of A. planci and A. rubens, suggests potential roles of TEs in genome expansion and size variation.
Comparative analyses of Kimura substitution levels indicate that Patiria species have experienced more recent transposition bursts compared to A. planci and A. rubens, suggesting recent amplification of TEs (Figure 2). Several TEs in Patiria species exhibit unique or higher expansion in their genomes compared to A. planci and A. rubens, including Academ-1, MULE-MuDR, PIF-Harbinger, Sola-2, Zator, hAT-Ac, hAT-Blackjack, hAT-Tip100, Helitron, Gypsy, and tRNA-Deu-CR1 (Figure 3). While information on the role of each TE in invertebrates is still scarce, in the case of Oikopleura dioica, which has a very small and compact genome, many absences in elements of the most ancient families of retrotransposons were observed [56,57]. Thus, the unique composition of certain TEs in Patiria species may be genus-specific contributors responsible for the variance in genome sizes in sea stars. A critical evaluation of the relationship between TE diversity and genome structure in sea stars would require an in-depth understanding of the molecular and evolutionary functions of TEs. Nonetheless, the information presented here on the genome and TEs in P. pectinifera serves as a useful reference for understanding genomic structure and genome evolution in sea stars.
In summary, we successfully constructed the genome of the sea star P. pectinifera by integrating Oxford Nanopore Technology and Illumina platforms. The CANU assembly yielded 2262 contigs, and the incorporation of Illumina RNA-Seq and Pacific Biosciences Iso-Seq techniques identified a total of 22,367 protein-coding genes. Furthermore, an analysis of repeat elements in the genome indicated their potential role in shaping the genomic structure of P. pectinifera. The availability of the sea star P. pectinifera genome is expected to enhance genetic monitoring efforts, providing a foundation for a deeper comprehension of evolutionary mechanisms and supporting further genetic investigations into the life history and ecological traits of sea stars.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d16020091/s1, Table S1. Statistics on sequencing data generated for Patiria pectinifera genome assembly; Table S2. Statistics for gene annotation for the Patiria pectinifera genome; Table S3. Statistics for repetitive elements identified in the Patiria pectinifera genome.
Author Contributions
Conceptualization, J.-S.R. and H.P.; methodology, J.-S.R.; software, S.J.L. and H.P.; validation, J.-S.R.; formal analysis, S.-E.N. and S.J.L.; investigation, J.-S.R.; resources, S.-E.N.; data curation, H.P.; writing—original draft preparation, J.-S.R.; writing—review and editing, J.-S.R. and H.P.; visualization, S.J.L.; supervision and project administration, J.-S.R. and H.P.; funding acquisition, J.-S.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2017R1A6A1A06015181). This work was also supported by the Korea Environment Industry & Technology Institute (KEITI) through the Aquatic Ecosystem Conservation Research Program (2022003050001), funded by Korea Ministry of Environment (MOE).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The Patiria pectinifera genome project was deposited at NCBI under BioProject number PRJNA1030923. The whole-genome sequence was deposited in the Sequence Read Archive (SRA) database under accession number SRR26462612.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gale, A.S. Phylogeny and classification of the Asteroidea (Echinodermata). Zool. J. Linn. Soc. 1987, 89, 107–132. [Google Scholar] [CrossRef]
- Mah, C.L.; Blake, D.B. Global diversity and phylogeny of the Asteroidea (Echinodermata). PLoS ONE 2012, 7, e35644. [Google Scholar] [CrossRef] [PubMed]
- Lafferty, K.D.; Suchanek, T.H. Revisiting Paine’s 1966 sea star removal experiment, the most-cited empirical article in the American naturalist. Am. Nat. 2016, 188, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Schiebelhut, L.M.; Puritz, J.B.; Dawson, M.N. Decimation by sea star wasting disease and rapid genetic change in a keystone species, Pisaster ochraceus. Proc. Natl. Acad. Sci. USA 2018, 115, 7069–7074. [Google Scholar] [CrossRef] [PubMed]
- Dickey, J.W.E.; Cuthbert, R.N.; Morón Lugo, S.C.; Casties, I.; Dick, J.T.A.; Steffen, G.T.; Briski, E. The stars are out: Predicting the effect of seawater freshening on the ecological impact of a sea star keystone predator. Ecol. Ind. 2021, 132, 108293. [Google Scholar] [CrossRef]
- Carnevali, M.C. Regeneration in Echinoderms: Repair, regrowth, cloning. Invertebr. Surviv. J. 2006, 3, 64–76. [Google Scholar]
- Lawrence, J.M. Energetic costs of loss and regeneration of arms in stellate echinoderms. Integr. Comp. Biol. 2010, 50, 506–514. [Google Scholar] [CrossRef]
- Chia, F.; Oguro, C.; Komatsu, M. Sea-star (asteroid) development. Oceanogr. Mar. Biol. 1993, 31, 223–257. [Google Scholar]
- Mercier, A.; Hamel, J.-F. Endogenous and exogenous control of gametogenesis and spawning in echinoderms. In Advances in Marine Biology; Elsevier: Amsterdam, The Netherlands, 2009; Volume 55, pp. 1–302. [Google Scholar]
- Hall, M.R.; Kocot, K.M.; Baughman, K.W.; Fernandez-Valverde, S.L.; Gauthier, M.E.A.; Hatleberg, W.L.; Krishnan, A.; McDougall, C.; Motti, C.A.; Shoguchi, E.; et al. The crown-of-thorns starfish genome as a guide for biocontrol of this coral reef pest. Nature 2017, 544, 231–234. [Google Scholar] [CrossRef]
- Hennebert, E.; Leroy, B.; Wattiez, R.; Ladurner, P. An integrated transcriptomic and proteomic analysis of sea star epidermal secretions identifies proteins involved in defense and adhesion. J. Proteomics 2015, 128, 83–91. [Google Scholar] [CrossRef]
- Cary, G.A.; Wolff, A.; Zueva, O.; Pattinato, J.; Hinman, V.F. Analysis of sea star larval regeneration reveals conserved processes of whole-body regeneration across the metazoa. BMC Biol. 2019, 17, 16. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.F.; Sherman, C.D. De novo assembly and characterization of the invasive Northern Pacific Seastar transcriptome. PLoS ONE 2015, 10, e0142003. [Google Scholar] [CrossRef] [PubMed]
- Gabre, J.L.; Martinez, P.; Sköld, H.N.; Ortega-Martinez, O.; Abril, J.F. The coelomic epithelium transcriptome from a clonal sea star, Coscinasterias muricata. Mar. Genom. 2015, 24, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.J.; Stewart, P.; Rivera-Posada, J. De novo assembly of the transcriptome of Acanthaster planci testes. Mol. Ecol. Resour. 2015, 15, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Bose, U.; Wang, T.; Zhao, M.; Motti, C.; Hall, M.; Cummins, S.F. Multiomics analysis of the giant triton snail salivary gland, a crown-of-thorns starfish predator. Sci. Rep. 2017, 7, 6000. [Google Scholar] [CrossRef]
- Musacchia, F.; Vasilev, F.; Borra, M.; Biffali, E.; Sanges, R.; Santella, L.; Chun, J.T. De novo assembly of a transcriptome from the eggs and early embryos of Astropecten aranciacus. PLoS ONE 2017, 12, e0184090. [Google Scholar] [CrossRef]
- Kim, C.-H.; Go, H.-J.; Oh, H.Y.; Jo, Y.H.; Elphick, M.R.; Park, N.G. Transcriptomics reveals tissue/organ-specific differences in gene expression in the starfish Patiria pectinifera. Mar. Genom. 2018, 37, 92–96. [Google Scholar] [CrossRef]
- Bates, L.; Wiseman, E.; Carroll, D.J. Analyzing gene expression in sea star eggs and embryos using bioinformatics. Methods Cell Biol. 2019, 150, 471–483. [Google Scholar]
- Gildor, T.; Cary, G.A.; Lalzar, M.; Hinman, V.F.; Ben-Tabou de-Leon, S. Developmental transcriptomes of the sea star, Patiria miniata, illuminate how gene expression changes with evolutionary distance. Sci.Rep. 2019, 9, 16201. [Google Scholar] [CrossRef]
- Byrne, M.; Koop, D.; Strbenac, D.; Cisternas, P.; Balogh, R.; Yang, J.Y.H.; Davidson, P.L.; Wray, G. Transcriptomic analysis of sea star development through metamorphosis to the highly derived pentameral body plan with a focus on neural transcription factors. DNA Res. 2020, 27, dsaa007. [Google Scholar] [CrossRef]
- Saotome, K.; Komatsu, M. Chromosomes of Japanese starfishes. Zool. Sci. 2002, 19, 1095–1103. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.-E.; Bae, D.-Y.; Ki, J.-S.; Ahn, C.-Y.; Rhee, J.-S. The importance of multi-omics approaches for the health assessment of freshwater ecosystems. Mol. Cell. Toxicol. 2023, 19, 3–11. [Google Scholar] [CrossRef]
- Marçais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Roach, M.J.; Schmidt, S.A.; Borneman, A.R. Purge Haplotigs: Allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinform. 2018, 19, 460. [Google Scholar] [CrossRef] [PubMed]
- Yeo, S.; Coombe, L.; Warren, R.L.; Chu, J.; Birol, I. ARCS: Scaffolding genome drafts with linked reads. Bioinformatics 2018, 34, 725–731. [Google Scholar] [CrossRef]
- Warren, R.L.; Yang, C.; Vandervalk, B.P.; Behsaz, B.; Lagman, A.; Jones, S.J.; Birol, I. LINKS: Scalable, alignment-free scaffolding of draft genomes with long reads. GigaScience 2015, 4, s13742-015-0076-3. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Krzywinski, M.I.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An in-formation aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Eddy, S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002, 12, 1269–1276. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Jones, N.C.; Pevzner, P.A. De novo identification of repeat families in large genomes. Bioinformatics 2005, 21, i351–i358. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Holt, C.; Yandell, M. MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinform. 2011, 12, 491. [Google Scholar] [CrossRef]
- Korf, I. Gene finding in novel genomes. BMC Bioinform. 2004, 5, 59. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Schöffmann, O.; Morgenstern, B.; Waack, S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinform. 2006, 7, 62. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA genes in genomic sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Dimmer, E.C.; Huntley, R.P.; Alam-Faruque, Y.; Sawford, T.; O′Donovan, C.; Martin, M.J.; Bely, B.; Browne, P.; Mun Chan, W.; Eberhardt, R.; et al. The UniProt-GO annotation database in 2011. Nucleic. Acids. Res. 2012, 40, D565–D570. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Brunk, B.P.; Chen, F.; Gao, X.; Harb, O.S.; Iodice, J.B.; Shanmugam, D.; Roos, D.S.; Stoeckert, C.J., Jr. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinform. 2011, 35, 6.12.1–6.12.19. [Google Scholar] [CrossRef] [PubMed]
- Kidwell, M.G. Transposable elements and the evolution of genome size in eukaryotes. Genetica 2002, 115, 49–63. [Google Scholar] [CrossRef]
- Feschotte, C.; Jiang, N.; Wessler, S.R. Plant transposable elements: Where genetics meets genomics. Nat. Rev. Genet. 2002, 3, 329–341. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Wang, H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 2014, 65, 505–530. [Google Scholar] [CrossRef]
- Chalopin, D.; Naville, M.; Plard, F.; Galiana, D.; Volff, J.-N. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol. Evol. 2015, 7, 567–580. [Google Scholar] [CrossRef]
- Szitenberg, A.; Cha, S.; Opperman, C.H.; Bird, D.M.; Blaxter, M.L.; Lunt, D.H. Genetic drift, not life history or RNAi, determine long-term evolution of transposable elements. Genome Biol. Evol. 2016, 8, 2964–2978. [Google Scholar] [CrossRef]
- Petersen, M.; Armisén, D.; Gibbs, R.A.; Hering, L.; Khila, A.; Mayer, G.; Richards, S.; Niehuis, O.; Misof, B. Diversity and evolution of the transposable element repertoire in arthropods with particular reference to insects. BMC Ecol. Evol. 2019, 19, 11. [Google Scholar] [CrossRef]
- Naville, M.; Henriet, S.; Warren, I.; Sumic, S.; Reeve, M.; Volff, J.-N.; Chourrout, D. Massive changes of genome size driven by expansions of non-autonomous transposable elements. Curr. Biol. 2019, 29, 1161–1168.e6. [Google Scholar] [CrossRef]
- Heckenhauer, J.; Frandsen, P.B.; Sproul, J.S.; Li, Z.; Paule, J.; Larracuente, A.M.; Maughan, P.J.; Barker, M.S.; Schneider, J.V.; Stewart, R.J.; et al. Genome size evolution in the diverse insect order Trichoptera. GigaScience 2022, 11, giac011. [Google Scholar] [CrossRef]
- Volff, J.-N.; Lehrach, H.; Reinhardt, R.; Chourrout, D. Retroelement dynamics and a novel type of chordate retrovirus-like element in the miniature genome of the tunicate Oikopleura dioica. Mol. Biol. Evol. 2004, 21, 2022–2033. [Google Scholar] [CrossRef]
- Denoeud, F.; Henriet, S.; Mungpakdee, S.; Aury, J.-M.; Da Silva, C.; Brinkmann, H.; Mikhaleva, J.; Olsen, L.C.; Jubin, C.; Cañestro, C.; et al. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 2010, 330, 1381–1385. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).