The RNA Viruses in Samples of Endemic Lake Baikal Sponges
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Sample Processing
2.2. Library Preparation and Sequencing
2.3. Initial Shotgun Metagenomic Datasets on RNA Viruses in Marine and Freshwater Samples
2.4. Primary Processing and Taxonomic Analysis of Initial Virome Reads
2.5. Assembly of Virome Reads
2.6. Viral Scaffolds Detection
2.7. Taxonomic Assignment of Viral Scaffolds
2.8. Functional Assignment of Viral Scaffolds
2.9. Statistical Analysis of Taxonomic and Functional Diversity
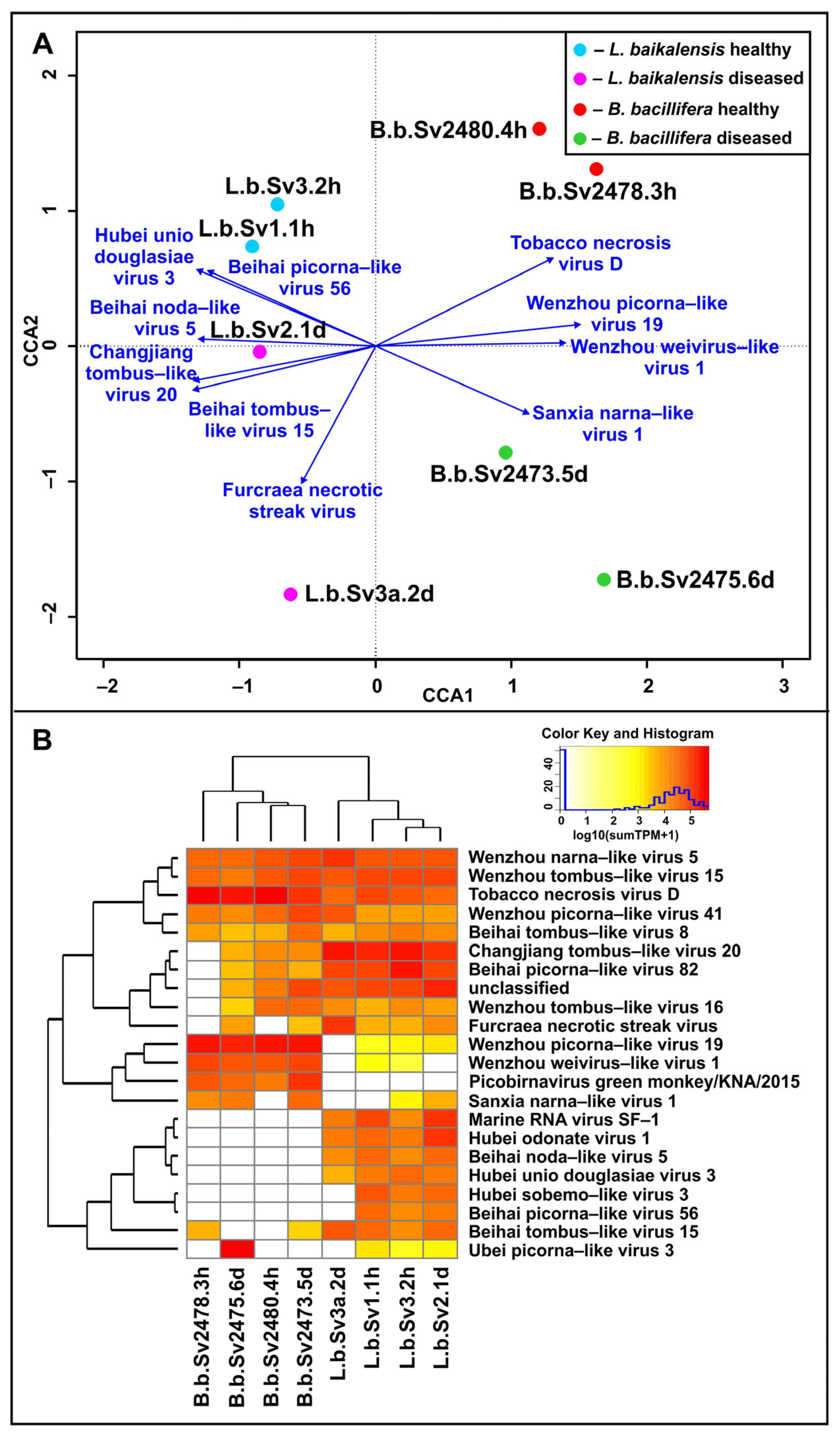
3. Results
3.1. Analysis of Virome Reads
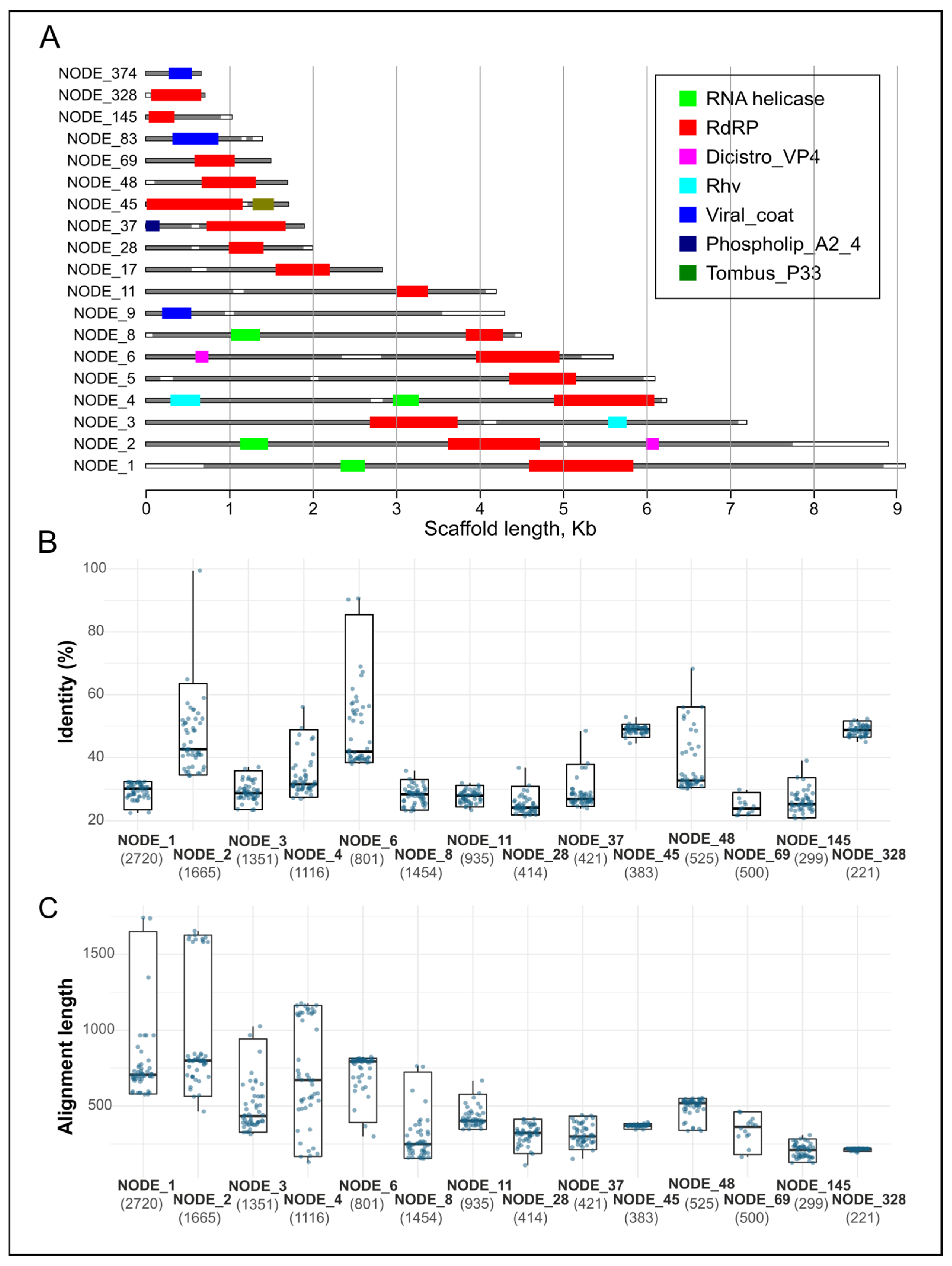
3.2. Assembly and Analysis of Viral Scaffolds
3.3. Potential Hosts for Identified Viruses
3.4. Diversity of RNA-Viral Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Webster, N.S.; Taylor, M.W. Marine sponges and their microbial symbionts: Love and other relationships. Environ. Microbiol. 2012, 14, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.C.; Rützler, K. Sponges: An essential component of Caribbean coral reefs. Bull. Mar. Sci. 2001, 69, 535–546. [Google Scholar]
- Bell, J.J. The functional roles of marine sponges. Estuar. Coast. Shelf Sci. 2008, 79, 341–353. [Google Scholar] [CrossRef]
- Wulff, J. Ecological Interactions and the Distribution, Abundance, and Diversity of Sponges. Adv. Mar. Biol. 2012, 61, 273–344. [Google Scholar] [PubMed]
- Wulff, J.L. Ecological interactions of marine sponges. Can. J. Zool. 2006, 84, 146–166. [Google Scholar] [CrossRef]
- Laffy, P.W.; Wood-Charlson, E.M.; Turaev, D.; Weynberg, K.D.; Botté, E.S.; Van Oppen, M.J.H.; Webster, N.S.; Rattei, T. HoloVir: A workflow for investigating the diversity and function of viruses in invertebrate holobionts. Front. Microbiol. 2016, 7, 822. [Google Scholar] [CrossRef] [PubMed]
- Batista, D.; Costa, R.; Carvalho, A.P.; Batista, W.R.; Rua, C.P.J.; de Oliveira, L.; Leomil, L.; Fróes, A.M.; Thompson, F.L.; Coutinho, R.; et al. Environmental conditions affect activity and associated microorganisms of marine sponges. Mar. Environ. Res. 2018, 142, 59–68. [Google Scholar] [CrossRef]
- Pascelli, C.; Laffy, P.W.; Botté, E.; Kupresanin, M.; Rattei, T.; Lurgi, M.; Ravasi, T.; Webster, N.S. Viral ecogenomics across the Porifera. Microbiome 2020, 8, 144. [Google Scholar] [CrossRef]
- Nguyen, M.; Wemheuer, B.; Laffy, P.W.; Webster, N.S.; Thomas, T. Taxonomic, functional and expression analysis of viral communities associated with marine sponges. PeerJ 2021, 9, e10715. [Google Scholar] [CrossRef]
- Waldron, F.M.; Stone, G.N.; Obbard, D.J. Metagenomic sequencing suggests a diversity of RNA interference-like responses to viruses across multicellular eukaryotes. PLoS Genet. 2018, 14, e1007533. [Google Scholar] [CrossRef]
- Laffy, P.W.; Wood-Charlson, E.M.; Turaev, D.; Jutz, S.; Pascelli, C.; Botté, E.S.; Bell, S.C.; Peirce, T.E.; Weynberg, K.D.; van Oppen, M.J.H.; et al. Reef invertebrate viromics: Diversity, host specificity and functional capacity. Environ. Microbiol. 2018, 20, 2125–2141. [Google Scholar] [CrossRef] [PubMed]
- Urayama, S.I.; Takaki, Y.; Hagiwara, D.; Nunoura, T. DsRNA-seq reveals novel RNA virus and virus-like putative complete genome sequences from Hymeniacidon sp. Sponge. Microbes Environ. 2020, 35, ME19132. [Google Scholar] [CrossRef]
- Jahn, M.T.; Arkhipova, K.; Markert, S.M.; Stigloher, C.; Lachnit, T.; Pita, L.; Kupczok, A.; Ribes, M.; Stengel, S.T.; Rosenstiel, P.; et al. A Phage Protein Aids Bacterial Symbionts in Eukaryote Immune Evasion. Cell Host Microbe 2019, 26, 542–550.e5. [Google Scholar] [CrossRef]
- Butina, T.V.; Bukin, Y.S.; Khanaev, I.V.; Kravtsova, L.S.; Maikova, O.O.; Tupikin, A.E.; Kabilov, M.R.; Belikov, S.I. Metagenomic analysis of viral communities in diseased Baikal sponge Lubomirskia baikalensis. Limnol. Freshw. Biol. 2019, 1, 155–162. [Google Scholar] [CrossRef]
- Butina, T.V.; Khanaev, I.V.; Kravtsova, L.S.; Maikova, O.O.; Bukin, Y.S. Metavirome datasets from two endemic Baikal sponges Baikalospongia bacillifera. Data Br. 2020, 29, 105260. [Google Scholar] [CrossRef] [PubMed]
- Butina, T.V.; Petrushin, I.S.; Khanaev, I.V.; Bukin, Y.S. Metagenomic Assessment of DNA Viral Diversity in Freshwater Sponges, Baikalospongia bacillifera. Microorganisms 2022, 10, 480. [Google Scholar] [CrossRef]
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The sponge holobiont in a changing ocean: From microbes to ecosystems. Microbiome 2018, 6, 46. [Google Scholar] [CrossRef]
- Khanaev, I.V.; Kravtsova, L.S.; Maikova, O.O.; Bukshuk, N.A.; Sakirko, M.V.; Kulakova, N.V.; Butina, T.V.; Nebesnykh, I.A.; Belikov, S.I. Current state of the sponge fauna (Porifera: Lubomirskiidae) of Lake Baikal: Sponge disease and the problem of conservation of diversity. J. Great Lakes Res. 2018, 44, 77–85. [Google Scholar] [CrossRef]
- Bondarenko, N.A.; Vorobyova, S.S.; Zhuchenko, N.A.; Golobokova, L.P. Current state of phytoplankton in the littoral area of Lake Baikal, spring. J. Great Lakes Res. 2020, 46, 17–28. [Google Scholar] [CrossRef]
- Kravtsova, L.; Vorobyeva, S.; Naumova, E.; Izhboldina, L.; Mincheva, E.; Potemkina, T.; Pomazkina, G.; Rodionova, E.; Onishchuk, N.; Sakirko, M.; et al. Response of aquatic organisms communities to global climate changes and anthropogenic impact: Evidence from Listvennichny bay of Lake Baikal. Biology 2021, 10, 904. [Google Scholar] [CrossRef] [PubMed]
- Khodzher, T.V.; Domysheva, V.M.; Sorokovikova, L.M.; Sakirko, M.V.; Tomberg, I.V. Current chemical composition of Lake Baikal water. Inland Waters 2017, 7, 250–258. [Google Scholar] [CrossRef]
- Onishchuk, N.A.; Netsvetaeva, O.G.; Tomberg, I.V.; Sakirko, M.V.; Domysheva, V.M.; Golobokova, L.P.; Khodzher, T.V. Seasonal dynamics of mineral forms of nitrogen in the river water, snow cover and precipitation near the Listvyanka settlement (west coast of the Southern Baikal). Limnol. Freshw. Biol. 2019, 3, 245–252. [Google Scholar] [CrossRef]
- Bukin, Y.S.; Bondarenko, N.A.; Rusanov, I.I.; Pimenov, N.V.; Bukin, S.V.; Pogodaeva, T.V.; Chernitsyna, S.M.; Shubenkova, O.V.; Ivanov, V.G.; Zakharenko, A.S.; et al. Interconnection of bacterial and phytoplanktonic communities with hydrochemical parameters from ice and under-ice water in coastal zone of Lake Baikal. Sci. Rep. 2020, 10, 11087. [Google Scholar] [CrossRef] [PubMed]
- Malnik, V.V.; Suturin, A.N.; Gorshkova, A.S.; Shtykova, Y.R.; Timoshkin, O.A. Water Quality in the Shallow Zone of Lake Baikal as Deduced from Sanitary and Microbiological Indicators. Geogr. Nat. Resour. 2022, 43, 141–148. [Google Scholar] [CrossRef]
- Maikova, O.; Bukshuk, N.; Kravtsova, L.; Nebesnyh, I.; Yakhnenko, A.; Butina, T.; Khanaev, I. Baikal endemic sponge disease and anthropogenic factor. IOP Conf. Ser. Earth Environ. Sci. 2021, 937, 22071. [Google Scholar] [CrossRef]
- Maldonado, M.; Sánchez-Tocino, L.; Navarro, C. Recurrent disease outbreaks in corneous demosponges of the genus Ircinia: Epidemic incidence and defense mechanisms. Mar. Biol. 2010, 157, 1577–1590. [Google Scholar] [CrossRef]
- Sweet, M.; Bulling, M.; Cerrano, C. A novel sponge disease caused by a consortium of micro-organisms. Coral Reefs 2015, 34, 871–883. [Google Scholar] [CrossRef]
- Maikova, O.O.; Bukshuk, N.A.; Kravtsova, L.S.; Onishchuk, N.A.; Sakirko, M.V.; Nebesnykh, I.A.; Lipko, I.A.; Khanaev, I.V. Sponge fauna of Lake Baikal in the monitoring system for six years of observations. Contemp. Probl. Ecol. 2023, 30, 11–24. [Google Scholar]
- Shi, M.; Lin, X.D.; Tian, J.H.; Chen, L.J.; Chen, X.; Li, C.X.; Qin, X.C.; Li, J.; Cao, J.P.; Eden, J.S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Chen, Y.; Wei, X.; Cui, J. Viromes in marine ecosystems reveal remarkable invertebrate RNA virus diversity. Sci. China Life Sci. 2022, 65, 426–437. [Google Scholar] [CrossRef]
- Johnson, P.T. Viral diseases of marine invertebrates. Helgoländer Meeresunters. 1984, 37, 65–98. [Google Scholar] [CrossRef]
- Ryabov, E.V. Invertebrate RNA virus diversity from a taxonomic point of view. J. Invertebr. Pathol. 2017, 147, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology 2015, 479–480, 2–25. [Google Scholar] [CrossRef]
- Dolja, V.V.; Koonin, E.V. Metagenomics reshapes the concepts of RNA virus evolution by revealing extensive horizontal virus transfer. Virus Res. 2018, 244, 36–52. [Google Scholar] [CrossRef]
- Shi, M.; Zhang, Y.Z.; Holmes, E.C. Meta-transcriptomics and the evolutionary biology of RNA viruses. Virus Res. 2018, 243, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 1993, 15, 532–537. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Bolduc, B.; Zayed, A.A.; Varsani, A.; Dominguez-Huerta, G.; Delmont, T.O.; Pratama, A.A.; Gazitúa, M.C.; Vik, D.; Sullivan, M.B.; et al. VirSorter2: A multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome 2021, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Nayfach, S.; Camargo, A.P.; Schulz, F.; Eloe-Fadrosh, E.; Roux, S.; Kyrpides, N.C. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 2021, 39, 578–585. [Google Scholar] [CrossRef]
- Zhao, S.; Ye, Z.; Stanton, R. Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. RNA 2020, 26, 903–909. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Mihara, T.; Nishimura, Y.; Shimizu, Y.; Nishiyama, H.; Yoshikawa, G.; Uehara, H.; Hingamp, P.; Goto, S.; Ogata, H. Linking virus genomes with host taxonomy. Viruses 2016, 8, 66. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- O’Hara, R.B. Species richness estimators: How many species can dance on the head of a pin? J. Anim. Ecol. 2005, 74, 375–386. [Google Scholar] [CrossRef]
- Colwell, R.K.; Coddington, J.A. Estimating terrestrial biodiversity through extrapolation. Biodivers. Meas. Estim. 1995, 345, 101–118. [Google Scholar]
- Hill, M.O. Diversity and Evenness: A Unifying Notation and Its Consequences. Ecology 1973, 54, 427–432. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. R Package, Version 2.6-4, Package ‘Vegan’: Community Ecology Package; 2022; pp. 1–297. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 12 October 2022).
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Liaw, W.H.A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; Schwartz, M.; et al. R Package, Version 2.17.0; Package “gplots”: Various R Programming Tools for Plotting Data; ScienceOpen: Berlin, Germany, 2015; pp. 1–68. [Google Scholar]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Conceição-Neto, N.; Zeller, M.; Lefrère, H.; De Bruyn, P.; Beller, L.; Deboutte, W.; Yinda, C.K.; Lavigne, R.; Maes, P.; Ranst, M.V.; et al. Modular approach to customise sample preparation procedures for viral metagenomics: A reproducible protocol for virome analysis. Sci. Rep. 2015, 5, 16532. [Google Scholar] [CrossRef] [PubMed]
- Kuhar, U.; Vengust, G.; Jamnikar-Ciglenecki, U. Complete genome sequence of roe deer picobirnavirus strain PBV/roe_deer/SLO/D38-14/2014. Genome Announc. 2017, 5, e01329-17. [Google Scholar] [CrossRef]
- Golobokova, L.P.; Sakirko, M.V.; Onishchuk, N.A.; Pogodaeva, T.V.; Sez’ko, N.P.; Dolya, I.N. Hydrochemical characteristics of littoral waters of North-Western coast of Southern Baikal. In Index of Animal Species Inhabiting Lake Baikal and Its Catchment Area II (1); Timoshkin, O.A., Ed.; Nauka: Novosibirsk, Russia, 2009; pp. 760–784. (In Russian) [Google Scholar]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E. (Eds.) Family—Tombusviridae. In Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Fregolente, M.C.D.; de Castro-Dias, E.; Martins, S.S.; Spilki, F.R.; Allegretti, S.M.; Gatti, M.S.V. Molecular characterization of picobirnaviruses from new hosts. Virus Res. 2009, 143, 134–136. [Google Scholar] [CrossRef]
- Delmas, B.; Attoui, H.; Ghosh, S.; Malik, Y.S.; Mundt, E.; Vakharia, V.N. ICTV virus taxonomy profile: Picobirnaviridae. J. Gen. Virol. 2019, 100, 133–134. [Google Scholar] [CrossRef]
- Lang, A.S.; Vlok, M.; Culley, A.I.; Suttle, C.A.; Takao, Y.; Tomaru, Y.; Siddell, S.G.; Lefkowitz, E.; Simmonds, P.; Zerbini, F.M.; et al. ICTV virus taxonomy profile: Marnaviridae. J. Gen. Virol. 2021, 102, 1633. [Google Scholar] [CrossRef]
- Obbard, D.J. Expansion of the metazoan virosphere: Progress, pitfalls, and prospects. Curr. Opin. Virol. 2018, 31, 17–23. [Google Scholar] [CrossRef]
- Chen, Y.M.; Sadiq, S.; Tian, J.H.; Chen, X.; Lin, X.D.; Shen, J.J.; Chen, H.; Hao, Z.Y.; Wille, M.; Zhou, Z.C.; et al. RNA viromes from terrestrial sites across China expand environmental viral diversity. Nat. Microbiol. 2022, 7, 1312–1323. [Google Scholar] [CrossRef]
- French, R.; Charon, J.; Lay, C.L.; Muller, C.; Holmes, E.C. Human land use impacts viral diversity and abundance in a New Zealand river. Virus Evol. 2022, 8, veac032. [Google Scholar] [CrossRef]
- Glasl, B.; Robbins, S.; Frade, P.R.; Marangon, E.; Laffy, P.W.; Bourne, D.G.; Webster, N.S. Comparative genome-centric analysis reveals seasonal variation in the function of coral reef microbiomes. ISME J. 2020, 14, 1435–1450. [Google Scholar] [CrossRef]
- Evseev, P.; Tikhonova, I.; Krasnopeev, A.; Sorokovikova, E.; Gladkikh, A.; Timoshkin, O.; Miroshnikov, K.; Belykh, O. Tychonema sp. BBK16 Characterisation: Lifestyle, Phylogeny and Related Phages. Viruses 2023, 15, 442. [Google Scholar] [CrossRef]
- Belikov, S.; Belkova, N.; Butina, T.; Chernogor, L.; Kley, A.M.-V.; Nalian, A.; Rorex, C.; Khanaev, I.; Maikova, O.; Feranchuk, S. Diversity and shifts of the bacterial community associated with Baikal sponge mass mortalities. PLoS ONE 2019, 14, e0213926. [Google Scholar] [CrossRef] [PubMed]
- Erwin, P.M.; Pita, L.; López-Legentil, S.; Turon, X. Stability of sponge-associated bacteria over large seasonal shifts in temperature and irradiance. Appl. Environ. Microbiol. 2012, 78, 7358–7368. [Google Scholar] [CrossRef] [PubMed]
- Slaby, B.M.; Franke, A.; Rix, L.; Pita, L.; Bayer, K.; Jahn, M.T.; Hentschel, U. Symbiotic Microbiomes of Coral Reefs Sponges and Corals; Springer: Dordrecht, The Netherlands, 2019; pp. 81–104. [Google Scholar]
- Webster, N.S. Sponge disease: A global threat? Environ. Microbiol. 2007, 9, 1363–1375. [Google Scholar] [CrossRef] [PubMed]
- Luter, H.M.; Webster, N.S. Sponge Disease and Climate Change. In Climate Change, Ocean Acidification and Sponges; Springer: Cham, Switzerland, 2017; pp. 411–428. [Google Scholar]
- Maldonado, M.; Aguilar, R.; Bannister, R.; Bell, J.; Conway, J.; Dayton, P.; Diaz, C.; Gutt, J.; Kelly, M.; Kenchington, E.; et al. Sponge Grounds as Key Marine Habitats: A Synthetic Review of Types, Structure, Functional Roles, and Conservation Concerns. In Marine Animal Forests: The Ecology of Benthic Biodiversity Hotspots; Springer International Publishing: Cham, Switzerland, 2017. [Google Scholar]
- Fan, L.; Liu, M.; Simister, R.; Webster, N.S.; Thomas, T. Marine microbial symbiosis heats up: The phylogenetic and functional response of a sponge holobiont to thermal stress. ISME J. 2013, 7, 991–1002. [Google Scholar] [CrossRef]
- Shimaraev, M.N.; Domysheva, V.M. Trends in Hydrological and hydrochemical Processes in Lake Baikal under Conditions of Modern Climate Change. In Climatic Change and Global Warming of Inland Waters: Impacts and Mitigation for Ecosystems and Societies; Goldman, C.R., Kumagai, M., Robarts, R.D., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; pp. 43–66. [Google Scholar]
- Izmest’eva, L.R.; Moore, M.V.; Hampton, S.E.; Ferwerda, C.J.; Gray, D.K.; Woo, K.H.; Pislegina, H.V.; Krashchuk, L.S.; Shimaraeva, S.V.; Silow, E.A. Lake-wide physical and biological trends associated with warming in Lake Baikal. J. Great Lakes Res. 2016, 42, 6–17. [Google Scholar] [CrossRef]
- Shimaraev, M.N.; Sinyukovich, V.N.; Sizova, L.N.; Troitskaya, E.S. Heat balance of Lake Baikal and the relationship of its ice-thermal and water regime with global atmospheric circulation in the Northern Hemisphere during the modern period. Limnol. Freshw. Biol. 2018, 2018, 10–14. [Google Scholar] [CrossRef]
- Valipour, M.; Bateni, S.M.; Jun, C. Global surface temperature: A new insight. Climate 2021, 9, 81. [Google Scholar] [CrossRef]
- Richmond, R.H.; Tisthammer, K.H.; Spies, N.P. The effects of anthropogenic stressors on reproduction and recruitment of corals and reef organisms. Front. Mar. Sci. 2018, 5, 226. [Google Scholar] [CrossRef]
- Tompkins-Macdonald, G.J.; Leys, S.P. Glass sponges arrest pumping in response to sediment: Implications for the physiology of the hexactinellid conduction system. Mar. Biol. 2008, 154, 973–984. [Google Scholar] [CrossRef]
- Bannister, R.J.; Battershill, C.N.; de Nys, R. Suspended sediment grain size and mineralogy across the continental shelf of the Great Barrier Reef: Impacts on the physiology of a coral reef sponge. Cont. Shelf Res. 2012, 32, 86–95. [Google Scholar] [CrossRef]
- Maldonado, M.; Giraud, K.; Carmona, C. Effects of sediment on the survival of asexually produced sponge recruits. Mar. Biol. 2008, 154, 631–641. [Google Scholar] [CrossRef]
- Smith, K.M.; Bald, J.G. A description of a necrotic virus disease affecting tobacco and other plants. Parasitology 1935, 27, 231–245. [Google Scholar] [CrossRef]
- Dabek, A.J.; Castano, J.J. The occurrence, symptomatology, transmission and virus aetiology of macana disease of fique (Furcraea spp.) in Colombia, South America. J. Phytopathol. 1978, 92, 57–69. [Google Scholar] [CrossRef]
- Roveta, C.; Annibaldi, A.; Afghan, A.; Calcinai, B.; Di Camillo, C.G.; Gregorin, C.; Illuminati, S.; Mantas, T.P.; Truzzi, C.; Puce, S. Biomonitoring of heavy metals: The unexplored role of marine sessile taxa. Appl. Sci. 2021, 11, 580. [Google Scholar] [CrossRef]
- Culley, A. New insight into the RNA aquatic virosphere via viromics. Virus Res. 2018, 244, 84–89. [Google Scholar] [CrossRef]
- Kolundžija, S.; Cheng, D.-Q.; Lauro, F.M. RNA Viruses in Aquatic Ecosystems through the Lens of Ecological Genomics and Transcriptomics. Viruses 2022, 14, 702. [Google Scholar] [CrossRef]


| Dataset Name 1,2 | Sample Description | Geographic Location | Latitude and Longitude | Data | Nucleic Acids | Experiments | Reference |
|---|---|---|---|---|---|---|---|
| L.b.Sv1.1h | Lubomirskia baikalensis, healthy branch | Russia: Lake Baikal | 53.02 N 106.93 E | March 2015 | RNA (V) | SRX19982199 | This study |
| L.b.Sv2.1d | Lubomirskia baikalensis, branch with bleaching | Russia: Lake Baikal | 53.02 N 106.93 E | March 2015 | RNA (V) | SRX19982200 | This study |
| L.b.Sv3.2h | Lubomirskia baikalensis, healthy branch | Russia: Lake Baikal | 53.02 N 106.93 E | March 2015 | RNA (V) | SRX19982201 | This study |
| L.b.Sv3a.2d | Lubomirskia baikalensis, branch with brown spots | Russia: Lake Baikal | 53.02 N 106.93 E | March 2015 | RNA (V) | SRX19982202 | This study |
| B.b.Sv2478.3h | Baikalospongia bacillifera, healthy | Russia: Lake Baikal | 51.90 N 105.10 E | June 2018 | RNA (V) | SRX19982203 | This study |
| B.b.Sv2480.4h | Baikalospongia bacillifera, healthy | Russia: Lake Baikal | 51.90 N 105.10 E | June 2018 | RNA (V) | SRX19982204 | This study |
| B.b.Sv2473.5d | Baikalospongia bacillifera, necrosis, brown spots | Russia: Lake Baikal | 51.90 N 105.10 E | June 2018 | RNA (V) | SRX19982205 | This study |
| B.b.Sv2475.6d | Baikalospongia bacillifera, necrosis, brown spots | Russia: Lake Baikal | 51.90 N 105.10 E | June 2018 | RNA (V) | SRX19982206 | This study |
| Ch.reniformis_1 | Chondrosia reniformis, m/p | Spain: Mediterranean Sea, RM | 42.08 N 3.20 E | July 2016 | RNA/DNA (V) | SRX5385349, SRX5385338 | [13] |
| Ch.reniformis_2 | Chondrosia reniformis, m/p | Spain: Mediterranean Sea, RM | 42.08 N 3.20 E | July 2016 | RNA/DNA (V) | SRX5385350, SRX5385337 | [13] |
| Ch.reniformis_3 | Chondrosia reniformis, m/p | Spain: Mediterranean Sea, RM | 42.08 N 3.20 E | July 2016 | RNA/DNA (V) | SRX5385353, SRX5385336 | [13] |
| Ap.aerophoba_1 | Aplysina aerophoba, m/p | Spain: Mediterranean Sea, PL | 42.30 N 3.29 E | July 2016 | RNA/DNA (V) | SRX5385319, SRX5385345 | [13] |
| Ap.aerophoba_2 | Aplysina aerophoba, m/p | Spain: Mediterranean Sea, PL | 42.30 N 3.29 E | July 2016 | RNA/DNA (V) | SRX5385320, SRX5385346 | [13] |
| Ap.aerophoba_3 | Aplysina aerophoba, m/p | Spain: Mediterranean Sea, PL | 42.30 N 3.29 E | July 2016 | RNA/DNA (V) | SRX5385347, SRX5385351 | [13] |
| P.ficiformis_1 | Petrosia ficiformis, m/p | Spain: Mediterranean Sea, RM | 42.08 N 3.20 E | July 2016 | RNA/DNA (V) | SRX5385334, SRX5385330 | [13] |
| P.ficiformis_2 | Petrosia ficiformis, m/p | Spain: Mediterranean Sea, RM | 42.08 N 3.20 E | July 2016 | RNA/DNA (V) | SRX5385333, SRX5385329 | [13] |
| P.ficiformis_3 | Petrosia ficiformis, m/p | Spain: Mediterranean Sea, RM | 42.08 N 3.20 E | July 2016 | RNA/DNA (V) | SRX5385332, SRX5385339 | [13] |
| Ag.oroides_1 | Agelas oroides, m/p | Spain: Mediterranean Sea, RM | 42.08 N 3.20 E | July 2016 | RNA/DNA (V) | SRX5385321, SRX5385325 | [13] |
| Ag.oroides_2 | Agelas oroides, m/p | Spain: Mediterranean Sea, RM | 42.08 N 3.20 E | July 2016 | RNA/DNA (V) | SRX5385322, SRX5385326 | [13] |
| Ag.oroides_3 | Agelas oroides, m/p | Spain: Mediterranean Sea, RM | 42.08 N 3.20 E | July 2016 | RNA/DNA (V) | SRX5385323, SRX5385327 | [13] |
| Med.sw.PL (2) | Seawater | Spain: Mediterranean Sea, PL | 42.30 N 3.29 E | July 2016 | RNA/DNA (V) | SRX5385341, SRX5385342 | [13] |
| Med.sw.RM (2) | Seawater | Spain: Mediterranean Sea, RM | 42.30 N 3.29 E | July 2016 | RNA/DNA (V) | SRX5385343, SRX5385344 | [13] |
| Hym.sp_1 | Hymeniacidon sp. | Japan: Tokyo bay | 35.34 N 139.64 E | April 2014 | RNA (T) | DRX171015 | [12] |
| Hym.sp_2 | Hymeniacidon sp. | Japan: Tokyo bay | 35.34 N 139.64 E | April 2015 | RNA (T) | DRX171017 | [12] |
| GBR.sw (3) | Seawater | Australia: Great Barrier Reef | 18.83 S 147.63 E | October 2014 | RNA (V) | SRX2883311, SRX2883316, SRX2883317 | [11] |
| Rh.odorabile_1 | Rhopaloeides odorabile | Australia: Great Barrier Reef | 18.83 S 147.63 E | October 2014 | RNA (V) | SRX2883299 | [11] |
| Rh.odorabile_2 | Rhopaloeides odorabile | Australia: Great Barrier Reef | 18.83 S 147.63 E | October 2014 | RNA (V) | SRX2883312 | [11] |
| Rh.odorabile_3 | Rhopaloeides odorabile | Australia: Great Barrier Reef | 18.83 S 147.63 E | October 2014 | RNA (V) | SRX2883313 | [11] |
| Hal.panicea_A (37) | Halichondria panacea | United Kingdom: North Sea, BN | 55.99 N–2.45 W | 2014 | RNA (T) | SRX4378335 | [10] |
| Hal.panicea-B (11) | Halichondria panacea | United Kingdom: North Sea, BN | 55.99 N–2.45 W | 2014 | RNA (T) | SRX4378332 | [10] |
| Species | Family * | Genus * | Host/Isolation Source | No. of Scaffolds |
|---|---|---|---|---|
| Tobacco necrosis virus D | Tombusviridae | Betanecrovirus | Plants | 45 |
| Changjiang tombus-like virus 20 | - | - | /Crustaceans | 15, 17, 37 |
| Wenzhou narna-like virus 5 | - | - | /Mollusk | 24, 9, 532 |
| Beihai picorna-like virus 82 | - | - | /Crustaceans | 69 |
| Wenzhou picorna-like virus 19 | - | - | /Mollusk | 3 |
| unclassified | - | - | 5, 25, 374 | |
| Picobirnavirus green monkey/KNA/2015 | Picobirnaviridae | Picobirnavirus | Vertebrates | 48 |
| Wenzhou tombus-like virus 15 | - | - | /Mollusk | 145, 173 |
| Marine RNA virus SF-1 | Marnaviridae | Locarnavirus | /Wasterwater | 6 |
| Ubei picorna-like virus 3 | - | - | /Insecta | 2 |
| Hubei odonate virus 1 | - | - | /Insecta | 1 |
| Furcraea necrotic streak virus | Tombusviridae | Macanavirus | Plants | 328 |
| Wenzhou picorna-like virus 41 | - | - | /Mollusk | 4 |
| Wenzhou weivirus-like virus 1 | - | - | /Mollusk | 10 |
| Beihai tombus-like virus 15 | - | - | /Crustaceans | 13 |
| Hubei sobemo-like virus 3 | - | - | /Mollusk | 167, 286 |
| Beihai noda-like virus 5 | - | - | /Crustaceans | 11 |
| Beihai tombus-like virus 8 | - | - | /Mollusk | 49 |
| Hubei unio douglasiae virus 3 | - | - | /Mollusk | 28 |
| Wenzhou tombus-like virus 16 | - | - | /Mollusk | 83 |
| Beihai picorna-like virus 56 | - | - | /Mollusk | 8 |
| Sanxia narna-like virus 1 | - | - | /Crustaceans | 168 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Butina, T.V.; Khanaev, I.V.; Petrushin, I.S.; Bondaryuk, A.N.; Maikova, O.O.; Bukin, Y.S. The RNA Viruses in Samples of Endemic Lake Baikal Sponges. Diversity 2023, 15, 835. https://doi.org/10.3390/d15070835
Butina TV, Khanaev IV, Petrushin IS, Bondaryuk AN, Maikova OO, Bukin YS. The RNA Viruses in Samples of Endemic Lake Baikal Sponges. Diversity. 2023; 15(7):835. https://doi.org/10.3390/d15070835
Chicago/Turabian StyleButina, Tatyana V., Igor V. Khanaev, Ivan S. Petrushin, Artem N. Bondaryuk, Olga O. Maikova, and Yurij S. Bukin. 2023. "The RNA Viruses in Samples of Endemic Lake Baikal Sponges" Diversity 15, no. 7: 835. https://doi.org/10.3390/d15070835
APA StyleButina, T. V., Khanaev, I. V., Petrushin, I. S., Bondaryuk, A. N., Maikova, O. O., & Bukin, Y. S. (2023). The RNA Viruses in Samples of Endemic Lake Baikal Sponges. Diversity, 15(7), 835. https://doi.org/10.3390/d15070835










