The Case for a Nuclear Barcode: Using the CAD CPS Region for Species and Genus Level Discrimination in Beetles
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon and Gene Selection
2.2. Gene Statistics
2.3. Tests of Specimen Placement
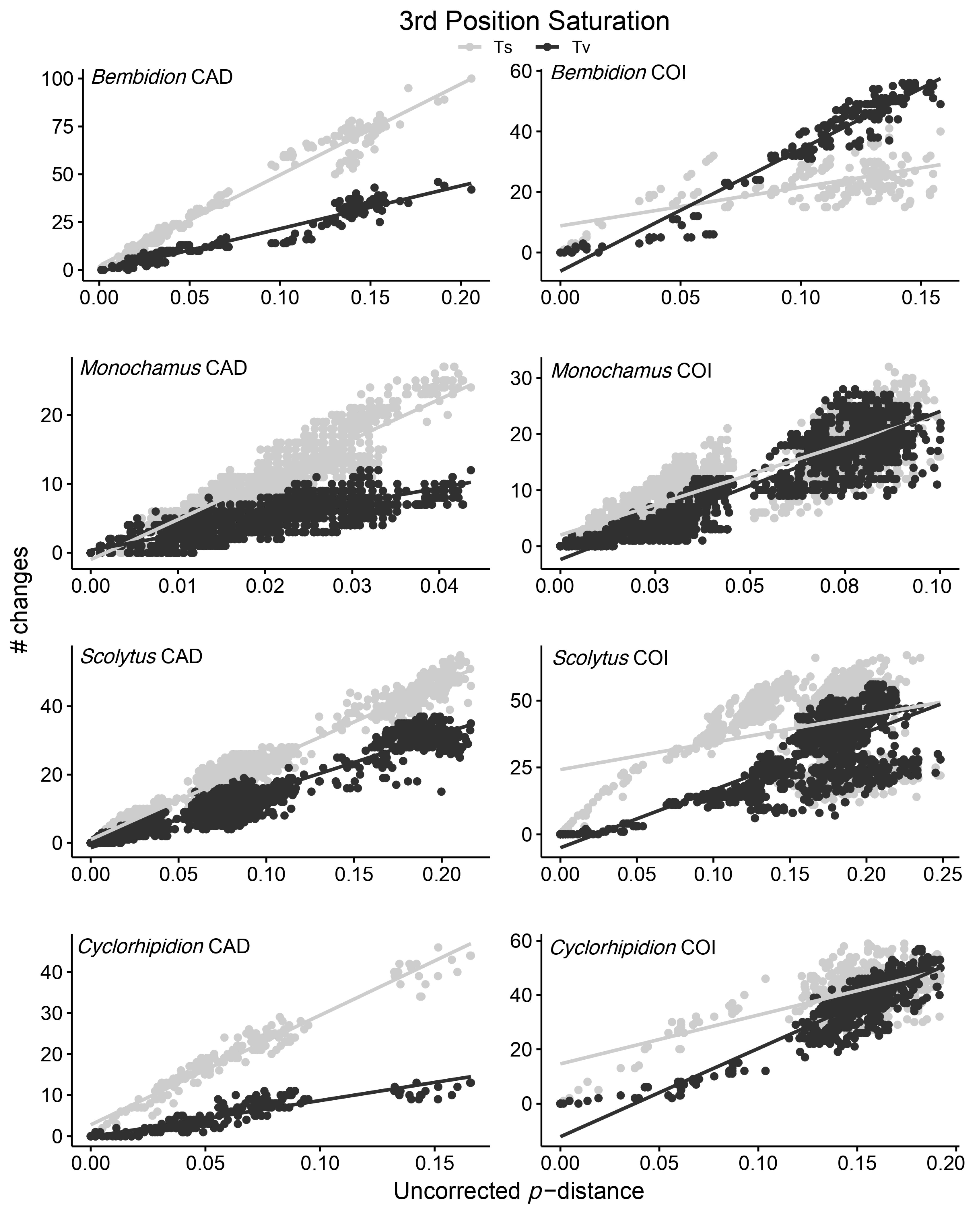
3. Results
3.1. Comparison of COI and CAD Gene Characteristics
| COI Barcode | Length | # PIS | % PIS | Conspecifics Properly Placed in NJ | Outgroups Properly Placed in NJ | Primers |
|---|---|---|---|---|---|---|
| Bembidion (Carabidae) | 658 | 146 | 22.19 | 4/6 | 3/4 | LCO1490, HCO2198 [51] |
| Monochamus (Cerambycidae) | 659 | 120 | 18.21 | 46/62 | 3/4 | LCO1490, HCO2198 [51] |
| Scolytus (Scolytinae) | 612 | 248 | 40.52 | 31/39 | 4/4 | 1495b, rev750, F215, Rev453 [35] |
| Cyclorhipidion (Scolytinae: Xyleborini) | 656 | 240 | 36.59 | 27/27 | 3/4 | LCO1490, HCO2198 [51]; 1495b, rev750 [35] |
| Xyleborus (Scolytinae: Xyleborini) | 649 | 175 | 26.96 | 22/22 | 3/4 | LCO1490, HCO2198 [51]; 1495b, rev750 [35] |
| CAD Barcode | ||||||
| Bembidion (Carabidae) | 854 | 174 | 20.37 | 8/8 | 2/4 | many [31,37] |
| Monochamus (Cerambycidae) | 943 | 94 | 9.97 | 60/66 | 3/4 | CD338, CD668, CD688 [33] |
| Scolytus (Scolytinae) | 471 | 157 | 33.33 | 48/50 | 4/4 | CADforB2, CADfor4, CADrev1mod [52] |
| Cyclorhipidion (Scolytinae: Xyleborini) | 594 | 70 | 11.78 | 11/11 | 2/4 | CADforB2, CADfor4, CADrev1mod [52] |
| Xyleborus (Scolytinae: Xyleborini) | 594 | 15 | 2.53 | 17/17 | 4/4 | CADforB2, CADfor4, CADrev1mod [52] |
3.2. Taxon Placement and Diagnostic Potential
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, S.E.; Hausmann, A.; Hallwachs, W.; Janzen, D.H. Advancing Taxonomy and Bioinventories with DNA Barcodes. Philos. Trans. R. Soc. B Biol. Sci. 2016, 20150339. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Sigsgaard, E.E. Environmental DNA Metabarcoding of Wild Flowers Reveals Diverse Communities of Terrestrial Arthropods. Ecol. Evol. 2019, 9, 1665–1679. [Google Scholar] [CrossRef]
- Caesar, R.M.; Sörensson, M.; Cognato, A.I. Integrating DNA Data and Traditional Taxonomy to Streamline Biodiversity Assessment: An Example from Edaphic Beetles in the Klamath Ecoregion, California, USA. Divers. Distrib. 2006, 12, 483–489. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten Species in One: DNA Barcoding Reveals Cryptic Species in the Neotropical Skipper Butterfly Astraptes Fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef]
- Dukes, C.D.; Janssens, F.; Recuero, E.; Caterino, M.S. Specific and Intraspecific Diversity of Symphypleona and Neelipleona (Hexapoda: Collembola) in Southern High Appalachia (USA). Diversity 2022, 14, 847. [Google Scholar] [CrossRef]
- Prendini, L. Comment on “Identifying Spiders through DNA Barcodes”. Can. J. Zool. 2005, 83, 498–504. [Google Scholar] [CrossRef]
- Vences, M.; Thomas, M.; Van Der Meijden, A.; Chiari, Y.; Vieites, D.R. Comparative Performance of the 16S RRNA Gene in DNA Barcoding of Amphibians. Front. Zool. 2005, 2, 5. [Google Scholar] [CrossRef]
- Siddappa, C.M.; Saini, M.; Das, A.; Doreswamy, R.; Sharma, A.K.; Gupta, P.K. Sequence Characterization of Mitochondrial 12S RRNA Gene in Mouse Deer (Moschiola indica) for PCR-RFLP Based Species Identification. Mol. Biol. Int. 2013, 2013, 783925. [Google Scholar] [CrossRef]
- Yacoub, H.A.; Fathi, M.M.; Sadek, M.A. Using Cytochrome b Gene of MtDNA as a DNA Barcoding Marker in Chicken Strains. Mitochondrial DNA 2015, 26, 217–223. [Google Scholar] [CrossRef]
- Foster, B.T.; Cognato, A.I.; Gold, R.E. DNA-Based Identification of the Eastern Subterranean Termite, Reticulitermes flavipes (Isoptera: Rhinotermitidae). J. Econ. Entomol. 2004, 97, 95–101. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- DeSalle, R.; Goldstein, P. Review and Interpretation of Trends in DNA Barcoding. Front. Ecol. Evol. 2019, 7, 302. [Google Scholar] [CrossRef]
- Doorenweerd, C.; San Jose, M.; Leblanc, L.; Rubinoff, D. Inadequate Molecular Identification Protocols for Invasive Pests Threaten Biosecurity. Syst. Entomol. 2022, 48, 355–360. [Google Scholar] [CrossRef]
- Chase, M.W.; Fay, M.F. Barcoding of Plants and Fungi. Science 2009, 325, 682–683. [Google Scholar] [CrossRef] [PubMed]
- Koutroumpa, F.A.; Lieutier, F.; Roux-Morabito, G. Incorporation of Mitochondrial Fragments in the Nuclear Genome (Numts) of the Longhorned Beetle Monochamus galloprovincialis (Coleoptera, Cerambycidae). J. Zool. Syst. Evol. Res. 2009, 47, 141–148. [Google Scholar] [CrossRef]
- Jordal, B.H.; Kambestad, M. DNA Barcoding of Bark and Ambrosia Beetles Reveals Excessive NUMTs and Consistent East-West Divergence across Palearctic Forests. Mol. Ecol. Resour. 2014, 14, 7–17. [Google Scholar] [CrossRef]
- Song, H.; Buhay, J.E.; Whiting, M.F.; Crandall, K.A. Many Species in One: DNA Barcoding Overestimates the Number of Species When Nuclear Mitochondrial Pseudogenes Are Coamplified. Proc. Natl. Acad. Sci. USA 2008, 105, 13486–13491. [Google Scholar] [CrossRef]
- Cognato, A.I.; Caesar, R.M.; Blaxter, M.; Vogler, A.P. Will DNA Barcoding Advance Efforts to Conserve Biodiversity More Efficiently than Traditional Taxonomic Methods? Front. Ecol. Environ. 2006, 4, 268–273. [Google Scholar] [CrossRef]
- Yao, H.; Song, J.; Liu, C.; Luo, K.; Han, J.; Li, Y.; Pang, X.; Xu, H.; Zhu, Y.; Xiao, P.; et al. Use of ITS2 Region as the Universal DNA Barcode for Plants and Animals. PLoS ONE 2010, 5, e13102. [Google Scholar] [CrossRef]
- Cognato, A.I.; Sari, G.; Smith, S.M.; Beaver, R.A.; Li, Y.; Hulcr, J.; Jordal, B.H.; Kajimura, H.; Lin, C.S.; Pham, T.H.; et al. The Essential Role of Taxonomic Expertise in the Creation of DNA Databases for the Identification and Delimitation of Southeast Asian Ambrosia Beetle Species (Curculionidae: Scolytinae: Xyleborini). Front. Ecol. Evol. 2020, 8, 27. [Google Scholar] [CrossRef]
- Sonnenberg, R.; Nolte, A.; Tautz, D. An Evaluation of LSU RDNA D1-D2 Sequences for Their Use in Species Identification. Front. Zool. 2007, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Gaut, B.S. Evolution of Genes and Taxa: A Primer. Plant Mol. Biol. 2000, 42, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Cognato, A.I.; Vogler, A.P. Exploring Data Interaction and Nucleotide Alignment in a Multiple Gene Analysis of Ips (Coleoptera: Scolytinae). Syst. Biol. 2001, 50, 758–780. [Google Scholar] [CrossRef] [PubMed]
- Bratzel, F.; Heller, S.; Cyrannek, N.; Paule, J.; Leme, E.M.C.; Loreth, A.; Nowotny, A.; Kiefer, M.; Till, W.; Barfuss, M.H.J.; et al. The Low-Copy Nuclear Gene Agt1 as a Novel DNA Barcoding Marker for Bromeliaceae. BMC Plant Biol. 2020, 20, 111. [Google Scholar] [CrossRef]
- Pillon, Y.; Johansen, J.B.; Sakishima, T.; Roalson, E.H.; Price, D.K.; Stacy, E.A. Gene Discordance in Phylogenomics of Recent Plant Radiations, an Example from Hawaiian Cyrtandra (Gesneriaceae). Mol. Phylogenet. Evol. 2013, 69, 293–298. [Google Scholar] [CrossRef]
- Caterino, M.S.; Cho, S.; Sperling, F.A. The Current State of Insect Molecular Systematics: A Thriving Tower of Babel. Annu. Rev. Entomol. 2000, 45, 1–54. [Google Scholar] [CrossRef]
- Cognato, A.I.; Taft, W.; Osborn, R.K.; Rubinoff, D. Multi-Gene Phylogeny of North American Clear-Winged Moths (Lepidoptera: Sesiidae): A Foundation for Future Evolutionary Study of a Speciose Mimicry Complex. Cladistics 2023, 39, 1–17. [Google Scholar] [CrossRef]
- Dowton, M.; Meiklejohn, K.; Cameron, S.L.; Wallman, J. A Preliminary Framework for DNA Barcoding, Incorporating the Multispecies Coalescent. Syst. Biol. 2014, 63, 639–644. [Google Scholar] [CrossRef]
- Foster, P.G.; Bergo, E.S.; Bourke, B.P.; Oliveira, T.M.P.; Nagaki, S.S.; Sant’Ana, D.C.; Sallum, M.A.M. Phylogenetic Analysis and DNA-Based Species Confirmation in Anopheles (Nyssorhynchus). PLoS ONE 2013, 8, e54063. [Google Scholar] [CrossRef]
- Che, L.; Zhang, S.; Li, Y.; Liang, D.; Pang, H.; Slipiński, A.; Zhang, P. Genome-Wide Survey of Nuclear Protein-Coding Markers for Beetle Phylogenetics and Their Application in Resolving Both Deep and Shallow-Level Divergences. Mol. Ecol. Resour. 2017, 17, 1342–1358. [Google Scholar] [CrossRef]
- Wild, A.L.; Maddison, D.R. Evaluating Nuclear Protein-Coding Genes for Phylogenetic Utility in Beetles. Mol. Phylogenet. Evol. 2008, 48, 877–891. [Google Scholar] [CrossRef] [PubMed]
- Karpiński, L.; Gorring, P.; Kruszelnicki, L.; Kasatkin, D.G.; Szczepański, W.T. A Fine Line between Species and Ecotype: A Case Study of Anoplistes halodendri and A. kozlovi (Coleoptera: Cerambycidae) Occurring Sympatrically in Mongolia. Arthropod Syst. Phylogeny 2021, 79, 1–23. [Google Scholar] [CrossRef]
- Gorring, P.S.; Farrell, B.D. Evaluating Species Boundaries Using Coalescent Delimitation in Pine-Killing Monochamus (Coleoptera: Cerambycidae) Sawyer Beetles. Mol. Phylogenet. Evol. 2023, 184, 107777. [Google Scholar] [CrossRef]
- Cognato, A.I.; Smith, S.M.; Jordal, B.H. Patterns of Host Tree Use within a Lineage of Saproxlic Snout-Less Weevils (Coleoptera: Curculionidae: Scolytinae: Scolytini). Mol. Phylogenet. Evol. 2021, 159, 107107. [Google Scholar] [CrossRef]
- Smith, S.M.; Cognato, A.I. A Taxonomic Monograph of Nearctic Scolytus Geoffroy (Coleoptera, Curculionidae, Scolytinae). Zookeys 2014, 450, 1–182. [Google Scholar] [CrossRef]
- Smith, S.M.; Cognato, A.I. New Non-Native Pseudocryptic Cyclorhipidion Species (Coleoptera: Curculionidae: Scolytinae: Xyleborini) Found in the United States as Revealed in a Multigene Phylogeny. Insect Syst. Divers. 2022, 6, 2. [Google Scholar] [CrossRef]
- Maddison, D.R. Phylogeny of Bembidion and Related Ground Beetles (Coleoptera: Carabidae: Trechinae: Bembidiini: Bembidiina). Mol. Phylogenet. Evol. 2012, 63, 533–576. [Google Scholar] [CrossRef]
- Grzywacz, A.; Wyborska, D.; Piwczyński, M. DNA Barcoding Allows Identification of European Fanniidae (Diptera) of Forensic Interest. Forensic Sci. Int. 2017, 278, 106–114. [Google Scholar] [CrossRef]
- Will, K.W.; Rubinoff, D. Myth of the Molecule: DNA Barcodes for Species Cannot Replace Morphology for Identification and Classification. Cladistics 2004, 20, 47–55. [Google Scholar] [CrossRef]
- Gorring, P.S. Gene to Genus: Systematics and Population Dynamics in Lamiini Beetles (Coleoptera: Cerambycidae) with Focus on Monochamus Dejean. Ph.D. Dissertation, Harvard University, Cambridge, MA, USA, 2019. Available online: http://nrs.harvard.edu/urn-3:HUL.InstRepos:42029751 (accessed on 30 May 2023).
- Horn, S. Target Enrichment via DNA Hybridization Capture. Methods Mol. Biol. 2012, 840, 177–188. [Google Scholar] [CrossRef]
- Peñalba, J.V.; Smith, L.L.; Tonione, M.A.; Sass, C.; Hykin, S.M.; Skipwith, P.L.; Mcguire, J.A.; Bowie, R.C.K.; Moritz, C. Sequence Capture Using PCR-Generated Probes: A Cost-Effective Method of Targeted High-Throughput Sequencing for Nonmodel Organisms. Mol. Ecol. Resour. 2014, 14, 1000–1010. [Google Scholar] [CrossRef] [PubMed]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. 2018. Available online: http://www.mesquiteproject.org (accessed on 30 May 2023).
- Heibl, C. PHYLOCH: R Language Tree Plotting Tools and Interfaces to Diverse Phylogenetic Software Packages. 2013. Available online: http://www.christophheibl.de/Rpackages.html (accessed on 30 May 2023).
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4; Sinauer Associates: Sunderland, MA, USA, 2003; Available online: http://phylosolutions.com/paup-test (accessed on 30 May 2023).
- Srivathsan, A.; Meier, R. On the Inappropriate Use of Kimura-2-Parameter (K2P) Divergences in the DNA-Barcoding Literature. Cladistics 2012, 28, 190–194. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble Species by Automatic Partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Modica, M.V.; Zhang, Y.; Sirovich, L.; Boisselier, M.C.; Cruaud, C.; Holford, M.; Samadi, S. Large-Scale Species Delimitation Method for Hyperdiverse Groups. Mol. Ecol. 2012, 21, 2671–2691. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Trepanowski, N.F.; Molongoski, J.J.; Reagel, P.F.; Lingafelter, S.W.; Nadel, H.; Myers, S.W.; Ray, A.M. Identification of Wood-Boring Beetles (Cerambycidae and Buprestidae) Intercepted in Trade Associated Solid Wood Packaging Material Using DNA Barcoding and Morphology. Sci. Rep. 2017, 7, 40316. [Google Scholar] [CrossRef] [PubMed]
- Koutroumpa, F.A.; Rougon, D.; Bertheau, C.; Lieutier, F.; Roux-Morabito, G. Evolutionary Relationships within European Monochamus (Coleoptera: Cerambycidae) Highlight the Role of Altitude in Species Delineation. Biol. J. Linn. Soc. 2013, 109, 354–376. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Jordal, B.H.; Sequeira, A.S.; Cognato, A.I. The Age and Phylogeny of Wood Boring Weevils and the Origin of Subsociality. Mol. Phylogenet. Evol. 2011, 59, 708–724. [Google Scholar] [CrossRef]
- Després, L. One, Two or More Species? Mitonuclear Discordance and Species Delimitation. Mol. Ecol. 2019, 28, 3845–3847. [Google Scholar] [CrossRef]
- Hinojosa, J.C.; Koubínová, D.; Szenteczki, M.A.; Pitteloud, C.; Dincă, V.; Alvarez, N.; Vila, R. A Mirage of Cryptic Species: Genomics Uncover Striking Mitonuclear Discordance in the Butterfly Thymelicus Sylvestris. Mol. Ecol. 2019, 28, 3857–3868. [Google Scholar] [CrossRef]
- Funk, D.J.; Omland, K. Species-Level Paraphyly and Polyphyly: Frequency, Causes, and Consequences, with Insights from Animal Mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 397–423. [Google Scholar] [CrossRef]
- Chan, K.M.A.; Levin, S.A. Leaky Prezygotic Isolation and Porous Genomes: Rapid Introgression of Maternally Inherited DNA. Evolution 2005, 59, 720–729. [Google Scholar] [PubMed]
- Linnen, C.R.; Farrell, B.D. Mitonuclear Discordance Is Caused by Rampant Mitochondrial Introgression in Neodiprion (Hymenoptera: Diprionidae) Sawflies. Evolution 2007, 61, 1417–1438. [Google Scholar] [CrossRef]
- Avise, J.C. Gene Trees and Organismal Histories: A Phylogenetic Approach to Population Biology. Evolution 1989, 43, 1192. [Google Scholar] [CrossRef]
- Bensasson, D.; Zhang, D.X.; Hartl, D.L.; Hewitt, G.M. Mitochondrial Pseudogenes: Evolution’s Misplaced Witnesses. Trends Ecol. Evol. 2001, 16, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Philippe, H.; Brinkmann, H.; Lavrov, D.V.; Littlewood, D.T.J.; Manuel, M.; Wörheide, G.; Baurain, D. Resolving Difficult Phylogenetic Questions: Why More Sequences Are Not Enough. PLoS Biol. 2011, 9, e1000602. [Google Scholar] [CrossRef]
- Duchêne, D.A.; Mather, N.; Van Der Wal, C.; Ho, S.Y.W. Excluding Loci with Substitution Saturation Improves Inferences from Phylogenomic Data. Syst. Biol. 2022, 71, 676–689. [Google Scholar] [CrossRef]
- McKenna, D.D.; Wild, A.L.; Kanda, K.; Bellamy, C.L.; Beutel, R.G.; Caterino, M.S.; Farnum, C.W.; Hawks, D.C.; Ivie, M.A.; Jameson, M.L.; et al. The Beetle Tree of Life Reveals That Coleoptera Survived End-Permian Mass Extinction to Diversify during the Cretaceous Terrestrial Revolution. Syst. Entomol. 2015, 40, 835–880. [Google Scholar] [CrossRef]
- Meiklejohn, K.A.; Damaso, N.; Robertson, J.M. Assessment of BOLD and GenBank—Their Accuracy and Reliability for the Identification of Biological Materials. PLoS ONE 2019, 14, e0217084. [Google Scholar] [CrossRef]
- Hudson, R.; Turelli, M. Stochasticity Overrules the “Three-Times Rule”: Genetic Drift, Genetic Draft, and Coalescence Times for Nuclear Loci versus Mitochondrial DNA. Evolution 2003, 57, 182–190. [Google Scholar]
- Maddison, W.P. Gene Trees in Species Trees. Syst. Biol. 1997, 46, 523–536. [Google Scholar] [CrossRef]
- Moore, W.S. Inferring Phylogenies From MtDNA Variation: Mitochondrial-Gene Trees Versus Nuclear-Gene Trees. Evolution 1995, 49, 718–726. [Google Scholar] [CrossRef] [PubMed]
- Karpiński, L.; Gorring, P.; Cognato, A.I. DNA vs. Morphology in Delineating Species Boundaries of Endemic Mongolian Eodorcadion Taxa (Coleoptera: Cerambycidae). Diversity 2023, 15, 662. [Google Scholar] [CrossRef]
- Andermann, T.; Fernandes, A.M.; Olsson, U.; Töpel, M.; Pfeil, B.; Oxelman, B.; Aleixo, A.; Faircloth, B.C.; Antonelli, A. Allele Phasing Greatly Improves the Phylogenetic Utility of Ultraconserved Elements. Syst. Biol. 2019, 68, 32–46. [Google Scholar] [CrossRef]
- Moulton, J.K.; Wiegmann, B.M. Evolution and Phylogenetic Utility of CAD (Rudimentary) among Mesozoic-Aged Eremoneuran Diptera (Insecta). Mol. Phylogenet. Evol. 2004, 31, 363–378. [Google Scholar] [CrossRef]
- Danforth, B.N.; Fang, J.; Sipes, S. Analysis of Family-Level Relationships in Bees (Hymenoptera: Apiformes) Using 28S and Two Previously Unexplored Nuclear Genes: CAD and RNA Polymerase II. Mol. Phylogenet. Evol. 2006, 39, 358–372. [Google Scholar] [CrossRef]

| # Species in Tree (Including Outgroups) | Best ASAP Score # Species | Difference (# spp.) | Threshold Value for Best ASAP Score | ASAP Rank of Score Matching True Species # | Species Threshold of Correct ASAP Delim. | Empirical Intra Q3 Score | Empirical Inter Q1 Score | Is Best Species Threshold within Q3–Q1 Gap? | |
|---|---|---|---|---|---|---|---|---|---|
| COI | |||||||||
| Monochamus | 21 | 3 | 18 | 0.108388 | 7th | 0.011198 | 0.027 | 0.031 | no |
| Scolytus | 38 | 35 | 3 | 0.04616 | 6th | 0.018987 | 0.016 | 0.158 | yes |
| Cyclorhipidion | 21 | 31 | 10 | 0.040572 | 8th | 0.109425 | 0.089 | 0.142 | yes |
| Xyleborus | 8 | 14 | 6 | 0.020031 | 2nd | 0.112481 | 0.086 | 0.16 | yes |
| Bembidion | 20 | 16 | 4 | 0.027204 | 7th (19 spp.) | 0.004669 | 0.003 | 0.096 | yes |
| CAD | |||||||||
| Monochamus | 21 | 3 | 18 | 0.126855 | 4th (19 spp.) | 0.003886 | 0.002 | 0.014 | yes |
| Scolytus | 44 | 19 | 25 | 0.044184 | 6th (48 spp.) | 0.003189 | 0.004 | 0.057 | no |
| Cyclorhipidion | 16 | 18 | 2 | 0.006112 | 3rd (17 spp.) | 0.009061 | 0.015 | 0.042 | no |
| Xyleborus | 8 | 3 | 5 | 0.143786 | 4th | 0.009514 | 0.004 | 0.015 | yes |
| Bembidion | 20 | 3 | 17 | 0.174192 | tied-2nd | 0.007185 | 0.009 | 0.029 | no |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorring, P.S.; Cognato, A.I. The Case for a Nuclear Barcode: Using the CAD CPS Region for Species and Genus Level Discrimination in Beetles. Diversity 2023, 15, 847. https://doi.org/10.3390/d15070847
Gorring PS, Cognato AI. The Case for a Nuclear Barcode: Using the CAD CPS Region for Species and Genus Level Discrimination in Beetles. Diversity. 2023; 15(7):847. https://doi.org/10.3390/d15070847
Chicago/Turabian StyleGorring, Patrick S., and Anthony I. Cognato. 2023. "The Case for a Nuclear Barcode: Using the CAD CPS Region for Species and Genus Level Discrimination in Beetles" Diversity 15, no. 7: 847. https://doi.org/10.3390/d15070847
APA StyleGorring, P. S., & Cognato, A. I. (2023). The Case for a Nuclear Barcode: Using the CAD CPS Region for Species and Genus Level Discrimination in Beetles. Diversity, 15(7), 847. https://doi.org/10.3390/d15070847








